Abstract
Assembly of H/ACA RNPs in yeast is aided by at least two accessory factors, Naf1p and Shq1p. Although the function of Naf1p and its human ortholog NAF1 has been delineated in detail, that of Shq1p and its putative human ortholog SHQ1 remains obscure. We demonstrate that SHQ1 indeed functions in the biogenesis of human H/ACA RNPs and we dissect its mechanism of action. Like NAF1, SHQ1 binds the major H/ACA core protein and pseudouridine synthase NAP57 (aka dyskerin) but precedes the assembly role of NAF1 at nascent H/ACA RNAs because the interaction of SHQ1 with NAP57 in vivo and in vitro precludes that of NAF1 and of the other H/ACA core proteins that are present at the sites of H/ACA RNA transcription. The N-terminal heat shock protein 20-like CS domain of SHQ1 is dispensable for NAP57 binding. Consistent with its role as an assembly factor, SHQ1 localizes to the nucleoplasm and is excluded from nucleoli and Cajal bodies, the sites of mature H/ACA RNPs. In an in vitro assembly system of functional H/ACA RNPs that is dependent on NAF1, excess recombinant SHQ1 interferes with assembly. Importantly, knockdown of cellular SHQ1 prevents accumulation of a newly synthesized H/ACA reporter RNA and generally reduces the levels of endogenous H/ACA RNAs including telomerase RNA. In summary, the sequential action of SHQ1 and NAF1 is required for functional assembly of H/ACA RNPs in vivo and in vitro. This step-wise process could serve as an efficient means of quality control during H/ACA RNP assembly.
Keywords: NAP57/dyskerin, H/ACA RNP, snoRNP, RNP assembly, telomerase
INTRODUCTION
In each mammalian cell, 150 or so H/ACA ribonucleoproteins (RNPs) function in diverse processes, including modification and processing of ribosomal RNAs, modification of spliceosomal small nuclear RNAs, and stabilization of telomerase. Each particle consists of a function-defining H/ACA RNA and the same four core proteins. H/ACA RNPs are conserved from archaea to man and their structure and function have been delineated in genetic, biochemical, and X-ray crystallographic studies. The pseudouridine synthase NAP57 (aka dyskerin or in yeast Cbf5p) together with NOP10 and NHP2 forms a core trimer that specifically recognizes and associates with an H/ACA RNA and with the fourth core protein GAR1. All proteins of the core trimer are essential for the metabolic stability of H/ACA RNPs. Importantly, NAP57 is the subject of the X-linked form of the bone marrow failure syndrome dyskeratosis congenita (for reviews, see Meier 2005, 2008; Matera et al. 2007; Ye 2007; Kirwan and Dokal 2008; Li 2008).
These simple five component particles have proven surprisingly resistant to functional reconstitution. Although H/ACA RNPs can be formed from recombinant archaeal components, eukaryotic ones have yet to yield (Baker et al. 2005; Charpentier et al. 2005). This may be ascribed to the inherent instability of NAP57 and to the need for at least two assembly factors, Naf1p and Shq1p, that seem missing from archaea (Wang and Meier 2004; Normand et al. 2006). These two essential factors were identified in yeast, mainly by virtue of binding to Cbf5p (yeast NAP57), and are required for the stability of all H/ACA RNAs without being part of the mature particles (Dez et al. 2002; Fatica et al. 2002; Yang et al. 2002). Naf1p (NAF1 in mammals) has a GAR1 homology domain and consequently binds NAP57 (and the core trimer) at the same site as GAR1 (Fatica et al. 2002; Wang and Meier 2004; Leulliot et al. 2007). Together with the core trimer, NAF1 associates with H/ACA RNAs at their site of transcription and is replaced by GAR1 in mature RNPs functioning in Cajal bodies and nucleoli (Ballarino et al. 2005; Yang et al. 2005; Darzacq et al. 2006; Hoareau-Aveilla et al. 2006). In contrast, nothing is known about SHQ1 the putative mammalian ortholog of Shq1p.
Here, we dissect in vitro and in vivo the interactions of SHQ1 with the H/ACA RNP proteins and show that it is required for the stable accumulation of H/ACA RNAs including telomerase RNA, apparently acting before NAF1 in assembly. Our studies take advantage of a reconstitution assay that depends on both assembly factors for the generation of functional mammalian H/ACA RNPs.
RESULTS
SHQ1 binds NAP57 alone
Shq1p was identified in yeast as a protein interacting with Cbf5p, the ortholog of mammalian NAP57 (Ito et al. 2001; Ho et al. 2002). Indeed, Shq1p proved essential for the quantitative accumulation of Cbf5p-associated H/ACA RNAs and was evolutionary conserved with putative homologs in man, fly, worm, and fission yeast (Yang et al. 2002). In order to characterize the putative human SHQ1 (NM_018130; 26% identity and 47% similarity over the first 445 amino acids), we tested if it also interacted with H/ACA RNP associated factors.
Both, human SHQ1 and NAP57 (as an N-terminal fusion with the maltose binding protein [MBP]) were expressed in bacteria with C-terminal histidine tags and affinity purified (Fig. 1A, lanes 2,3). Equal amounts of recombinant SHQ1 were added to MBP-NAP57 and to a control MBP-fusion, the MS2 bacteriophage coat protein (MCP). The mixtures were bound to amylose resin through the MBP moieties of the fusion proteins (Fig. 1A, lanes 4–7). Only MBP-NAP57 retained SHQ1 (Fig. 1A, lane 6), but not MBP-MCP (Fig. 1A, lane 7), demonstrating a direct interaction between SHQ1 and NAP57.
FIGURE 1.
