Abstract
Proteins of the GW182 family are essential components of the miRNA pathway in animal cells. Vertebrate genomes encode three GW182 paralogs (TNRC6A, TNRC6B, and TNRC6C), which may be functionally redundant. Here, we show that the N-terminal GW-repeat-containing regions of all three TNRC6s interact with the four human Argonaute proteins (AGO1–AGO4). We also show that TNRC6A, TNRC6B, and TNRC6C silence the expression of bound mRNAs. This activity is mediated by their C-terminal silencing domains, and thus, is independent of the interaction with AGO1–AGO4. Silencing by TNRC6A, TNRC6B, and TNRC6C is effected by changes in protein expression and mRNA stability that can, in part, be attributed to deadenylation. Our findings indicate that TNRC6A, TNRC6B, and TNRC6C are recruited to miRNA targets through an interaction between their N-terminal domain and an Argonaute protein; the TNRC6s then promote translational repression and/or degradation of miRNA targets through a C-terminal silencing domain.
Keywords: Argonaute, GW182, GW-repeats, miRNAs, mRNA decay, P-bodies
INTRODUCTION
Proteins of the GW182 family are essential components of mRNA-processing bodies (P-bodies) and are required for miRNA-mediated gene silencing in animal cells (Eystathioy et al. 2002, 2003; for review, see Ding and Han 2007; Eulalio et al. 2007; 2008a). There are three GW182 paralogs in vertebrates (TNRC6A/GW182, TNRC6B, and TNRC6C), a single ortholog in insects, and no orthologs in fungi (Behm-Ansmant et al. 2006a,b; Ding and Han 2007). The vertebrate and insect proteins share a common domain organization characterized by two annotated structural domains: a central ubiquitin associated (UBA)-like domain and a C-terminal RNA recognition motif (RRM). These domains are embedded into regions predicted to be unstructured. These unstructured regions include both two to three distinctive blocks of glycine-tryptophan repeats (referred to as: the N-terminal, middle, and C-terminal GW-repeats), as well as a glutamine-rich (Q-rich) region located between the UBA-like domain and the RRM (Fig. 1A; Eystathioy et al. 2002; Behm-Ansmant et al. 2006a; Ding and Han 2007). The N-terminal GW-repeat-containing regions of vertebrate and insect GW182 bear similarity to two proteins in C. elegans (AIN-1 and AIN-2); however, AIN-1 and AIN-2 do not contain a Q-rich region, a UBA-like, or RRM domains (Ding et al. 2005; Behm-Ansmant et al. 2006a; Zhang et al. 2007).
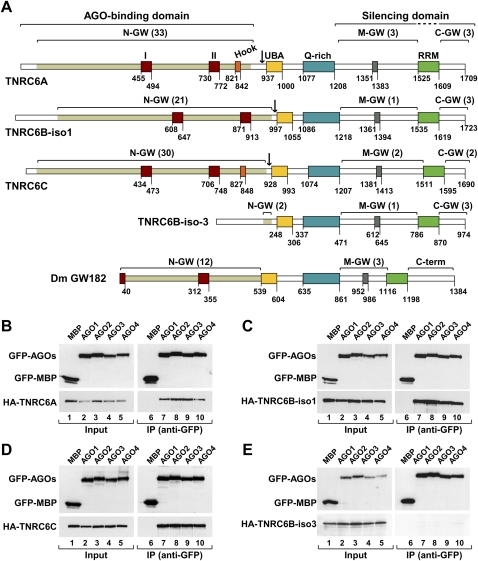
FIGURE 1.
TNRC6A, TNRC6B, and TNRC6C interact with human AGO1–AGO4. (A) Domain organization of human TNRC6A–TNRC6C and D. melanogaster GW182. N-GW, M-GW, and C-GW: N-terminal, middle, and C-terminal GW-repeat-containing regions, respectively (the number of GW-repeats for each region is indicated in brackets). The white sectors within the N-terminal regions do not contain GW-repeats. UBA (yellow box): ubiquitin associated-like domain; Q-rich (cyan box): region rich in glutamine; RRM (green box): RNA recognition motif. Red boxes I and II: two conserved motifs within the N-terminal GW repeats. (Orange box) Ago-hook motif; (gray box) conserved motif III in the middle region. Numbers underneath the protein outline represent amino acid positions at fragment boundaries for each protein. Black arrows indicate the boundaries of the N and C-terminal protein fragments used in this study. The protein domains sufficient for the interaction with AGO1 and for silencing are indicated. (B–E) GFP-AGO1–AGO4 and HA-TNRC6A–TNRC6C were coexpressed in human HEK293 cells as indicated. Cell lysates were immunoprecipitated using anti-GFP antibodies. GFP-MBP served as a negative control. Inputs (2%) and immunoprecipitates (30%) were analyzed by Western blotting using anti-GFP or anti-HA antibodies.
