Abstract
tRNA anticodon damage inflicted by secreted ribotoxins such as Kluyveromyces lactis γ-toxin and bacterial colicins underlies a rudimentary innate immune system that distinguishes self from nonself species. The intracellular expression of γ-toxin (a 232-amino acid polypeptide) arrests the growth of Saccharomyces cerevisiae by incising a single RNA phosphodiester 3′ of the modified wobble base of tRNAGlu. Fungal γ-toxin bears no primary structure similarity to any known nuclease and has no plausible homologs in the protein database. To gain insight to γ-toxin's mechanism, we tested the effects of alanine mutations at 62 basic, acidic, and polar amino acids on ribotoxin activity in vivo. We thereby identified 22 essential residues, including 10 lysines, seven arginines, three glutamates, one cysteine, and one histidine (His209, the only histidine present in γ-toxin). Structure–activity relations were gleaned from the effects of 44 conservative substitutions. Recombinant tag-free γ-toxin, a monomeric protein, incised an oligonucleotide corresponding to the anticodon stem–loop of tRNAGlu at a single phosphodiester 3′ of the wobble uridine. The anticodon nuclease was metal independent. RNA cleavage was abolished by ribose 2′-H and 2′-F modifications of the wobble uridine. Mutating His209 to alanine, glutamine, or asparagine abolished nuclease activity. We propose that γ-toxin catalyzes an RNase A-like transesterification reaction that relies on His209 and a second nonhistidine side chain as general acid–base catalysts.
Keywords: RNA damage, general acid–base catalysis, innate immune system
INTRODUCTION
Target-specific endoribonuclease toxins (“ribotoxins”) of bacteria and fungi play important roles in defending the organism against nonself species or viruses (for review, see Masaki and Ogawa 2002; Lacadena et al. 2007) and as mediators of programmed cell death (Nariya and Inouye 2008). Secreted ribotoxins comprise an RNA-based innate immune system in which killing or growth arrest of the “foreign” cell that takes up the toxin relies on that cell's inability to repair the toxin-induced damage to essential RNAs. Other ribotoxins are maintained intracellularly in a latent state until the nuclease is activated by stress or virus infection (Amitsur et al. 2003). Transfer RNAs figure prominently as the specific targets of bacterial ribotoxins, exemplified by the bacterial tRNA anticodon nucleases PrrC (and its homolog RloC), colicin D, and colicin E5 (Ogawa et al. 1999; Tomita et al. 2000; Graille et al. 2004; Blanga-Kanfi et al. 2006; Davidoff and Kaufmann 2008). PrrC and colicin E5 incise specific tRNAs at a single phosphodiester flanking the wobble base (position 34) of the anticodon via a transesterification reaction that yields 2′,3′ cyclic phosphate and 5′-OH ends. Their target preferences are determined by base modifications within the anticodon loop. The specificity of colicin E5 for Tyr, His, Asn, and Asp tRNAs is dictated by presence of the wobble base queosine (Yajima et al. 2006). PrrC cleaves at the mnm5s2U (5-methylaminomethyl-2-thiouridine) wobble base of tRNALys, aided by recognition of the t6A (6-threonylcarbamoyl adenosine) nucleoside at position 37 (Jiang et al. 2002).
Fungi also produce and secrete tRNA-directed ribotoxins (Lin et al. 2005; Lu et al. 2005, 2008; Jablonowski et al. 2006; Klassen et al. 2008). Kluyveromyces lactis and Pichia acaciae harbor cytoplasmic episomes, called “killer” plasmids, that encode secreted protein toxins known as zymocin and PaT, respectively (Stark and Boyd 1986; Klassen et al. 2004; Jablonowski and Schaffrath 2007). Zymocin is a heterotrimer of α, β, and γ subunits that arrests growth of the non-self-yeast species Saccharomyces cerevisiae by exposure from without. The α and β subunits interact with the target cell surface to effect the transport of the γ subunit (the toxin) into the cytoplasm of the target cell. Induced expression in S. cerevisiae of only the γ-toxin polypeptide suffices to arrest cell growth (Butler et al. 1991). γ-toxin is an anticodon nuclease that specifically cleaves tRNAGlu at a single phosphodiester 3′ of the modified wobble base mcm5s2U (5-methoxycarbonylmethyl-2-thiouridine) of the UUC anticodon to generate 2′,3′ cyclic phosphate and 5′-OH termini at the break site. γ-toxin can cleave the two other yeast tRNAs (Gln and Lys) that have an mcm5s2U wobble base, albeit less efficiently than it incises tRNAGlu, which is γ-toxin's principal target in vivo. γ-toxin arrests yeast growth by depleting the pool of functional tRNAGlu and its toxicity can be ameliorated by overexpression of tRNAGlu (Lu et al. 2005; Jablonowski et al. 2006). The mcm5s2U modification is required for tRNA cleavage by γ-toxin; yeast mutants that lack any of the enzymes responsible for synthesizing the mcm5 moiety are resistant to exogenous zymocin and endogenous γ-toxin (Huang et al. 2005; Lu et al. 2005; Jablonowski et al. 2006). Resistance to γ-toxin can be achieved as well by expression of heterologous tRNA repair enzymes that heal and seal the ends of the broken anticodon loop (Nandakumar et al. 2008).