In vitro, SHQ1 only binds NAP57, to the exclusion of other proteins. (A) The bacterially expressed fusion protein of the maltose binding protein (MBP) with NAP57 (lane 3), but not that with the MS2 bacteriophage coat protein (MCP; lane 1) retains recombinant SHQ1 (lane 2) on amylose resin (lanes 6,7, respectively). Ten percent of the inputs were loaded (lanes 4,5) on this Coomassie blue-stained SDS-PAGE, and the proteins used for amylose resin binding are indicated (bold). Note, for unknown reasons, MBP-NAP57 migrates as two mass spectrometrically identical bands (lane 3). (B) In an experiment analogous to A, MBP-NAP57 (lane 3) only retains SHQ1 missing its N-terminal CS domain (SHQ-ΔCS; lane 2), but not its CS domain alone (SHQ1-CS; lane 1) on amylose resin (lanes 7,6, respectively). (C–F) Fluorographs of immunoprecipitations of in vitro translated and 35S-methionine labeled proteins separated by SDS-PAGE. The proteins targeted for precipitation are indicated (bold), and the input lanes (I) contained 10% of the material used for immunoprecipitation (IP). (C) HA-tagged SHQ1 is precipitated by anti-HA antibodies (lane 2) and coprecipitates NAP57 (lane 6) but not NAF1 (lane 4). When both NAP57 and NAF1 are present, HA-SHQ1 only precipitates NAP57 (lane 8). (D) NAP57 antibodies only precipitate SHQ1 and NAF1 in the presence of NAP57 (lanes 2,5) but not in its absence (lanes 1,4). If specifically the amount of NAF1 is increased in the input (cf. lanes 2 and 3), NAP57 only precipitates NAF1 (lane 6), suggesting a competition between SHQ1 and NAF1 for NAP57 binding. (E) HA-NHP2 fails to coprecipitate SHQ1 alone (lanes 1,2) or in the presence of the core trimer (lanes 3,4) although precipitating NAP57 and NOP10. (F) HA-NOP10 fails to coprecipitate SHQ1 in the presence of NHP2 (lanes 1,2) and in addition of NAP57 (lanes 3,4) although precipitating NAP57 and NHP2. For unknown reasons and as noted before (Wang and Meier 2004; Darzacq et al. 2006), precipitated NAP57 occasionally migrated anomalously (vertical bar in E and F). (G) Schematic summarizing the results that SHQ1 only binds NAP57 alone but not when associated with NAF1 or the core trimer (and not with NAF1, NOP10, or NHP2 alone).
To map the NAP57-interacting domain of SHQ1, its sequence was scanned for conserved domains using the NCBI conserved domain architecture retrieval tool (CDART) and the UCSC genome browser. In addition to a central SHQ1-unique domain (pfam04925), only the first 90 amino acids contain a conserved domain, termed CS for CHORD-containing proteins and Sgt1 (Shirasu et al. 1999). The CS domain is homologous to the HSP90 cochaperone p23 and to HSP20, whose main function is to interact with other proteins and which has been demonstrated to simultaneously interact with HSP90 and another protein (Lee et al. 2004; Boter et al. 2007). Therefore, it was surprising when the recombinant CS domain of SHQ1 (SHQ1-CS) failed binding to MBP-NAP57 (Fig. 1B, lane 6), whereas the other part of SHQ1, SHQ1-ΔCS, did (Fig. 1B, lane 7). Hence, SHQ1-NAP57 interaction occurred independent of the CS domain, which does not preclude an essential role for this domain in SHQ1 function (Singh et al. 2009).
To dissect the interactions of SHQ1 with other proteins, we employed a previously established in vitro translation-immunoprecipitation assay (Wang and Meier 2004; Darzacq et al. 2006). Reported crystal structures of homologous complexes subsequently validated interactions identified by this assay (Li and Ye 2006; Manival et al. 2006; Rashid et al. 2006; Leulliot et al. 2007). In vitro translated, HA-tagged SHQ1 failed to coprecipitate NAF1 (Fig. 1C, lane 4) but precipitated NAP57 (Fig. 1C, lane 6). Interestingly, no NAF1 was associated with the coprecipitating NAP57 (Fig. 1C, lane 8), indicating a possible competition for NAP57 between SHQ1 and NAF1. Indeed, increasing the amount of NAF1 in the input, while keeping that of SHQ1 constant (Fig. 1D, lanes 2,3), was accompanied by a loss of SHQ1 coprecipitation (Fig. 1D, cf. lanes 5 and 6). HA-tagged NHP2 alone and in the context of the H/ACA core trimer failed to precipitate SHQ1 (Fig. 1E, lanes 2,4). Similarly, HA-NOP10 in the presence of NHP2 and additionally NAP57 (i.e., the core trimer) failed to precipitate SHQ1 (Fig. 1F, lanes 2,4). In summary, SHQ1 in vitro directly interacted with NAP57 alone but not in the context of the core trimer or NAF1 (Fig. 1G). Moreover, SHQ1 failed to interact with NOP10, NHP2, or NAF1 alone.
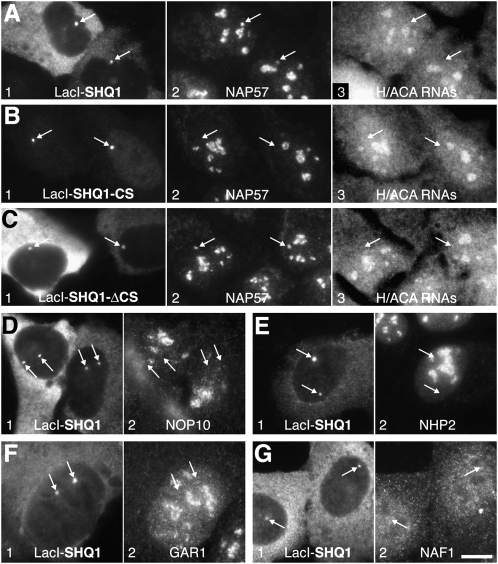
We previously documented a NAF1-RNP association in cell lysates that is not observed in intact cells (Kittur et al. 2006). Therefore, we aimed to confirm these interactions of SHQ1 in vivo using our nuclear tethering assay (Darzacq et al. 2006). Specifically, SHQ1 constructs fused to the lac repressor (LacI) were transfected into a cell line with arrays of the lac operator stably integrated in its genome. The resulting nuclear sites of LacI-SHQ1 concentration were interrogated for enrichment of endogenous proteins using indirect immunofluorescence (Fig. 2). In this manner, SHQ1 recruited endogenous NAP57 (Fig. 2A, panels 1,2, arrows), which otherwise was mostly confined to H/ACA RNPs in nucleoli. However, unlike nucleolar NAP57, that associated with SHQ1 was free of H/ACA RNAs (Fig. 2A, panel 3, arrows). SHQ1 recruitment of NAP57 was specific because the CS domain of SHQ1 alone (consistent with the in vitro results) failed to do so (Fig. 2B, arrows), whereas SHQ1 lacking the CS domain recruited NAP57 without H/ACA RNAs (Fig. 2C, arrows). Finally, full-length SHQ1 recruited neither NOP10, NHP2, GAR1, nor NAF1 (Fig. 2D–G, arrows). Therefore, the nuclear tethering data corroborated the in vitro findings that SHQ1 binding of NAP57 precluded association with any other H/ACA RNP core component. In agreement with this, GFP-tagged SHQ1, unlike NAF1 (Kittur et al. 2006), failed to coprecipitate NAP57 associated H/ACA RNA E3 (data not shown). The mutually exclusive interaction of SHQ1 and NAF1 with NAP57 suggests a sequential action of these assembly factors.