The N-terminal GW-repeat-containing region of GW182 proteins is required for the interaction with the Argonaute proteins (Behm-Ansmant et al. 2006a,b; Till et al. 2007). This region contains two highly conserved motifs, motif I and motif II (Fig. 1A; Behm-Ansmant et al. 2006a), and downstream from motif II, an additional region (termed the AGO-hook) that is sufficient for the interaction between human TNRC6B and AGO2 in vitro (Till et al. 2007). Apart from the role of the N-terminal region in interacting with the Argonaute proteins, it is not known what contribution the other domains make to the silencing activity of the human TNRC6A–TNRC6C. Furthermore, it is unclear whether TNRC6C localizes to P-bodies and whether it exhibits silencing activity.
To gain insight into the role of all three human TNRC6A, TNRC6B, and TNRC6C in the miRNA pathway, we introduced into them a series of deletions. We then tested whether these mutant proteins interact with Argonaute proteins and localize to P-bodies. We show that the N-terminal GW-repeat-containing regions of TNRC6A, TNRC6B, and TNRC6C interact with all four human Argonaute proteins. This N-terminal AGO-binding region is sufficient for TNRC6A to accumulate in P-bodies. In contrast, the accumulation of TNRC6B and TNRC6C in P-bodies requires, in addition, the Q-rich region. We further show that TNRC6A, TNRC6B, and TNRC6C trigger silencing of bound transcripts by inhibiting protein expression and promoting mRNA decay via deadenylation. This activity is mediated by a bipartite C-terminal region of the proteins, referred to as silencing domain, which includes the middle and C-terminal GW-repeat-containing regions. The C-terminal silencing domains do not interact with AGO1–AGO4, indicating that TNRC6A–TNRC6C can trigger silencing of bound mRNAs independently of the Argonaute proteins. Furthermore, the silencing domains do not localize to P-bodies. Thus, P-body localization is not required for silencing activity. Together, our results indicate that TNRC6A–TNRC6C proteins interact with the Argonaute proteins via their N-terminal domains and trigger silencing of bound transcripts via their C-terminal silencing domains.
RESULTS AND DISCUSSION
The N-terminal GW-repeats of TNRC6A–TNRC6C interact with AGO1–AGO4
The N-terminal GW-repeat-containing region of the GW182 family proteins is necessary and sufficient for the interaction with the Argonaute proteins (Behm-Ansmant et al. 2006a; Ding and Han 2007; Till et al. 2007). This region comprises two highly conserved motifs (motifs I and II), present in all members of the protein family, and a variable number of glycine-tryptophane (GW) repeats (Fig. 1A; Eystathioy et al. 2002; Behm-Ansmant et al. 2006a). GW-repeats represent specificity determinants in the interaction of various proteins with the Argonaute proteins (El-Shami et al. 2007; Till et al. 2007; Eulalio et al. 2009a). An additional protein sequence downstream from motif II, known as the AGO-hook was shown to be sufficient for the interaction between human TNRC6B (isoform 2) and AGO2 in vitro (Till et al. 2007); it is not known, however, whether the AGO-hook is required for AGO binding in the context of full-length TNRC6B. Furthermore, the AGO-hook is not conserved in all members of the GW182 family: It is missing from isoform 1 of TNRC6B (TNRC6B-iso1), and isoform 3 of TNRC6B (TNRC6B-iso3) lacks the entire N-terminal GW-repeat-containing region (Fig. 1A).
To investigate whether TNRC6A–TNRC6C interact equally with AGO1–AGO4 and to define more precisely the interaction domains, we coexpressed GFP-tagged AGO1–AGO4 with HA-tagged TNRC6A–TNRC6C in all combinations in human HEK293 cells and performed coimmunoprecipitation assays using anti-GFP antibodies. We also tested for the interaction between TNRC6B-iso3 and AGO1–AGO4. We observed that TNRC6A, TNRC6B, and TNRC6C coimunoprecipitated with each of the four human Argonaute proteins (Fig. 1B–D, AGO1–AGO4), suggesting that the interactions are redundant. These results are consistent with those reported by Landthaler et al. (2008). In contrast, isoform 3 of TNRC6B did not coimmunoprecipitate with any of the Argonaute proteins (Fig. 1E). Because TNRC6B-iso3 is identical to the C-terminal half of TNRC6B-iso1, but lacks most of the N-terminal GW-repeats, these results indicate the N-terminal repeats are required for the interaction between TNRC6s and AGO1–AGO4; this is in agreement with previous studies (Behm-Ansmant et al. 2006a; Till et al. 2007; Eulalio et al. 2009a).
