Abstract
Two pools of small RNAs were cloned from inflorescences of Petunia hybrida using a 5′-ligation dependent and a 5′-ligation independent approach. The two libraries were integrated into a public website that allows the screening of individual sequences against 359,769 unique clones. The library contains 15 clones with 100% identity and 53 clones with one mismatch to miRNAs described for other plant species. For two conserved miRNAs, miR159 and miR390, we find clear differences in tissue-specific distribution, compared with other species. This shows that evolutionary conservation of miRNA sequences does not necessarily include a conservation of the miRNA expression profile. Almost 60% of all clones in the database are 24-nucleotide clones. In accordance with the role of 24mers in marking repetitive regions, we find them distributed across retroviral and transposable element sequences but other 24mers map to promoter regions and to different transcript regions. For one target region we observe tissue-specific variation of matching 24mers, which demonstrates that, as for 21mers, 24mer concentrations are not necessarily identical in different tissues. Asymmetric distribution of a putative novel miRNA in the two libraries suggests that the cloning method can be selective for the representation of certain small RNAs in a collection.
Keywords: microRNA, siRNA, small RNA, database, petunia
INTRODUCTION
Small noncoding RNAs play important roles in plant development, environmental response, antiviral defense, and genome structure. The most important classes of small RNAs are microRNAs (miRNAs) and the different forms of small interfering (si)RNAs, trans-acting (ta) siRNAs, natural antisense (nat) siRNAs, and heterochromatin siRNAs.
Mature ∼21-nucleotide (nt)-long miRNAs control regulatory genes, especially transcription factors (Rhoades et al. 2002). They are processed by RNAseIII-like enzymes from noncoding primary precursor transcripts with local stem–loop structures (Chen 2005). miRNAs guide the degradation of target transcripts via endonucleolytic slicer activity (Baumberger and Baulcombe 2005) or act to translationally repress mRNAs (Brodersen et al. 2008). miRNAs and their targets are usually evolutionarily conserved, which suggests an ancient origin of miRNAs in the plant lineage (Bartel and Bartel 2003).
miRNAs share many features with siRNAs, but in contrast to miRNAs, siRNAs are processed from double-stranded RNAs, frequently requiring RNA-dependent RNA polymerase activity and different RNAseIII-type enzymes. In Arabidopsis thaliana, miRNAs are predominantly processed by DICER-LIKE 1 (DCL1), viral siRNAs, and nat-siRNAs by DCL2, heterochromatin siRNAs by DCL3, and tasiRNAs by DCL4 (Xie et al. 2004; Gasciolli et al. 2005). tasiRNAs derive from single-stranded tasiRNA precursor RNAs (TAS) that were converted into a double-strand by RNA-dependent RNA polymerase 6 (RDR6) (Peragine et al. 2004), and which are cleaved in a phased manner by DCL4. DCL1 is also required for tasiRNA production, as certain miRNAs set the phase for the production of tasiRNAs (Allen et al. 2005). DCL1 is also involved in nat-si RNA production. Primary 24-nt nat-siRNAs derive from DCL2 cleavage of dsRNAs formed between a sense and an overlapping natural antisense transcript. The 24-nt RNA directs the cleavage of the sense transcript and sets the phase for production of secondary 21-nt nat-siRNAs by DCL1 in combination with RDR6 (Borsani et al. 2005).
Twenty-four-nucleotide heterochromatin siRNAs are components of the RNA directed DNA methylation (RdDM) pathway that regulate histone modification and heterochromatin formation. An atypical RNA polymerase, NUCLEAR RNA POLYMERASE D 1A (NRPD1A), produces RNA single strands as substrates for RNA-dependent RNA polymerase2 (RDR2). The resulting dsRNA is cleaved by DICER-like3 (DCL3) into 24mers of sense and antisense orientation, which are incorporated into a silencing complex that directs modification of cytosine and histone marks to homologous regions (Pontes et al. 2006) (Li et al. 2006).
While 21–24mers represent the dominant class of small RNAs, larger RNAs have also been described. An antisense-specific 30nt RNA was detected in the 3′ region of the Arabidopsis FLC gene (Swiezewski et al. 2007), and a ∼35-nt antisense RNA was found in the 3′ region of the petunia Sho gene (Zubko and Meyer 2007).