FIGURE 2.
In vivo, SHQ1 only binds NAP57 alone. Indirect immunofluorescence of transfected LacI-SHQ1 constructs tethered to lac operator repeats stably integrated into the genome of U2OS cells (arrows) and triple labeled for LacI (panel 1), the indicated endogenous H/ACA RNP associated factors (panel 2), and by fluorescent in situ hybridization for simultaneously four H/ACA RNAs, E3, ACA8, ACA18, and ACA25 (panel 3). Note, LacI-SHQ1 only recruits NAP57 (A) but none of the other H/ACA associated proteins (D–G) nor RNAs (A, panel 3). The NAP57 recruitment excludes the CS domain of SHQ1 (B) and is confined to its C-terminal SHQ1-ΔCS domain (C). Bar, 10 μm.
SHQ1 localization and dynamics
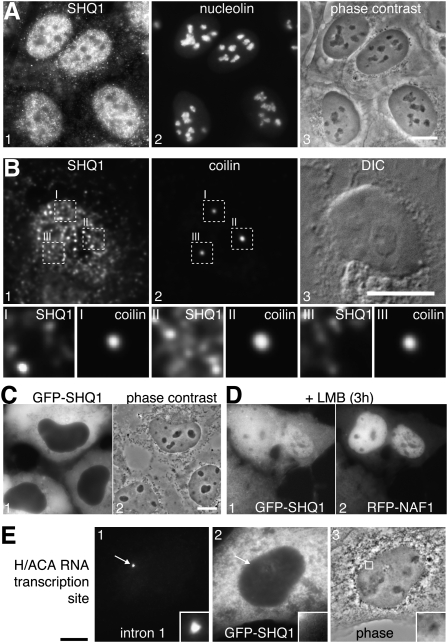
To localize endogenous SHQ1, we raised polyclonal peptide antibodies for indirect immunofluorescence experiments. SHQ1 localized to the nucleoplasm in a granular pattern but was excluded from nucleoli identified by nucleolin (Fig. 3A) and from Cajal bodies identified by coilin (Fig. 3B). Due to the small size of Cajal bodies, 0.2-μm optical sections through an entire nucleus were acquired and deconvolved using a Huygens algorithm. Maximum projections of 22 sections are depicted in Figure 3B, panels 1 and 2, whereas only the single 0.2-μm-thick plane with the maximum intensity of each Cajal body is shown magnified underneath (Fig. 3B, panels I–III). Overall, this localization further supported that SHQ1 was absent from mature H/ACA RNPs in nucleoli and Cajal bodies.
FIGURE 3.
Localization of endogenous and exogenous SHQ1. (A,B) Indirect double immunofluorescence of endogenous SHQ1 (panel 1) and nucleolin and coilin (A,B, respectively, panel 2), and corresponding phase contrast and differential interference contrast images (panel 3). Due to the small size of the coilin labeled Cajal bodies, the images in B were acquired in 22 consecutive optical planes of 0.2 μm step size and deconvolved using a Huygens algorithm. Panels 1 and 2 show maximum projections highlighting each Cajal body (squares), which are depicted enlarged in their different individual 0.2 μm planes below, demonstrating that SHQ1 is not enriched (II) and excluded from Cajal bodies (I and III). (C) Overexpressed GFP tagged SHQ1 accumulates in the cytoplasm. (D) Partial shuttling of GFP-SHQ1. Cells cotransfected with GFP-SHQ1 and RFP-NAF1 were treated for 3 h with the CRM1-dependent protein export inhibitor Leptomycin B (LMB), which caused all of RFP-NAF1 and about half of GFP-SHQ1 to accumulate in nuclei, indicating that both proteins shuttle SHQ1, at least to some extent, between nucleus and cytoplasm. Note that even when concentrated in the nucleoplasm both proteins are still excluded from nucleoli, the sites of mature H/ACA RNPs. (E) GFP-SHQ1 is not recruited to the transcription site of an H/ACA RNA. GFP-SHQ1 (panel 2) transfected into our cell line that harbors an array of an H/ACA RNA whose induction is detected by in situ hybridization using a fluorescent probe complementary to an intron of the pre-mRNA containing the H/ACA RNA (panel 1) (Darzacq et al. 2006) is not recruited to the site of transcription (arrow). Bars, 10 μm. Width of enlarged panels, 3.2 μm (B) and 2.6 μm (E).
Surprisingly, exogenously expressed, GFP-tagged SHQ1 was primarily cytoplasmic (Fig. 3C). This apparently inconsistent localization of endogenous and exogenous SHQ1 was reminiscent of that of NAF1, which shuttles between the nucleus and cytoplasm (Kittur et al. 2006). To test if SHQ1 also shuttled, GFP-SHQ1 and RFP-NAF1 were coexpressed and the cells incubated with Leptomycin B, an inhibitor of CRM1-mediated export. After 3 h, all of NAF1 accumulated in the nucleoplasm, whereas SHQ1 equilibrated between the nucleus and cytoplasm (Fig. 3D). This differential localization of GFP-SHQ1 and RFP-NAF1 was independent of their relative expression levels, suggesting that, consistent with our biochemical data (Fig. 1C), they were not complexed. The only partial nuclear accumulation of GFP-SHQ1 in the presence of Leptomycin B indicated that not all of it shuttled or that its export occurred by a pathway in addition to that mediated by CRM1. For example, there could be a limiting cytosolic factor for SHQ1 import or part of SHQ1 was actively retained. Significantly, even when concentrated in the nucleoplasm, SHQ1 was excluded from nucleoli, the sites of mature H/ACA RNPs (Fig. 3D, panel 1).
We previously identified NAF1 (together with NAP57, NOP10, and NHP2) at the site of transcription of an inducible H/ACA RNA (Darzacq et al. 2006) and tested here in the same cell system, if SHQ1 was also recruited to nascent H/ACA RNAs. SHQ1 was absent from most (98.4%, n = 123) H/ACA RNA transcription sites (Fig. 3E). This was not due to an inability to enter nuclei because SHQ1 accumulated at nuclear arrays of lac operators (Fig. 2A,D–G) and shuttled (Fig. 3D). These data are consistent with the lack of cotranscriptional association of yeast Shq1p with H/ACA RNA genes in chromatin immunoprecipitation assays (Yang et al. 2005). The absence of SHQ1 from nascent H/ACA RNA was consistent with its interaction with NAP57 precluding that of other H/ACA components (Figs. 1, 2) because NOP10, NHP2, and NAF1 together with NAP57 associate with H/ACA RNA transcription sites (Darzacq et al. 2006). Therefore, SHQ1 appears to act prior to NAF1 in H/ACA RNP biogenesis.