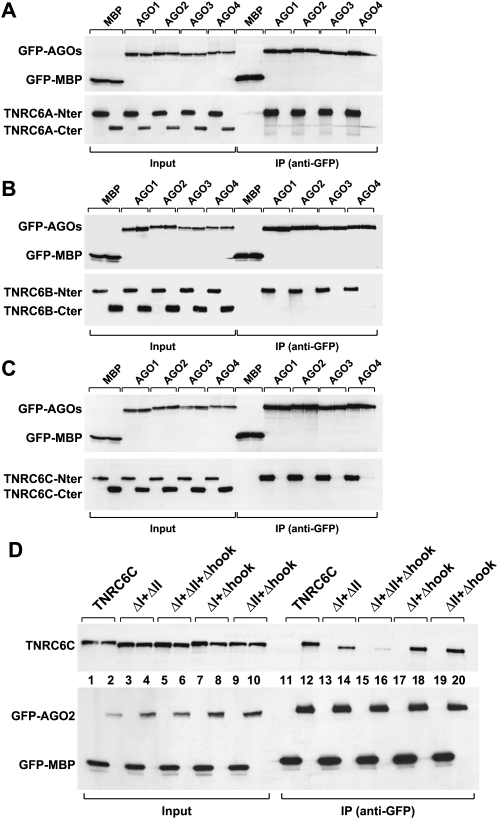
To test whether the N-terminal GW-repeat-containing region was sufficient for the interaction with AGO1–AGO4, we generated TNRC6A–TNRC6C deletion mutants containing either the N-terminal portion of the proteins (i.e., upstream of the UBA-like domain) or the corresponding C-terminal fragments (Fig. 1A, black arrows indicate fragment boundaries). We observed that all N-terminal fragments interacted with AGO1–AGO4 (Fig. 2A–C), whereas the C-terminal fragments did not interact with AGO1–AGO4 despite the fact that these fragments also contain GW-repeats (Figs. 1A, 2A–C). Together, our results indicate that the N-terminal GW-repeats of TNRC6s are required for the interaction with AGO1–AGO4, whereas the middle and C-terminal GW-repeats do not contribute to AGO1–AGO4 binding.
FIGURE 2.
The N-terminal GW-repeat-containing regions of TNRC6A–TNRC6C are required for the interaction with AGO1–AGO4. (A–C) GFP-AGO1–AGO4 and HA-TNRC6A–TNRC6C N- and C-terminal fragments were coexpressed in human HEK293 cells as indicated. Cell lysates were immunoprecipitated using anti-GFP antibodies. GFP-MBP served as a negative control. Inputs and immunoprecipitates were analyzed by Western blot as described in Figure 1, B–E. (D) Wild-type HA-TNRC6C or mutants were coexpressed with GFP-AGO2 or GFP-MBP in human HEK293 cells as indicated. Cell lysates were immunoprecipitated using anti-GFP antibodies. Inputs (2%) and immunoprecipitates (50%) were analyzed as described in A–C.
The N-terminal GW-repeats provide multiple binding sites for the interaction with AGO1–AGO4
To further investigate the sequences required for the interaction between TNRC6A–TNRC6C and the Argonaute proteins, we deleted motif I or II in each of the three TNRC6A–TNRC6C proteins. We also deleted the AGO-hook in TNRC6A and TNRC6C. Deleting each of these motifs individually did not prevent TNRC6A, TNRC6B, or TNRC6C from interacting with AGO2 (Supplemental Fig. 1A–C). In contrast, deleting motifs I and II from TNRC6C impaired the interaction with AGO2 (Fig. 2D, lane 14). The interaction was further inhibited when, in addition to motifs I and II, the AGO-hook was deleted (Fig. 2D, lane 16). However, deleting the AGO-hook in combination with either motif I or II did not interfere with AGO2 binding (Fig. 2D, lanes 18,20). Thus, the conserved motifs I and II in TNRC6C provide major binding sites for the interaction with AGO1–AGO4.