Small RNAs have been sequenced for a number of plant species, especially those for which a fully sequenced genome or an extensive DNA database was available (Rajagopalan et al. 2006; Fahlgren et al. 2007; Pilcher et al. 2007; Yao et al. 2007; Moxon et al. 2008). Genomic information for Petunia hybrida is rather limited, but comparison of a petunia small RNA database with small RNAs in other species and with sequence data for petunia genes and ESTs allowed us to identify conserved and putative new microRNAs as well as an unusual class of 24mers associated with nonheterochromatic regions.
RESULTS
Cloning of small RNAs
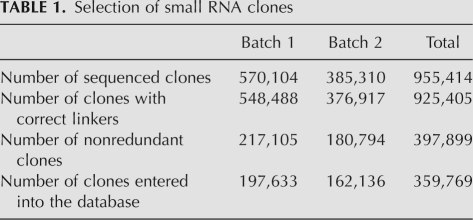
We prepared two types of small RNA libraries from floral bud RNA. Clones in batch 1 were prepared after adapter ligation to the 5′ and 3′ end of the RNA. As this method requires target RNAs with a 5′monophosphate, we used a 5′-ligation-independent method for batch 2 to include RNAs with a modified 5′ end. For both batches, RNA fractions in size ranges around 21–25 nt and 33–37 nt were excised and combined. From a total of 925,405 sequences for which adaptor sequences could be identified, 397,899 unique clones were extracted, from which 38,130 were removed, which represented rRNA, tRNA, snRNA, mitochondrial, or chloroplast gene sequences (Table 1).
TABLE 1.
Selection of small RNA clones
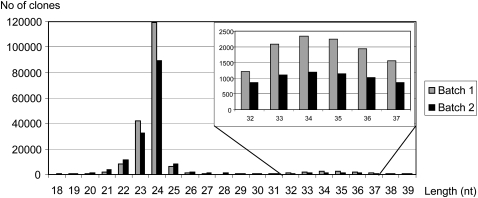
Both batches contain a similar size distribution of small RNAs, with 24mers representing the largest class in batch 1 (60%) and batch 2 (54%). A total of 2.3% of batch 2 clones and 1.1% of batch 1 clones are 21mers. Seven percent of batch 2 clones and 4% of batch 1 clones are 22mer. With 2.3%, 34mers and 35mers are more frequent in batch 1 compared with batch 2 with 1.4% (Fig. 1).
FIGURE 1.
Size distribution of small RNA clones in batches 1 and 2.
The petunia small RNA BLAST website
The 359,769 small RNA sequences of batch 1 and batch 2 were entered into a website that allows the screening of DNA sequences for perfect or partial matches to small RNA sequences. Users can submit DNA sequences of any length that can be compared with batch 1 and batch 2 clones, either separately or jointly. Search parameters include maximum and minimum length of the matching region, number of aligned clones to be displayed, orientation of the aligned clones, and selection of clones with 100% identity or less (http://www.petunia_smrna.leeds.ac.uk/).
The output graphic displays all small RNAs that fulfill the search criteria. Along the query sequence, each hit is illustrated as a box in one of four colors that define its batch origin and its orientation. For each box, the name of the clone, its length and its representation frequency in the small RNA library are listed. The graphic display is followed by a list of matching clones, their normalized bit score and expect value (E-value), and by an alignment for each clone with the query sequence.