SHQ1- and NAF1-dependent H/ACA RNP assembly in vitro
Having established SHQ1 interactions and localization, we tested if, like its yeast counterpart, it functioned in H/ACA RNP biogenesis. For this purpose, we used a previously developed reconstitution system of functional H/ACA RNPs (Wang and Meier 2004). Specifically, HeLa cytosolic S-100 extract was incubated with an in vitro transcribed H/ACA RNA and, after pre-incubation, with its cognate target RNA. Only in the presence of its specific guide RNA was the uridine of the site-specifically labeled target RNA isomerized to pseudouridine (Fig. 4A). This indicated that the H/ACA RNA was packaged into an RNP and that the extract contained all factors required for functional RNP assembly. Therefore, pseudouridylase activity of the E3 H/ACA RNP was used as a measure for functional RNP assembly (Fig. 4B, solid bars, lanes 1,2). Addition of increasing amounts of NAP57 antibodies before or simultaneously with the E3 H/ACA RNA inhibited RNP assembly (Fig. 4B, solid bars, lanes 3,4). Antibody addition after RNP assembly impacted activity less (Fig. 4B, open bars, lanes 6,7), consistent with the fact that H/ACA RNPs bound to NAP57 antibodies were still active (Wang et al. 2002). In subsequent experiments, the assembly of functional RNPs was expressed relative to the pseudouridylation activity observed after reconstitution in the absence of exogenous factors.
FIGURE 4.
SHQ1 functions in H/ACA RNP assembly in vitro (A–E) and in vivo (F–H). (A) Autoradiograph of a thin layer chromatogram on which uridine (U) was separated from pseudouridine (Ψ) after liberation from a site-specifically 32P-labeled substrate RNA that was incubated in HeLa cytosolic S-100 extracts. Only when the cognate H/ACA RNA E3 was added to the extract for 30 min before substrate addition (lane 2), but not in its absence (lane 1), was the substrate RNA pseudouridylated, indicating that the in vitro transcribed H/ACA RNA was incorporated into a functional RNP during pre-incubation. Therefore, the pseudouridylase activity was used as a read-out for functional H/ACA RNP assembly. (B) Quantification of A (bars 1,2, respectively). Addition of increasing amounts of NAP57 antibodies prior to or concomitant with the H/ACA RNA (black bars) gradually inhibited activity (bars 3–6), indicating that they interfered with the incorporation of NAP57 into a functional RNP. Even if added after RNP assembly (open bars), the NAP57 antibodies were somewhat inhibitory. (C) Similarly, NAF1 antibodies only interfered with activity when added before (bars 1–3) but not after assembly (bars 3–6), showing that, as in vivo, functional assembly in this extract depended on NAF1. (D) SHQ1 antibodies did not interfere with activity whether added before (bars 1–3) or after assembly (bars 4–6). (E) However, addition of excess recombinant SHQ1 inhibited activity when added before (bars 1–4) but not after (bars 5–8) assembly, suggesting competition for a limiting factor, likely NAP57 itself. Indeed, this inhibitory activity resides in the NAP57-interacting SHQ1-ΔCS domain (bar 10) but not the SHQ1-CS domain (bar 9). Increasing amounts of antibodies were added as 1, 2, and 4 μL of the respective polyclonal anti-sera (B–D). Approximately 0.25, 0.5, 1.0, and 2.0 μg of recombinant proteins (Fig. 1B, lanes 1–3) were added (E). One hundred percent activity, i.e., functional assembly, corresponded to 34.6% (C), 31.0% (D), and 29.6% (E) of pseudouridine produced. The mean of duplicate values (B–D) and of one of several experiments (E) are shown. (F) Our inducible H/ACA RNA U2OS cell line (Fig. 3E) was treated for 3 d with siRNAs complementary to SHQ1 and the control nucleolar gene fibrillarin, induced for expression of the gene harboring the E3 H/ACA RNA in one of its intron, and the expression was analyzed using an RNase protection assay. Note, only in the case of SHQ1 knockdown (lane 3), but not that of fibrillarin (lane 2) or mock-treated cells (lane 1), did newly synthesized E3 fail to be stabilized, whereas the mRNA (exemplified by exon 2) was stably expressed in all three cases. Protection of SRP RNA with a separate probe served as a loading control. (G) Knockdown of the various proteins in HeLa (top) and U2OS cells (bottom) after treatment with siRNAs (targets indicated on top) was confirmed on Western blots with antibodies to the indicated antigens (right side). SHQ1 and the reference protein tubulin were extracted under low-salt conditions, whereas extraction of H/ACA RNPs (NAP57 and NHP2) and the nucleolar reference Nopp140 required 0.5 M salt. The antibodies were visualized using an infrared imaging system. Note in the case of fibrillarin knockdown, insufficient HeLa cells were harvested to analyze a separate gel for NHP2. (H) Autoradiographs of Northern blots of total RNA extracted from the siRNA treated cells in G and probed for the RNAs indicated on the right. The amounts of snoRNAs were quantified relative to those of U1 snRNA and expressed relative to those in mock-treated cells. The values for the H/ACA RNAs represent the means from two independent experiments. hTR could not be quantified because of interfering signal from spots, and levels of U17/E1 in HeLa cells appear too high due to the dark background. Finally, note that the H/ACA RNA E3 and the C/D RNA snR39B reside in different introns of the same gene.
In this in vitro reconstitution system, antibodies to NAF1 inhibited RNP assembly but failed to interfere with pseudouridylase activity if added after RNP biogenesis was allowed to proceed (Fig. 4C). These results were consistent with NAF1 being a bona fide assembly factor and indicated that our extracts replicated assembly factor-dependent H/ACA RNP biogenesis. Antibodies to SHQ1, although recognizing SHQ1 in the extracts on Western blots (data not shown), failed to interfere with assembly and pseudouridylase activity, i.e., were not inhibitory (Fig. 4D). However, addition of recombinant SHQ1 (Fig. 1A, lane 2) did interfere with assembly but not activity, consistent with being required during assembly but not for activity (Fig. 4E, lanes 1–8). This inhibitory effect was apparently related to its interaction with NAP57 because only SHQ1 without its CS domain (which is responsible for NAP57 interaction), but not the CS domain alone, behaved like the full-length protein (Fig. 4E, lanes 9–12).