In contrast, deleting motifs I and II and the AGO-hook in the context of full-length TNRC6A only slightly reduced AGO2 binding (Supplemental Fig. 2D). A small reduction in AGO binding was also observed by deleting motifs I and II in TNRC6B-iso1 (Supplemental Fig. 2E). Thus, most of the N-terminal GW-repeats in TNRC6A and TNRC6B appear to contribute to the interaction with the Argonaute proteins. These results differ from those reported for D. melanogaster GW182, where motif I alone provides the major binding site for the interaction with AGO1 (Eulalio et al. 2009a).
The AGO-binding domains and the Q-rich regions are required for the localization of TNRC6B and TNRC6C in P-bodies
While it was shown that TNRC6A and TNRC6B colocalize with AGO1 and AGO2 in P-bodies (Jakymiw et al. 2005; Liu et al. 2005a,b; Meister et al. 2005; Pillai et al. 2005; Till et al. 2007), it has not been established whether TNRC6C localizes to P-bodies. Moreover, the domains of TNRC6A–TNRC6C required for P-body localization have not been defined. To identify what domains are required by TNRC6A–TNRC6C to accumulate in P-bodies, we examined the subcellular localization of various deletion mutants.
First, we confirmed that all three HA-tagged TNRC6A–TNRC6C colocalized with GFP-AGO1–AGO4 in P-bodies (Fig. 3A; Supplemental Fig. 2A,B). In contrast, TNRC6B-iso3 did not localize to P-bodies and spread throughout the cytoplasm (Supplemental Fig. 2C). These results suggest that the N-terminal GW-repeat-containing regions are required for TNRC6A–TNRC6C to accumulate in P-bodies.
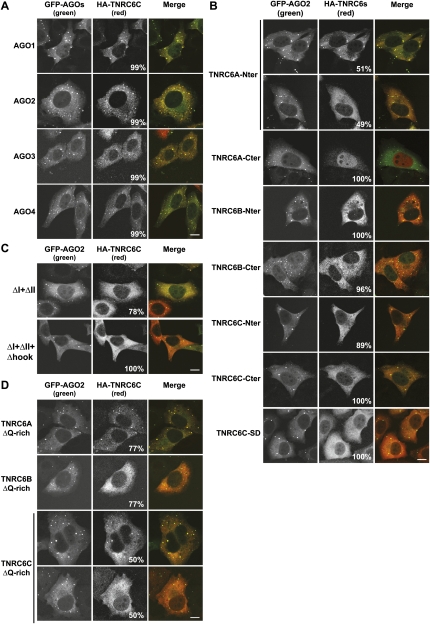
FIGURE 3.
The AGO-binding domains and the Q-rich regions are required for P-body localization. (A) Localization of HA-TNRC6C in HeLa cells expressing GFP-AGO1–AGO4. (B) Localization of HA-tagged N- or C-terminal fragments of HA-TNRC6A–TNRC6C in HeLa cells expressing GFP-AGO2. (C) Localization of TNRC6 mutants in HeLa cells expressing GFP-AGO2. (D) Localization of HA-TNRC6A, TNRC6B, and TNRC6C lacking the Q-rich regions in HeLa cells expressing GFP-AGO2. In all panels, the merged images show the GFP signal in green and the HA signal in red. The fraction of cells exhibiting a staining identical to that shown in the representative panel was determined by scoring at least 100 cells (showing Argonaute-containing foci) in two independent transfections performed per protein. Scale bar, 10 μm.