Matches to known microRNAs
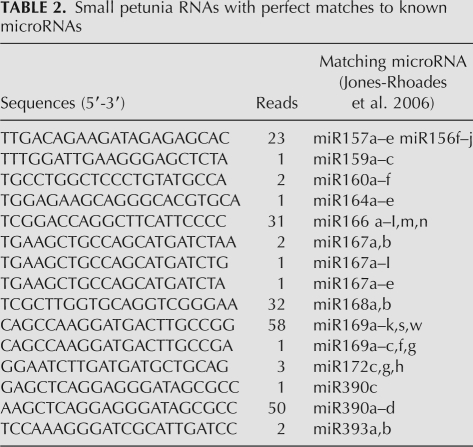
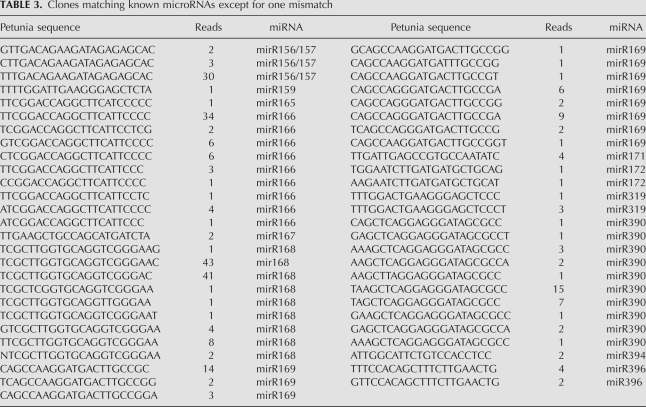
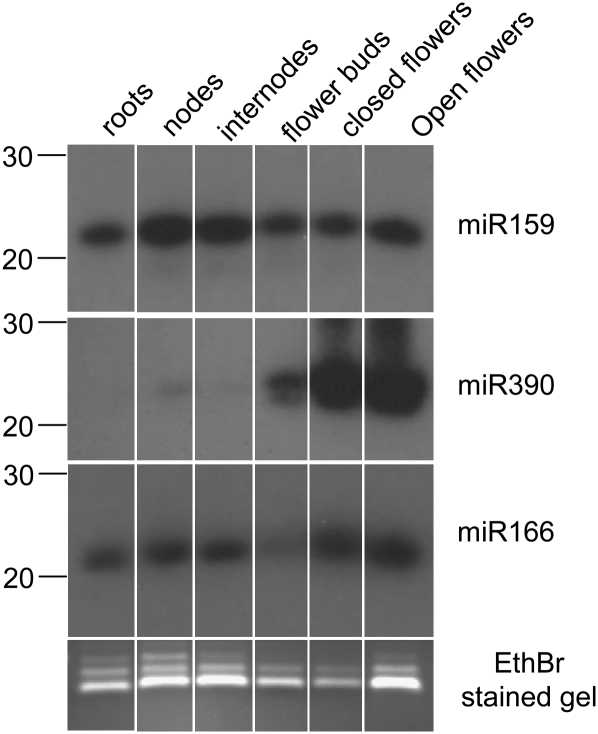
We identified 15 clones with perfect matches to microRNAs found in other species (Table 2; Supplemental File 1) and 53 clones with one mismatch to known microRNAs (Table 3; Supplemental File 2). A similarity screen of these clones against petunia sequences identified a hit for mirBL locus AM489765, which encodes a conserved miR169 microRNA (Cartolano et al. 2007). No other homologous petunia loci could be identified for the other 21mer sequences listed in Tables 2 and 3. We tested the distribution of three conserved microRNAs in different tissues. miR159 is relatively uniformly expressed in all tissue types, expression of miR166 is significantly reduced in flower buds and miR390 is mainly detectable in floral tissue (Fig. 2).
TABLE 2.
Small petunia RNAs with perfect matches to known microRNAs
TABLE 3.
Clones matching known microRNAs except for one mismatch
FIGURE 2.
Expression patterns of three conserved miRNA in different tissues of Petunia hybrida. Ethidium bromide stained fragments of <100 nt serve as loading control.