SHQ1 is required for H/ACA RNA accumulation in mammalian cells
To test for the involvement of SHQ1 in H/ACA RNP biogenesis in vivo, we used siRNAs to knock it down. Knockdown of SHQ1 and subsequent induction of the β-globin construct harboring the H/ACA RNA E3 in its second intron in our inducible U2OS cell line (Darzacq et al. 2006) prevented stable accumulation of E3 but not of β-globin mRNA, as assessed by RNase protection analysis (Fig. 4F, lane 3). Previous experiments demonstrated that both NAP57 and the assembly factor NAF1 were required for stable accumulation of E3 in those cells (Darzacq et al. 2006). In control experiments, mock siRNA treatment or knockdown of fibrillarin had no effect on H/ACA RNA accumulation (Fig. 4F, lanes 1,2). These data indicated that SHQ1 was required for integration of newly transcribed E3 into stable H/ACA RNPs.
Western blots confirmed that transfection of the respective siRNAs significantly reduced the levels of SHQ1 and NAP57 relative to mock-treated cells, both HeLa and U2OS (Fig. 4G). The amounts of SHQ1 in detergent whole cell extracts were compared to those of α-tubulin, whereas those of NAP57 and NHP2 in 0.5 M salt extracts were compared to those of the nucleolar protein Nopp140. The requirement for differential extraction of SHQ1 compared with NAP57 and NHP2 further supported a lack of association of SHQ1 with mature H/ACA RNPs in the cell. Knockdown of NAP57 led to a concomitant loss of NHP2 but left SHQ1 levels unaffected (Fig. 4G, lane 4). In contrast, knockdown of SHQ1 caused a slight but reproducible reduction of both NAP57 and NHP2 (Fig. 4G, lane 3). Interestingly, knockdown of the C/D box core protein fibrillarin specifically increased the cellular levels of NAP57 and NHP2 but not of SHQ1 (Fig. 4G, lane 2). Therefore, a loss of C/D RNPs may have caused stabilization rather than increased synthesis of H/ACA RNPs.
To assess the impact of the knockdown of SHQ1 on various snoRNAs, total RNA was extracted from the transfected cells and analyzed by Northern blot (Fig. 4H). Because telomerase RNA (hTR) was not detectable in U2OS cells, which maintain their telomeres through an alternative (ALT) pathway (Bryan et al. 1997), we included HeLa cells in all our analyses. Knockdown of SHQ1 caused a loss of hTR and a moderate but clear reduction of two H/ACA RNAs, the essential U17/E1 and E3 (Fig. 4H, lane 3). In contrast, the C/D RNA, snR39B, which is expressed from a different intron of the same gene as E3 (the eukaryotic translation initiation factor 4A, isoform 2, eIF4A2), remained unaffected. These data indicated that eIF4A2 was properly expressed but E3 specifically failed to be stabilized, apparently due to lack of association with the reduced levels of H/ACA core trimer. Surprisingly, therefore, both excess SHQ1 (Fig. 4E) and SHQ1 reduction (Fig. 4G,H) impaired H/ACA RNP biogenesis, indicating that specific levels of SHQ1 are essential to the cell. Knockdown of NAP57 had a similar but generally more pronounced effect on the H/ACA RNAs, as that of SHQ1 (Fig. 4H, lane 4). It is possible that this difference is due to a lower knockdown efficiency of SHQ1, which required a combination of three siRNAs. Interestingly, whereas knockdown of fibrillarin led to the expected loss of the C/D RNA snR39B, it increased the levels of all H/ACA RNAs (Fig. 4H, lane 2), mirroring the increase in NAP57 and NHP2 (Fig. 4G, lane 2). Although the reason for this observation is not clear, similar effects were observed previously (Hoareau-Aveilla et al. 2006). This increase included hTR, which is expressed from its own promoter, and E3, which is expressed from of the same gene as snR39B. Therefore, a loss of C/D RNPs caused not only an apparent increase of H/ACA RNAs, due to lower levels of ribosomal RNAs in total RNA extracts, but an increase of entire H/ACA RNPs. The increase was apparently caused by stabilization rather than synthesis of H/ACA RNPs because only the core proteins, but not the assembly factor SHQ1, were increased (Fig. 4G, lane 2). Finally, none of the knockdowns affected the levels of the spliceosomal snRNA U1 (Fig. 4H). In summary, these data showed that SHQ1 is required for the quantitative accumulation of all H/ACA RNPs tested, including telomerase, apparently through stabilization of NAP57 and thereby of the H/ACA core trimer.
DISCUSSION
We dissected the function and molecular interactions of human SHQ1, the mammalian counterpart to the yeast H/ACA RNP assembly factor Shq1p. SHQ1 is required for the stable accumulation of all H/ACA RNPs tested, including telomerase. Unlike all other H/ACA components, SHQ1 only binds to the free pseudouridine synthase NAP57, i.e., in the absence of other H/ACA proteins or RNAs. This is consistent with the nucleoplasmic localization of SHQ1 but its exclusion from nucleoli and Cajal bodies, the sites of mature H/ACA RNPs, and from H/ACA RNA transcription sites. Moreover, it implicates SHQ1 in an early step of RNP biogenesis because all H/ACA core proteins, except GAR1, and the assembly factor NAF1 join NAP57 for cotranscriptional association with H/ACA RNAs. Specifically, we propose that SHQ1 functions as a NAP57 chaperone accompanying NAP57 from the time of synthesis until association with NOP10 and NHP2 (forming the core trimer), and with NAF1 at the nascent H/ACA RNA.
Several lines of evidence support such a mode of SHQ1 function. First, NAP57 alone is unstable. For example, when it is expressed in mammalian, insect, or bacterial cells, it fails to fold properly, as attested by mislocalization and precipitation (Darzacq et al. 2006; Normand et al. 2006). In contrast, NAP57 is stable and soluble when fused to the maltose binding protein (Fig. 1A). Second, SHQ1, although nuclear at steady state, shuttles between nucleus and cytoplasm but lacks a classical nuclear localization signal. Therefore, it may depend on those of other proteins, such as the multiple nuclear localization signals of NAP57 (Meier and Blobel 1994; Heiss et al. 1999; Youssoufian et al. 1999). Third, SHQ1 has to act before cotranscriptional association of NAP57 with H/ACA RNA and other core proteins because SHQ1 only binds NAP57 alone and because, once assembled with the core trimer, H/ACA RNAs will no longer exchange (Wang and Meier 2004; Kittur et al. 2006). Therefore, SHQ1 may function as a NAP57 chaperone and/or licensing factor marking it ready for RNP assembly.