We next tested the subcellular localization of the N- and C-terminal TNRC6A–TNRC6C fragments described above. The C-terminal fragments (C-ter; i.e., starting at the UBA-like domain) did not localize to P-bodies as expected from the results obtained with TNRC6B-iso3 (Fig. 3B). Surprisingly, however, the N-terminal GW-repeat-containing fragments (N-ter) of TNRC6B and TNRC6C also failed to localize to P-bodies, despite that GFP-AGO2 did accumulate in P-bodies in these cells (Fig. 3B). The exception was the N-terminal fragment of TNRC6A, which was detected in AGO2-containing foci in about 50% of the cells (Fig. 3B). The observation that the N-terminal fragments do not efficiently accumulate in P-bodies despite the fact that they interact with AGO1–AGO4, reveals that the interaction with AGO1–AGO4 is not sufficient to direct TNRC6A–TNRC6C to P-bodies. Nevertheless, an interaction with AGO1–AGO4 is required for P-body localization because the TNRC6C mutants (ΔI+II and ΔI+II+hook) that were impaired in the interaction with AGO1–AGO4 also failed to accumulate in P-bodies (Fig. 3C)
Localization of D. melanogaster GW182 in P-bodies requires both the N-terminal GW-repeats and the Q-rich region (Behm-Ansmant et al. 2006a; Eulalio et al. 2009a). We therefore tested whether the Q-rich region was important for the localization of TNRC6A–TNRC6C in P-bodies. We observed that in the context of full-length TNRC6B and TNRC6C, deleting the Q-rich region but not the UBA-like domain impaired P-body localization with about 77% and 50% of cells, respectively, showing a diffuse staining (Fig. 3D). These results indicate that the Q-rich regions contribute to P-body localization; however, these regions on their own are not sufficient, because TNRC6B-iso3 and the C-terminal TNRC6A–TNRC6C fragments described above do not localize to P-bodies (Fig. 3B; Supplemental Fig. 2C). We conclude that the N-terminal AGO-binding domains and the Q-rich regions contribute to the localization of TNRC6B and TNRC6C to P-bodies, though none of these suffice on their own. In contrast, the accumulation of a TNRC6A protein lacking the Q-rich region in P-bodies was only slightly impaired, with about 77% of cells showing P-body staining (Fig. 3D), whereas full-length TNRC6A accumulated in P-bodies in about 99%–100% of the cell population (Supplemental Fig. 2A).
The requirements of the human proteins for P-body localization slightly differ from those observed for D. melanogaster GW182, where deletion of the Q-rich region completely abolished P-body localization (Eulalio et al. 2009a). Nevertheless, in all cases, the interaction with the Argonaute proteins is required for P-body localization, whereas the Q-rich regions either contribute (human TNRC6A and TNRC6C) or play an essential role (D. melanogaster GW182 and human TNRC6B). In the case of C. elegans AIN-1 and AIN-2, which lack a defined Q-rich region (Ding et al. 2005; Zhang et al. 2007), P-body localization may only require the interaction with the Argonaute proteins.
TNRC6A–TNRC6C silences the expression of bound mRNAs
D. melanogaster GW182 and human TNRC6A can silence the expression of bound transcripts independently of the Argonaute proteins (Behm-Ansmant et al. 2006a; Li et al. 2008). We therefore investigate whether TNRC6C and the two TNRC6B isoforms exhibited a similar activity. To this end, we made use of the tethering assay described by Pillai et al. (2004). This assay involves the expression of λN-fusion proteins that bind with high affinity to five BoxB sites (5BoxB) in the 3′UTR of a Renilla luciferase reporter mRNA (R-Luc-5BoxB reporter).
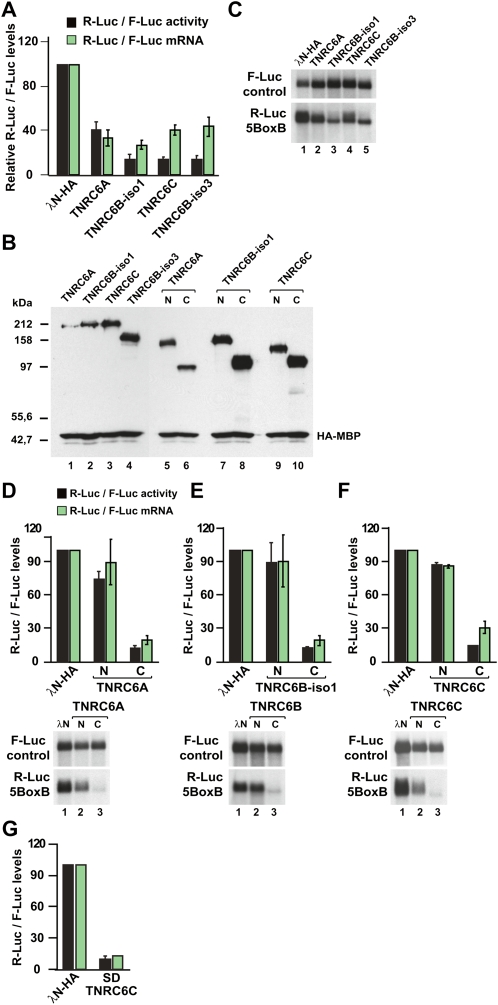
Human HEK293 cells were cotransfected with three plasmids: a R-Luc-5BoxB reporter, a plasmid-expressing TNRC6A, TNRC6B, or TNRC6C fused to the λN-peptide (λN-HA-TNRC6A–C), and a transfection control plasmid encoding firefly luciferase (F-Luc). When TNRC6A was tethered to the reporter luciferase, expression fell threefold relative to that measured in cells expressing the λN-HA-peptide alone (Fig. 4A, black bars). A stronger inhibitory effect was observed for TNRC6C and the two TNRC6B isoforms (Fig. 4A,B). The lower activity of TNRC6A is probably due to its lower level of expression (Fig. 2B). TNRC6A–TNRC6C did not affect the expression of a Renilla luciferase reporter lacking the 5BoxB binding sites (data not shown).