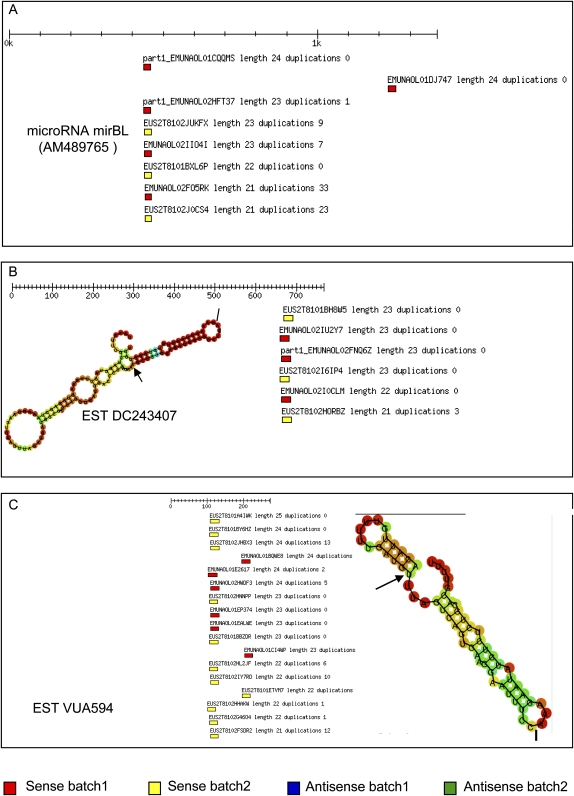
To test if our database contained additional, petunia-specific microRNAs, we searched for petunia clones with similar features to the mirBL locus (Fig. 3A). The small RNA database contains 231 hits for 21mer clones with 100% identity to a petunia sequence (Supplemental File 3). A screen for 21mer matches in sense orientation and for an RNA loop structure identified two ESTs which resembled features of the MirBL locus (Fig. 3B,C). A hybridization experiment confirmed the presence of homologous 21mers for one of these putative MIR loci, EST VUA594 (Fig. 4).
FIGURE 3.
Search for novel MIR loci. (A) The petunia mirBL locus AM489765 with matching clones in sense orientation. (B,C) Two candidate genes with 21mer matches in sense orientation and their RNA-fold structure. Start and stop of the miRNA are marked by an arrow and a bar, respectively.
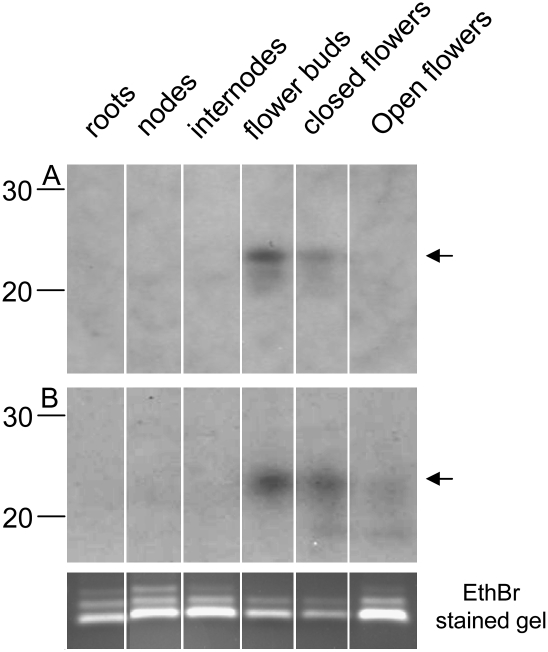
FIGURE 4.
Experimental verification of potential novel microRNAs. Expression pattern of two RNA clones in different tissues of Petunia hybrida. (A) Clone EUS2T8102FSDR2, which matches EST VUA594; (B) clone EMUNAOL02FR3EL, for which no matching ESTs were detected.
Only a fraction of small RNA clones match known petunia sequences, and these can be examined for secondary structure features. Without a matching petunia sequence, miRNA candidates can only be tested experimentally. Assuming that the frequent representation of 21mer or 22mer clones in the database is an indicators for a potential microRNA, we selected the 21mer EMUNAOL02FR3EL as a candidate. In the database, we find seven copies of this clone as well as 61 22mers that contain the 21mer sequence plus an additional nucleotide. Hybridization to an antisense probe of EMUNAOL02FR3EL confirmed the presence of a class of homologous 21–22mer in floral tissue and leaves (Fig. 4).