The presence of a CS domain, which is homologous to the HSP90 cochaperone p23 and to the HSP90-interacting domain of Sgt1 (Lee et al. 2004), at the N terminus of SHQ1 was interesting given the recent implication of HSP90 in the biogenesis of box C/D and H/ACA RNPs, including telomerase (Boulon et al. 2008; Venteicher et al. 2008; Zhao et al. 2008). Since the C-terminal part of SHQ1 bound to NAP57, was it possible that its CS domain recruited HSP90 to aid in RNP biogenesis? Apparently not, because geldanamycin, an inhibitor of ATP binding to HSP90 (Stebbins et al. 1997), had no effect on SHQ1-dependent H/ACA RNP assembly in our cell free reconstitution system and in our H/ACA RNA-inducible cell line (data not shown). Moreover, excess amounts of SHQ1 without its CS domain, but not the CS domain alone, negatively impacted H/ACA RNP assembly (Fig. 4E). Perhaps, the CS domain of SHQ1 does not bind to HSP90 or does so in a distinct manner, like the differential binding of the CS domains of Sgt1 and p23 (Zhang et al. 2008). Indeed, a very recent study indicates that the CS domain of SHQ1 fails to bind HSP90 (Singh et al. 2009). Regardless, SHQ1 and NAF1 are the only bona fide assembly factors for H/ACA RNPs.
Although the function of the CS domain of SHQ1 and a possible associated role of HSP90 in the biogenesis of H/ACA RNPs remains elusive, it is tempting to speculate that they were instrumental in the identification of Cbf5p (yeast NAP57) as a low affinity centromere binding factor (Jiang et al. 1993). Specifically, the founding CS domain containing protein Sgt1p is required for the assembly of the yeast kinetochore complex CBF3 (Kitagawa et al. 1999). CBF3 includes Cbf2p/Ndc10p, and a temperature-sensitive mutation in cbf2/ndc10 is rescued by overexpression of CBF5 (Jiang et al. 1993). Perhaps this rescue is mediated indirectly through the CS domain of Shq1p (instead of that of Sgt1p) while bound to Cbf5p.
Unexpectedly, no interaction of human SHQ1 with NHP2 or NAF1 was observed, although such interactions had been reported for their yeast counterparts (Fatica et al. 2002; Yang et al. 2002). Possibly the different approaches used to identify these interactions or species differences account for this discrepancy. Nevertheless, the fact that we failed to observe any of these interactions in vitro and in vivo suggests that at least the mammalian SHQ1 only binds to NAP57 but not to NHP2 and NAF1 or to any other H/ACA component. Moreover, the finding that SHQ1 only binds NAP57 alone but not in the context of other proteins or mature RNPs also contradicts a recent study reporting recruitment of Cajal body components, including H/ACA RNPs, to SHQ1 tethered in the nucleus (Kaiser et al. 2008). However, based on our inherent consistent biochemical (Fig. 1) and tethering data (Fig. 2), and, importantly, on the absence of endogenous SHQ1 from Cajal bodies (Fig. 3B), SHQ1 only binds to NAP57 alone.
Establishing a role for SHQ1 in H/ACA RNP biogenesis before that of NAF1 raises the question as to the function of NAF1 shuttling. NAF1 binds NAP57 at the site of H/ACA RNA transcription and subsequently is replaced by GAR1 to form mature H/ACA RNPs (Darzacq et al. 2006). Before this role, NAF1 might be involved in the import of NOP10 and NHP2, which do not possess classical nuclear localization signals. However, NOP10 and NHP2 are sufficiently small for passive diffusion into the nucleus, and in nuclear tethering assays, neither protein was recruited to NAF1 lacking its NAP57-interacting domain (data not shown). Alternatively, NAF1 may be involved in the nuclear export of the very recently discovered class of H/ACA RNAs that harbors microRNAs (Ender et al. 2008). Obviously, there is plenty more work ahead to dissect the detailed function of the two (and perhaps more) H/ACA RNP assembly factors.
MATERIALS AND METHODS
DNA/RNA constructs, transfections, and translations
Most constructs are as previously described (Wang and Meier 2004; Darzacq et al. 2006). The human SHQ1 clone, number 4840343, was obtained from American Type Culture Collection (Manassas, VA). The following constructs were for transient expression in tissue culture cells: GFP-SHQ1 (pNK37, SHQ1 fused to mGFP in mGFP-C1); RFP-SHQ1 (pNK38, SHQ1 fused to monomeric RFP in monomeric RFP-C1) (Campbell et al. 2002); SHQ1-LacI (pSR60, SHQ1 was fused to the lac-repressor, LacI, in pSR59, which is pcDNA3 containing LacI); SHQ1-CS-LacI (pPG10, SHQ1-CS was fused to LacI in pSR59); SHQ1-ΔCS-LacI (pPG11, SHQ1-ΔCS in pSR59); for in vitro translation: SHQ1 (pSR32, SHQ1 under T7 promoter in pBSII SK+; Stratagene); HA-SHQ1 (pSR38, SHQ1 with a single HA tag in pTM93) (Isaac et al. 1998). For bacterial expression, SHQ1 (pPG18), SHQ1-CS (pPG14), and SHQ1-ΔCS (pPG15) were fused to N- and C-terminal hexa-histidine tags in a pET22 (Novagen, EMD Chemicals, Inc.) derivative containing an additional N-terminal and cleavable (TEV protease) hexa-histidine tag, pET22HT (a kind gift from Jeffrey A. Chao, Albert Einstein College of Medicine of Yeshiva University); MBP-NAP57 (pPG17, NAP57 fused to N-terminal maltose binding protein and C-terminal hexa-histidine tag in a modified version of pMAL-c; New England BioLabs from Jeffrey A. Chao); MBP-MCP (MS2 bacteriophage coat protein fused to the maltose binding protein; a kind gift from Charles Query, Albert Einstein College of Medicine of Yeshiva University).
siRNAs were as previously described (Darzacq et al. 2006) except for siSHQ1, which was a combination of siSHQ1.1, GCUACGAAAAUUUGUCAAUTT; siSHQ1.2, GGAAGUAGUUGACGAUGAATT; and siSHQ1.3, GGACAGCAAAACCACUUGUTT (Ambion). Cells were maintained, treated, and transfected as we previously described (Darzacq et al. 2006; Kittur et al. 2006). Similarly, the inducible H/ACA RNA cell line and related analyses (Darzacq et al. 2006) and the cell line containing integrated Lac operator arrays (Janicki et al. 2004) have been described. Coupled in vitro transcription/translation and subsequent immunoprecipitations were performed according to our previous protocols (Wang and Meier 2004; Darzacq et al. 2006).