FIGURE 4.
TNRC6A–TNRC6C silence the expression of bound transcripts independently of the Argonaute proteins. (A–C) Human HEK293 cells were transfected with a mixture of three plasmids: the R-Luc-5BoxB reporter or the R-Luc control lacking the BoxB sequences, a plasmid expressing firefly luciferase as a transfection control, and vectors expressing the λN-HA-peptide or λN-HA-TNRC6A–TNRC6C. Renilla luciferase activity and mRNA levels were normalized to those of the firefly luciferase and set to 100 in cells expressing the λN-HA-peptide alone. Mean values ± standard deviations from three to four independent experiments are shown for the R-Luc-5BoxB reporter. Data corresponding to the R-Luc control (lacking the BoxB sequences) are not shown. In B, the expression levels of the λN-HA-TNRC6A–TNRC6C proteins and fragments were analyzed by Western blot. HA-MBP served as a transfection control. C shows a representative Northern blot. (D–F) Human HEK293 cells were transfected with a mixture of three plasmids: the R-Luc-5BoxB reporter or the R-Luc control reporter lacking the BoxB sequences, a plasmid expressing firefly luciferase as a transfection control, and vectors expressing the λN-HA-peptide or N- and C-terminal fragments of λN-HA-TNRC6A–TNRC6C. Renilla luciferase activity and mRNA levels were normalized to those of the firefly luciferase and analyzed as described in A. Representative Northern blots are shown in the lower panels. (G) Silencing activity of TNRC6C silencing domain (SD) measured as described in A.
We then determined whether TNRC6A–TNRC6C silenced Renilla luciferase expression either by inhibiting translation directly or by reducing mRNA levels. Using Northern blots, we analyzed the steady-state levels of the R-Luc-5BoxB mRNA and normalized them to the levels of the transfection control F-Luc mRNA. We found expression of the λN-HA-TNRC6A–TNRC6C proteins reduced reporter mRNA levels (Fig. 4B). In the case of TNRC6A, the reduction in R-Luc mRNA fully accounted for the decrease of R-Luc activity (Fig. 4A, cf. black and green bars). In contrast, the decrease in mRNA levels observed for tethered TNRC6C and the isoforms of TNRC6B did not fully account for the strong reduction in Renilla luciferase activity (Fig. 4A). These results suggest that TNRC6B and TNRC6C silence the expression of bound transcripts by two mechanisms, one involving an inhibition of protein expression and the other involving a reduction of mRNA levels, whereas TRNC6A directs bound transcripts to degradation.
Our results for TNRC6A differ from those reported by Li et al. (2008) in that in our conditions, the silencing activity of TNRC6A can be fully explained by changes in mRNA levels, whereas Li et al. (2008) observed a stronger contribution from translational repression. These differences might be caused by the use of different reporters, differences in cell lines, or in methods used to quantify mRNA levels (Northern blot in our study versus quantitative RT-PCR in Li et al. 2008).
The C-terminal fragments of TNRC6A–TNRC6C are sufficient for silencing
TNRC6B-iso3 is active in the tethering assay (Fig. 4A,C), yet this protein lacks the AGO-binding domain, suggesting that the interaction between TNRC6s and AGOs is not required for silencing. To test this assumption further, we performed tethering assays using the N- and C-terminal protein fragments described above. We observed that the N-terminal AGO-binding domains of TNRC6A–TNRC6C were inactive in the tethering assay, whereas the C-terminal fragments reduced both R-Luc activity and mRNA levels (Fig. 4D–F,N,C, respectively). Moreover, in the presence of the C-terminal fragments, the R-Luc-5BoxB mRNA migrated slightly faster, suggesting that the transcripts were deadenylated (Fig. 4D–F, lower panels, cf. lanes 3 and lanes 1).