Analysis of 24mer clones
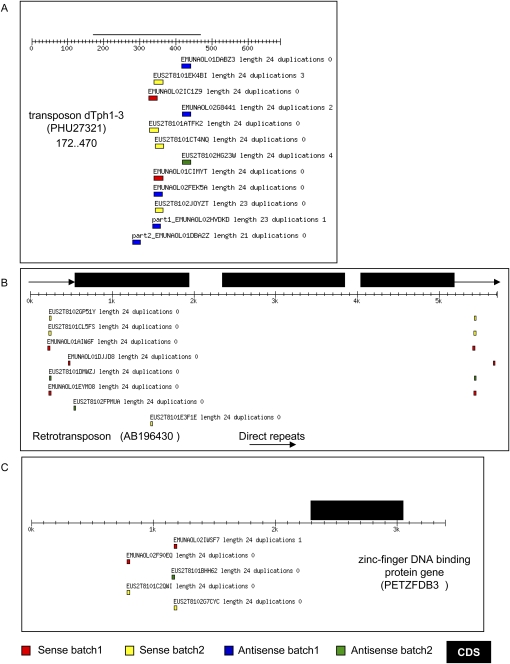
A BLAST search for the 208,721 24mer present in the library identified 3757 hits with a 100% identity to petunia sequences (Supplemental File 4). As expected, 24mer clones match sense and antisense sequences of moderately repeated sequences such as transposable elements, retrotransposons, and viral sequences (Fig. 5A,B; Supplemental File 6). However, 24mer identities can also be identified in promoter regions (Fig. 5C), 5′ or 3′ UTRs, introns, exons, and ESTs (Supplemental File 6). These 24mers often match to distinct regions, reminiscent of miRNA loci, but in contrast to miRNA, they represent sense and antisense matches to the region.
FIGURE 5.
Examples of petunia loci with matches to 24mer clones. (A) Transposon dTph1-3 inserted into exon 3 of the nia gene. (B) Direct repeat regions of retrotransposon rTph1. (C) Promoter region of a zinc-finger DNA binding protein gene.
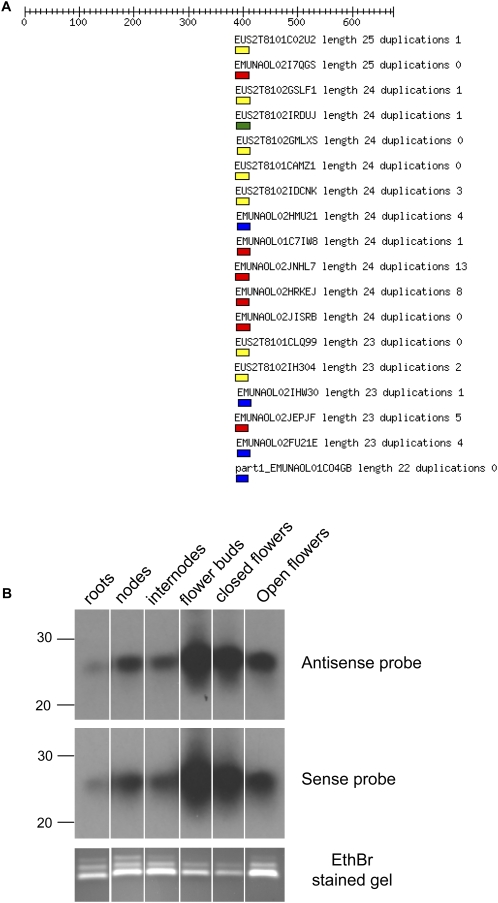
To test the presence of local 24mers experimentally, a 27-nt region between position 386 and 412 of EST887452 was selected, for which the library contains 41 24mer matches in sense or antisense orientation (Fig. 6A). Hybridization to sense and antisense specific probes confirmed the presence of 24-nt RNAs in different petunia tissues with significant overrepresentation in flower buds and very low levels in roots (Fig. 6B).
FIGURE 6.
Tissue-specific accumulation of 24mer clones matching EST887452. (A) Alignment of small RNA clones with EST887452. All matching clones are restricted to the region between position 386 and 412. (B) Presence of sense and antisense-specific 24mers in different tissues.
DISCUSSION
We used a 5′-ligation-dependent and a 5′-ligation-independent method to clone small RNAs from petunia flower buds. RNA cleavage by DICER generates primary small RNAs with 5′ monophosphates that can be cloned by 5′-adaptor ligation, while secondary RNAs that are synthesized by an RNA-directed RNA Polymerase (RdRP) have a triphosphate 5′ terminus and require a 5′-ligation-independent cloning method (Pak and Fire 2007). The two libraries were combined into one database that can be screened for perfect or near-perfect matches to sequences submitted to a public website. A color code allows the identification of the origin and orientation of each clone.