Protein expression, purification, and pull-down
The bacterial strain BL21 AI (Invitrogen) was used for expression of the SHQ1 constructs and Rosetta 2 (Novagen) for MBP-NAP57. The bacteria were grown at 37°C and expression induced for 3 h with 1 mM IPTG, and 0.2% arabinose in case of BL21 AI; cell pellets were resuspended in 1/20 culture volume of 0.8 M NaCl, 20 mM Tris-HCl at pH 7.4, 1 mM EDTA, 1 mM DTT, supplemented with protease inhibitors (Sigma) and disrupted by nitrogen cavitation through an EmulsiFlex-C5 (Avestin Inc.). The cleared cell suspension (27000g for 20 min at 4°C) was passed over a 10 mL amylose resin column (New England Biolabs) and the proteins eluted by 10 mM maltose in 0.2 M NaCl, 20 mM Tris-HCl at pH 7.4, 1 mM EDTA, 1mM DTT. MBP-NAP57 was further affinity purified by virtue of its hexa-histidine tag on a HIS select HF Ni Affinity gel (Sigma) after dialysis against 0.3 M NaCl, 50 mM sodium phosphate at pH 8.0, 10 mM imidazole, and was eluted by 0.5 M imidazole in 0.3 M NaCl, 50 mM sodium phosphate at pH 8.0. The SHQ1 constructs were similarly purified by nickel affinity chromatography except that the cells were directly suspended in 0.3 M NaCl, 50 mM sodium phosphate at pH 8.0, 10 mM imidazole, protease inhibitors. Additionally, the purified proteins were dialyzed against buffer used to assay pseudouridylation, 20 mM HEPES at pH 7.9, 120 mM KCl, 2 mM MgCl2. For pull-downs, MBP-fusion proteins were immobilized on 25 μL amylose resin in 120 mM NaCl, 20 mM Tris-HCl at pH 7.4, 0.02% TritonX-100; incubated with 2 μg of SHQ1 protein constructs; washed; and eluted with SDS sample buffer for analysis by SDS-polyacrylamide electrophoresis.
Pseudouridylation and H/ACA RNP reconstitution assay
In vitro assembly of H/ACA RNP was carried out as described previously (Wang and Meier 2004). Briefly, a typical 25 μL reaction contained 5 μL of HeLa cytoplasmic S-100 extracts (Dignam et al. 1983), reconstitution buffer (20 mM HEPES-KOH at pH 7.9, 120 mM KCl, and 2 mM MgCl2), 20 U RNaseOUT (Invitrogen), 1 mM ATP, 2 μg Escherichia coli tRNA, and ∼0.6 pmol of rat guide E3 snoRNA. The reaction mixture was pre-incubated for 30 min at 30°C to allow RNP assembly prior to adding the site-specifically and 32P-labeled substrate rRNA (Wang et al. 2002). After incubation for additional 90 min at 30°C, RNA was recovered, digested with RNase T2 (purified according to the method of Lichtler et al. [1992]), and the resulting 3′ nucleotide monophosphates were separated by thin layer chromatography (Wang et al. 2002). Where indicated increasing amounts of anti-sera and recombinant SHQ1 protein constructs were added either before the pre-incubation step or concomitant with the substrate RNA.
Fluorescence and immunochemical approaches
Unless stated otherwise the antibodies, equipment, and procedures were identical to those previously employed (Darzacq et al. 2006). Specifically for the data in Figure 3B, three-dimensional stacks of images were acquired with a 200-nm step size using a BX61 microscope with a 100×, 1.35-NA objective (Olympus) and CoolSNAP-HQ CCD camera (Photometrics) using IPLab software (version 3, BD Biosciences Bioimaging) using filters for Cy2 (model no. 41001) and Cy5 (model no. 41008, Chroma Technology). Deconvolutions were performed using the CMLE algorithm in Huygens Professional (version 3.3, Scientific Volume Imaging) set to perform 400 iterations to quality factor 0.01. The images were adjusted using Photoshop CS4 (Adobe) and Image J (NIH) software.
The following additional primary antibodies were used (dilutions in parentheses for Western blots and, where indicated, indirect immunofluorescence): SHQ1 antibodies (BA10 IgG at 0.7 μg/mL) were raised in rabbits (Covance Research Products) against a synthetic peptide GC472GSDSEQDELKDSPSE486 where the two N-terminal amino acids are not part of SHQ1 (Biosynthesis) and affinity purified (Meier and Blobel 1992); mouse monoclonal α-tubulin antibodies (Sigma; at 0.3 μg/mL); rabbit antibodies against recombinant human Nopp140 (RS8 serum, 1:1000) (Kittur et al. 2007); NHP2 antibodies (CG2 serum at 1:1000) were raised in rabbits against the synthetic peptide 125CVIMVKPHEEYQEAY139 (Covance Research Products); NOP10 antibodies (CF3 serum at 1:1000 for fluorescence) were raised in rabbits against the synthetic peptide 51RFKVLMTQQPRPVL64 with an additional N-terminal cysteine (Covance Research Products). Slides for indirect immunofluorescence were mounted in ProLong Gold Antifade medium (Invitrogen).
Cell and RNA extracts
For protein extraction, cells from one 10-cm dish were lysed in 150 mM NaCl, 20 mM HEPES at pH 7.4, 2mM EGTA, 2 mM EDTA, 1% TritonX-100 (v/v), 5% glycerol (v/v), 1 mM DTT, 25 μL/mL protease inhibitors (Sigma) and pelleted. The supernatant represented the low-salt extract, and the pellet was further extracted in 150 μL high-salt buffer (0.5M NaCl, 20 mM HEPES at pH 7.4, 2 mM MgCl2, 10% glycerol [v/v], 1 mM DTT, protease inhibitors). The protein concentration was determined using protein assay reagent (Bio-Rad), and 80 μg and 50 μg of the low- and high-salt extracts per lane, respectively, were analyzed by SDS-polyacrylamide electrophoresis. Proteins on immunoblot were detected using an infrared imaging system (Odyssey; LI-COR Biosciences) as described previously (Darzacq et al. 2006).