The N-terminal fragments of TNRC6A, TNRC6B, and TNRC6C lack activity in the tethering assay; this cannot be attributed to the lower expression levels relative to the C-terminal fragments, because the full-length proteins are active even though they are expressed at lower levels (Fig. 4B). We conclude that TNRC6A–TNRC6C silence the expression of bound mRNAs independently of the Argonaute proteins.
In D. melanogaster GW182, the middle and C-terminal GW-repeat-containing regions are essential for silencing, whereas the Q/N-rich region, the UBA-like, and the RRM domains are dispensable (Eulalio et al. 2009a,b). We therefore tested, in the tethering assay, whether fragments of TNRC6A, TNRC6B, and TNRC6C comprising sequences downstream from the Q-rich regions were sufficient for silencing. We observed that these fragments, referred to as silencing domains (SDs), indeed retained full silencing activity (Fig. 4G; SD; data not shown). These fragments did not localize to P-bodies (Fig. 3B; data not shown), revealing that neither AGO interaction nor P-body localization are required for the silencing activity of TNRC6 proteins.
In summary, we showed functional redundancy for human TNRC6A, TNRC6B, and TNRC6C; each interact with all four Argonaute proteins (AGO1–AGO4) via their N-terminal GW-repeats. Furthermore, TNRC6A, TNRC6B, and TNRC6C can silence the expression of bound transcripts independently of the Argonaute proteins. The silencing activity resides in the C-terminal end of the proteins, which we referred to as the silencing domains (Fig. 1A), and includes the middle and C-terminal GW-repeat-containing regions. The silencing domains do not accumulate in P-bodies, indicating a lack of correlation between silencing activity and the ability to accumulate in P-bodies. Together with the results reported on D. melanogaster GW182 (Eulalio et al. 2009a,b), our findings indicate that GW182 proteins are recruited to miRNA targets via interactions with Argonaute proteins and repress the expression of bound transcripts through their silencing domains.
MATERIALS AND METHODS
DNA constructs
Renilla luciferase reporters used in the tethering assay were described before (Pillai et al. 2004). To generate the transfection control (pEGFP-N3-F-Luc), the open reading frame of firefly luciferase was amplified by PCR using plasmid pGL3 (Promega) as template, and cloned into the NheI and BamHI sites of the pEGFP-N3 plasmid (Clontech). Plasmids allowing the expression of λN-HA-TNRC6B-iso1 and λN-HA-TNRC6C were kindly provided by G. Meister (Max Planck Institute for Biochemistry) and W. Filipowicz (Friedrich Miescher Institute for Biomedical Research), respectively. A plasmid for expression of λN-HA-TNRC6A was obtained by inserting the HA-TNRC6A cDNA (kindly provided by E.K.L. Chan, University of Florida) into the XhoI and NotI sites of the pCIneo-λN vector (Gehring et al. 2003). The sequence encoding amino acids 1–624 of TNRC6B-iso3 was amplified using an oligo(dT)15-primed HeLa cDNA library as a template. The amplified cDNA was cloned into the pCIneo-λN-HA-TNRC6B-iso1 vector, using the SalI and XmaI sites, thus replacing amino acids 1–1318 of TNRC6B-iso-1.
The N- and C-terminal fragments of the TNRC6A, TNRC6B, and TNRC6C were cloned into the pCIneo-λN vector using the SalI and NotI sites, except for the C-terminal fragment of TNRC6A, which was cloned into the MluI and XbaI sites. The boundaries of the fragments are as follows: TNRC6A N-term (amino acids 1–932), TNRC6A C-term (925–1709), TNRC6B-iso1 N-term (amino acids 1–994), TNRC6B-iso1 C-term (amino acids 987–1793), TNRC6C N-term (amino acids 1–926), and TNRC6C C-term (amino acids 919–1690). TNRC6C silencing domain includes amino acids 1248–1690.
AGO1–AGO4 cDNAs were amplified by PCR using as templates pCIneo-λN-HA-AGO1, pCIneo-λN-HA-AGO2, pCIneo-HA-AGO3, and pCIneo-HA-AGO4 kindly provided by W. Filipowicz. The corresponding cDNAs were cloned into the pEGFP-C1 vector (Clontech) using the following restriction sites: XhoI/BamHI (AGO1 and AGO3), BamHI/XbaI (AGO2), and SalI/XbaI (AGO4). cDNAs encoding HA-MBP and MBP were amplified by PCR and inserted into the pCIneo-λN and pEGFP-C1 vectors using EcoRI/NotI sites and XhoI/BamHI, respectively.