Compared with other genomes, petunia sequence data are relatively limited, which restricts the search for MIR loci or miRNA targets. For the 68 clones with perfect or near-perfect matches to known microRNAs, only one matching MIR locus, mirBL, could be identified. The mirBL locus served as a model for the identification of putative novel MIR loci. Like mirBL, two EST sequences contain a distinct region with a potential stem–loop structure and matches to small RNAs, which are all in sense orientation and predominantly 21-nt long. However, the presence of a homologous 21mer could only be demonstrated for one of the two candidates. This confirms that in silico prediction of MIR loci or small RNA target loci can still have a high false-positive rate and must be validated experimentally (Moxon et al. 2008).
Experimental validation also demonstrated that the tissue-specific expression profile for conserved microRNAs can vary among different species. At least two of the three conserved microRNAs that we tested show a tissue-specific distribution. MiR390 is strongly expressed in floral tissues but not detectable in root tissue. This distribution is a marked contrast to miR390 expression in rice, which is much stronger in roots than in inflorescences (Sunkar et al. 2005). This illustrates that, although certain microRNAs are conserved among different species, their tissue-specific expression level can differ significantly.
MiR390 has been cloned from Arabidopsis (Gustafson et al. 2005) and rice (Sunkar et al. 2005), and is present in fern and moss species as well (Axtell and Bartel 2005). Two Arabidopsis miRNAs, miR173 and miR390, target primary TAS transcripts encoding ta-siRNAs. Cleavage of TAS transcripts recruits RDR6 that synthesises a dsRNA substrate for ta-siRNA biogenesis (Allen et al. 2005). The TAS3 transcript upon which miR390 acts is conserved in Arabidopsis and rice, along with the corresponding mRNA targets of the miR390-dependent TAS3 siRNAs. This conservation suggests that setting the phasing of ta-siRNAs is an ancient function of miRNA in plants (Allen et al. 2005). However, rice contains a conventional mRNA target for miR390 (encoding a receptor-like kinase) (Sunkar et al. 2005), while in Arabidopsis an auxin response factor mRNA (ARF3/ETTIN and ARF4) is targeted by the ta-siRNAs generated after miR390-directed cleavage of the siRNA precursor transcript. It has therefore been suggested that miR390 may have dual roles regulating both mRNAs and loci encoding ta-siRNAs (Bartel 2005). The differences in tissue-specific expression of miR390 in petunia and other species indicate an additional level of flexibility in the evolution of miR390.
The tissue-specific expression profile of miR159 also differs from its expression in Arabidopsis. While in Arabidopsis miR159 is strongest in flowers but hardly detectable in roots (Achard et al. 2004), miR159 levels in roots and floral tissues are similar in petunia. Tissue-specific variation of miRNA expression in different species may therefore be a more common phenomenon than expected.
We identified two potentially new miRNA with strongest representation in flower buds, the tissue from which small RNAs had been cloned. One of these microRNA candidates matches VUA594 (gene bank gi91064442), a 272-bp EST from petals extracted from buds at maximum elongation. The database contains a total of 74 21–25-nt clones matching the 100–132-bp region of the VUA594 sense transcript. Thirteen clones are 21mer and 22 clones are 22mer, all of which exclusively derive from batch 2 (Fig. 3C). The uneven representation of clones in the two libraries confirms that the cloning methods can influence the representation of certain microRNAs.
Twenty-four mers represent the largest group of small RNAs in both batches, and the majority of these clones very likely mark target regions for RNA directed DNA methylation. The RdDM pathway controls methylation of many, although not all, transposons and DNA repeats located in constitutive or facultative heterochromatin, or dispersed within euchromatic regions (Pontier et al. 2005). Retrotransposon promoters and LTRs are also preferential RdDM targets (Huettel et al. 2006). It is therefore not surprising to detect matches to 24mer RNAs in petunia transposon and retrotransposon, or in a copy of an integrated viral genome. More surprising is the match of 24mers to individual small regions in EST sequences, and to coding regions or UTRs.