For Northern blots, total RNA was extracted using Trizol reagent (Invitrogen), 4–5 μg per lane were loaded on 6%–8% denaturing (8 M urea) polyacrylamide gels, transferred to positively charged nylon membranes (Immobilon NY+, Millipore), and UV cross-linked using a GS Gene Linker UV chamber (Bio-Rad) preset to 150 mJ/cm2. For detection, 32P-end-labeled oligonucleotides were used, except in the case of hTR, where the probe was generated by random prime labeling of the full-length sequence. Final washes of the blots were at 37°C with 7× SSC, 1% SDS and, in the case of hTR, at room temperature with 0.1× SSC, 50 mM sodium phosphate at pH 7.0, 1% SDS. The signals were recorded by autoradiography on BioMax MS film (Eastman Kodak.) and quantified using a PhosphorImager (Molecular Dynamics).
ACKNOWLEDGMENTS
We thank Jeffrey Chao, Charles Query, Witek Filipowicz, Maria Carmo-Fonseca, and Serafin Pinol-Roma for reagents. We thank Shailesh Shenoy for help in acquiring and analyzing the images in Figure 3B and the Einstein Analytical Imaging Facility for use of their microscopes. This work was supported by a grant from the National Institutes of Health (HL079566) to U.T.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1532109.
REFERENCES
- Baker D.L., Youssef O.A., Chastkofsky M.I., Dy D.A., Terns R.M., Terns M.P. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes & Dev. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M., Morlando M., Pagano F., Fatica A., Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Amigues B., Peart J., Breuer C., Kadota Y., Casais C., Moore G., Kleanthous C., Ochsenbein F., Shirasu K., et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jady B.E., Rothe B., Pescia C., Robert M.C., Kiss T., et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B., Muller S., Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R.H., Meier U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Noaillac-Depeyre J., Caizergues-Ferrer M., Henry Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Fatica A., Dlakic M., Tollervey D. Naf1p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Heiss N.S., Girod A., Salowsky R., Wiemann S., Pepperkok R., Poustka A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum. Mol. Genet. 1999;8:2515–2524. doi: 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hoareau-Aveilla C., Bonoli M., Caizergues-Ferrer M., Henry Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA. 2006;12:832–840. doi: 10.1261/rna.2344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C., Yang Y., Meier U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., et al. From silencing to gene expression: Real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Middleton K., Yoon H.-J., Fouquet C., Carbon J. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kirwan M., Dokal I. Dyskeratosis congenita: A genetic disorder of many faces. Clin. Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa K., Skowyra D., Elledge S.J., Harper J.W., Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Kittur N., Darzacq X., Roy S., Singer R.H., Meier U.T. Dynamic association and localization of human H/ACA RNP proteins. RNA. 2006;12:2057–2062. doi: 10.1261/rna.249306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittur N., Zapantis G., Aubuchon M., Santoro N., Bazett-Jones D.P., Meier U.T. The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol. Biol. Cell. 2007;18:2296–2304. doi: 10.1091/mbc.E07-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.T., Jacob J., Michowski W., Nowotny M., Kuznicki J., Chazin W.J. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 2004;279:16511–16517. doi: 10.1074/jbc.M400215200. [DOI] [PubMed] [Google Scholar]
- Leulliot N., Godin K.S., Hoareau-Aveilla C., Quevillon-Cheruel S., Varani G., Henry Y., Van Tilbeurgh H. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J. Mol. Biol. 2007;371:1338–1353. doi: 10.1016/j.jmb.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Li H. Unveiling substrate RNA binding to H/ACA RNPs: One side fits all. Curr. Opin. Struct. Biol. 2008;18:78–85. doi: 10.1016/j.sbi.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- Lichtler A., Barrett N.L., Carmichael G.G. Simple, inexpensive preparation of T1/T2 ribonuclease suitable for use in RNase protection experiments. Biotechniques. 1992;12:231–232. [PubMed] [Google Scholar]
- Manival X., Charron C., Fourmann J.B., Godard F., Charpentier B., Branlant C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 2006;34:826–839. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Meier U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U.T. Studying and working with ribonucleoproteins that catalyze H/ACA guided RNA modification. In: Smith H.C., editor. RNA and DNA editing: Molecular mechanisms and their integration into biological systems. Wiley; Hoboken, NJ: 2008. pp. 162–174. [Google Scholar]
- Meier U.T., Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier U.T., Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell. Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. (correction appeared in 140: 447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand C., Capeyrou R., Quevillon-Cheruel S., Mougin A., Henry Y., Caizergues-Ferrer M. Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA. RNA. 2006;12:1868–1882. doi: 10.1261/rna.141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R., Liang B., Baker D.L., Youssef O.A., He Y., Phipps K., Terns R.M., Terns M.P., Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Shirasu K., Lahaye T., Tan M.W., Zhou F., Azevedo C., Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans . Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Singh M., Gonzales F.A., Cascio D., Heckmann N., Chanfreau G., Feigon J. Structure and functional studies of the CS domain of the essential H/ACA RNP assembly protein Shq1. J. Biol. Chem. 2009;284:1906–1916. doi: 10.1074/jbc.M807337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Russo A.A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. Crystal structure of an Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., Artandi S.E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Query C.C., Meier U.T. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 2002;22:8457–8466. doi: 10.1128/MCB.22.24.8457-8466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.K., Hoareau C., Froment C., Monsarrat B., Henry Y., Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.K., Rotondo G., Porras T., Legrain P., Chanfreau G. The Shq1p•Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- Ye K. H/ACA guide RNAs, proteins and complexes. Curr. Opin. Struct. Biol. 2007;17:287–292. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Gharibyan V., Qatanani M. Analysis of epitope-tagged forms of the dyskeratosis congenital protein (dyskerin): Identification of a nuclear localization signal. Blood Cells Mol. Dis. 1999;25:305–309. doi: 10.1006/bcmd.1999.0258. [DOI] [PubMed] [Google Scholar]
- Zhang M., Boter M., Li K., Kadota Y., Panaretou B., Prodromou C., Shirasu K., Pearl L.H. Structural and functional coupling of Hsp90- and Sgt1-centred multi-protein complexes. EMBO J. 2008;27:2789–2798. doi: 10.1038/emboj.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R., et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]