Coimmunoprecipitations and Western blots
For coimmunoprecipitations, HEK293 cells were grown in 10-cm plates and transfected using Lipofectamine 2000 (Invitrogen). Two days after transfection, cells were washed with PBS and lysed for 15 min on ice in NET buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) supplemented with protease inhibitors (1 mL NET buffer/plate). Cell lysates were spun at 14,000 rpm for 15 min at 4°C. Polyclonal anti-GFP antibodies were added to the supernatants (dilution 1:100). Samples were incubated for 1 h at 4°C, 25 μL of Protein G-agarose (Roche) were added, and the mixtures were rotated for an additional hour at 4°C. Beads were washed three times with NET buffer and once with NET buffer without Triton X-100. Bound proteins were eluted with 100 μL of protein sample buffer. Western blotting was performed with monoclonal anti-HA antibodies (Covance Research Products, catalog number MMS-101P, dilution 1:1000) or monoclonal anti-GFP antibodies (Roche, catalog number 11814460001; dilution 1:2,000) using the CDP-Star chemiluminescent immunoblot system (PE Biosystems).
Fluorescence microscopy
For fluorescence microscopy, human HeLa cells were grown on coverslips in 24-well plates and transfected using Lipofectamine 2000 (Invitrogen). The transfection mixtures contained either 0.4 μg (for the full-length proteins) or 0.2 μg (for the N- and C-terminal fragments) of plasmids expressing λN-HA-protein fusions. Where indicated, plasmids encoding GFP-AGO1–AGO4 (0.4 μg) were also included.
Two days after transfection, cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized for 10 min with PBS containing 0.5% Triton-X100. HA-tagged proteins were detected using a monoclonal anti-HA antibody (Covance Research Products) diluted 1:1000 in PBS containing 0.1% Tween-20 and 10% fetal bovine serum (FBS). Alexa Fluor 594-coupled goat-anti-mouse secondary antibody (Molecular probes) was used at a dilution 1:1000. Cells were mounted using Fluoromount-G (Southern Biotechnology Associates, Inc.). Images were acquired using a Leica TCS SP2 confocal.
Tethering assays
For the tethering assay, human HEK293 cells were seeded in 6-well plates and transfected using Lipofectamine 2000 transfection reagent (Invitrogen). The transfection mixtures contained 0.2 μg of R-Luc reporter plasmid (with or without 5BoxB), 0.3 μg of the pEGFP-N3-F-Luc transfection control, and either 2 μg (for the full-length proteins) or 0.5 μg (for the N- and C-terminal fragments) of plasmids expressing λN-HA-protein fusions. For Western blots, 2.5 μg of plasmids expressing recombinant proteins were transfected, and a plasmid encoding HA-MBP was also included (1.5 μg) as a transfection control. R-Luc and F-Luc activities were measured 2 d after transfection using the Dual-Luciferase Reporter Assay System (Promega). Total RNA was isolated using Trifast Reagent (Peqlab) and analyzed by Northern blot as described before (Eulalio et al. 2008b).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to E.K.L. Chan for the kind gift of TNRC6A cDNA; W. Filipowicz for providing the R-Luc reporters (with and without 5BoxB), AGO1–AGO4, TNRC6A, and TNRC6C expression vectors; and G. Meister for the kind gift of TNRC6B-iso1 cDNA. This study was supported by the Max Planck Society, by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855, and the Gottfried Wilhelm Leibniz Program awarded to E.I.), and by the Sixth Framework Programme of the European Commission through the SIROCCO Integrated Project LSHG-CT-2006-037900. I.T. is the recipient of a fellowship from the Association pour la Recherche sur le Cancer (ARC).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1606309.
REFERENCES
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Ding L., Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans . Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009a;15 doi: 10.1261/rna.1605509. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Tritschler F., Buettner R., Weichenrieder O., Izaurralde E., Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucl. Acids Res. 2009b doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW-bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik G., Schell T., Hentze M.W., Kulozik A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1167–1174. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lian S.L., Moser J.J., Fritzler M.L., Fritzler M.J., Satoh M., Chan E.K. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T.H., Dong M.-Q., Chen J., Sewell A.K., Liu X., Yates J.R., Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]