There is no indication for the presence of repeat elements at these 24mer matches, but in the absence of a complete petunia genome analysis, we cannot exclude that at least some of the 24mer sequences have been generated from unspecified repetitive regions that share small homologies with the nonrepetitive loci. But even if the local 24mer target regions were false-positive matches with 24mers originating from heterochromatic regions, one would not expect a tissue-specific variation as it was observed for EST887452-specific 24mers. At least for 24mer clones with variable representation in different tissues, it is more likely that these derive from transcripts of the matching euchromatic regions. Other examples for 24-nt siRNAs matching euchromatic loci, include the NaCl-inducible nat-siRNA SRO5-P5CDH (Borsani et al. 2005) and a 24-nt RNA corresponding to the 3′ reverse strand of the FLOWERING LOCUS C (FLC) (Swiezewski et al. 2007). The SRO5–P5CDH 24-nt siRNA sets the phase for the production of secondary 21-nt nat-siRNAs by DCL1, and the FLC 24-nt siRNA is part of a RNAi-mediated chromatin modification process under the control of RdRM pathway functions. The lack of phased 21mers, makes it very unlikely that the EST887452-specific 24mers represent nat-siRNAs. Tissue-specific variation is a common feature between EST887452-specific 24mers and the FLC 24-nt siRNA, but while FLC siRNA is specific for the complementary strand, EST887452-specific 24mers comprise both sense and antisense specific 24mers. In this respect, they resemble a pool of 24 nt-long sense and antisense RNAs associated with the petunia Sho locus (Zubko and Meyer 2007). However, unlike Sho-specific 24mers, EST887452-specific 24mers map to a very small transcript region.
Despite the limited number of petunia sequences that small RNAs can be compared with, our analysis has provided some surprising results about the tissue-specific representation of some small RNA types. The petunia small RNA website will hopefully stimulate a wider comparison of our database with unpublished and novel petunia sequences.
MATERIALS AND METHODS
Cloning and sequencing of small RNA
Low molecular-weight RNAs were isolated from petunia flower buds as described (Hamilton and Baulcombe 1999). Cloning of small RNAs followed a published protocol (Arazi et al. 2005) with some modification. Small RNAs were size fractionated by 15% denaturing polyacrylamide gel, eluted from gel in 0.3 M NaCl, purified, and sequentially ligated to 5′ and 3′ RNA–DNA chimeric oligonucleotide adapters. Reverse transcription was performed using Superscript II RT and antisense PCR3′ primer. Resulting cDNAs were amplified by PCR using PCR5′ and PCR3′ primers tailed with sequence A and B, which are required for 454 sequencing (Supplemental File 5). PCR fragments were purified and sequenced by 454 Life Sciences (Margulies et al. 2005) using the Amplicon library preparation procedure (http://www.454.com/downloads/protocols/Guide_To_Amplicon_Sequencing.pdf). For 5′ independent cloning, small RNAs were first ligated to a 3′RNA–DNA adapter. The ligation products were reverse transcribed and the cDNA was ligated to a cDNA adapter, followed by PCR amplification.
Small RNA expression analysis using Northern blot
For hybridization analysis, 25 μg of small RNA was separated on a 15% denaturing polyacrylamide gel, and transferred onto a Hybond Nx membrane (Amersham). The Decade Marker (Ambion) radiolabeled by γ32P[ATP] was used as a size control. Cross-linking of RNA to a nylon membrane was performed using a soluble carbodiimide (Pall et al. 2007). Hybridization was done in ULTRAhyb-Oligo hybridization buffer (Ambion) according to the manufacturer's recommendation.
Production of final petunia RNA sequences
Deep sequencing was done by 454 Life Sciences (www.454.com). Raw sequence data for batch 1 and batch 2 were processed in standard fasta format files using Perl scripts and the BioPerl SearchIO package (http://www.bioperl.org/wiki/Module:Bio∷SearchIO). Known adaptor sequences were located contiguously to the petunia RNA sequence at the beginning and end of the sequence. Adaptor sequences were found to have a far lower sequence variability at the ends nearest the petunia RNA (referred to below as the “petunia RNA end”). Therefore, sequences that showed a perfect alignment to the last 10 bases of the petunia RNA end of both of the adaptors were deemed to have been detected and the adaptors were removed. Otherwise, the last 19 bases of the petunia RNA end of the adaptor were aligned using dynamic programming (Needleman and Wunsch 1970), and adaptors were deemed to be detected if the alignment had no more than two mismatches to the known adaptor. A small minority of sequences in batch 1 was found to be concatemers of two RNA sequences flanked by adaptors at each end and containing a central pair. These sequences were split at the junction between the two central adaptors and the resulting sequences were then identified by the additional suffixes part1 and part2. Adaptor sequences were removed and the resulting sequences saved to fasta format files. Nonredundant sequence sets were then created by identifying groups of identical sequences and removing duplicated sequences by the use of an associative array in Perl.
The nonredundant sequences were then aligned using the BLAST algorithm with sequences in the rRNA database SILVA (Pruesse et al. 2007), Arabidopsis thaliana tRNA (http://gtrnadb.ucsc.edu/), mitochondrial, and chloroplast genes (http://www.arabidopsis.org/). Petunia RNA sequences with >90% sequence identity to any of the above sequences were removed. Finally, to remove snRNA and any sequences missed by the above processes, the remaining sequences were aligned with sequences in GenBank (Benson et al. 2007), and any perfect matches to rRNA, tRNA, snRNA, mitochondrial, or chloroplast genes were removed.
This produced the final sets of petunia RNA sequences, which were used for the rest of this study.
Identification of known miRNAs in petunia RNAs
Known miRNAs were downloaded from miRBASE (http://microrna.sanger.ac.uk/sequences/ftp.shtml) and petunia RNA sequences that showed perfect matches or one mismatch to the known miRNA were extracted using the BLAST algorithm.
Secondary structure analysis
For secondary structure analysis, sequences were submitted to the RNA fold webserver (Gruber et al. 2008).
Website development
The website is based on the NCBI blastall program (ftp://ftp.ncbi.nih.gov/blast/) with graphics in the website being produced using the BioPerl Graphics package (http://www.bioperl.org/wiki/Module:Bio∷Graphics).
100% matches of 21 and 24 base pair petunia RNAs to petunia genes
Petunia sequences were extracted from GenBank and aligned using BLAST with the petunia RNA sequences of length 21 and 24 base pairs. Petunia RNA sequences that showed a 100% match to any of the petunia genes were then recorded.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by the Epigenome Network of Excellence (LSHG-CT-2004-503433). We thank Binbin Liu for IT support. EST VUA594 was submitted to the petunia EST collection by Francesca Quattrocchio, Vrije Universiteit, Amsterdam, Netherlands.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1517209.
REFERENCES
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. Cloning and characterization of microRNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B. MicroRNAs directing siRNA biogenesis. Nat. Struct. Mol. Biol. 2005;12:569–571. doi: 10.1038/nsmb0705-569. [DOI] [PubMed] [Google Scholar]
- Bartel B., Bartel D.P. MicroRNAs: At the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 2007;39:901–905. doi: 10.1038/ng2056. [DOI] [PubMed] [Google Scholar]
- Chen X. microRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Bio. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L. The Vienna RNA website. Nucleic Acids Res. 2008;36:w70–w74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D. ASRP: The Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J.M., Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis . EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana . Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., Jing R., Szittya G., Schwach F., Rusholme Pilcher R.L., Moulton V., Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pall G.S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by Northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm/112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher R., Moxon S., Pakseresht N., Moulton V., Manning K., Seymour G., Dalmay T. Identification of novel small RNAs in tomato (Solanum lycopersicum) Planta. 2007;226:709–717. doi: 10.1007/s00425-007-0518-y. [DOI] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis . Genes & Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glockner F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.-K. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Crevillen P., Liu F., Ecker J.R., Jerzmanowski A., Dean C. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. 2007;104:3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Guo G., Ni Z., Sunkar R., Du J., Zhu J.-K., Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E., Meyer P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J. 2007;52:1131–1139. doi: 10.1111/j.1365-313X.2007.03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]