The Pichia acaciae toxin (PaT) also cleaves yeast tRNAs that contain a modified mcm5s2U wobble base (Klassen et al. 2008). Whereas the tRNase subunit of PaT can break all three mcm5s2U-containing yeast tRNAs in vitro, its toxicity in vivo is attributed to specific cleavage and depletion of tRNAGln, insofar as PaT toxicity can be reversed by overexpression of tRNAGln, but not tRNAGlu (Klassen et al. 2008). Although PaT and γ-toxin are functionally analogous and have overlapping tRNase activities, PaT's cytotoxicity in S. cerevisiae is less stringently dependent than γ-toxin's on the presence of the mcm5-modified wobble uridine in the target tRNA (Klassen et al. 2008).
Perhaps the most remarkable features of the two known fungal tRNA anticodon nucleases is that they have no discernible primary structure similarity to one another, to any of the bacterial tRNA ribotoxins, or to any known ribonucleases or phosphotransferases. Indeed, K. lactis γ-toxin is effectively sui generis with respect to any known protein or predicted polypeptide in public databases. The goal of this study was to delineate structure–activity relations for γ-toxin that might illuminate its active site and catalytic mechanism. Initial efforts in this direction by Jablonowski et al. (2006) entailed selection in S. cerevisiae of nontoxic mutants of γ-toxin from a pool of randomly mutagenized plasmid clones. Although they identified nine different inactive mutants, the results were not immediately informative, because the inactive proteins contained either: (1) multiple missense changes; or (2) single missense changes that were drastic structurally (e.g., charge inversions or ionic-hydrophobic swaps). Here we conducted a systematic alanine scan of 62 residues of the intracellular form of K. lactis γ-toxin (Fig. 1A), covering 28% of the nonalanine side chains, focusing on the functional groups likely to be involved in catalysis or RNA recognition. We identified 22 essential residues and determined their relevant properties by introducing conservative substitutions. Biochemical characterization of recombinant γ-toxin highlighted its RNase A-like chemical mechanism and a likely role of His209 in catalysis of transesterification.
FIGURE 1.
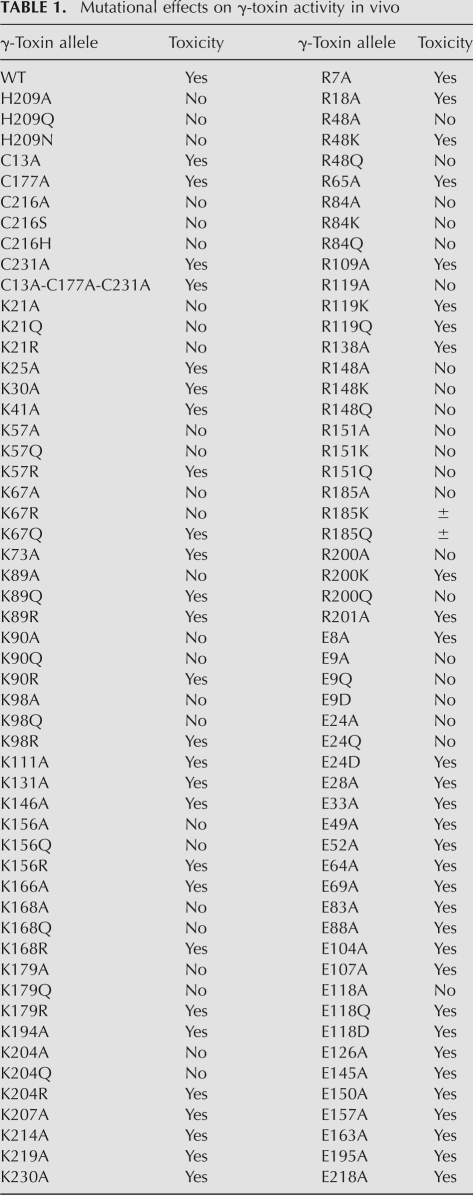
Mutational analysis of γ-toxin and essential catalytic role of His209. (A) The amino acid sequence of the intracellular version of K. lactis γ-toxin used in this study is shown. The amino acids subjected to alanine scanning are highlighted in green (essential residues) and yellow (nonessential residues). The lone histidine is denoted by an arrowhead. (B) Aliquots of serial dilutions of S. cerevisiae cells with a plasmid-borne galactose-regulated wild-type (WT) γ-toxin gene or H209A, H209N, or H209Q mutants thereof were spotted on glucose- or galactose-containing agar medium, in parallel with cells bearing an empty plasmid vector. The plates were photographed after incubation at 30°C for 2 d (glucose) or 3 d (galactose). (C) Aliquots (3 μg) of the recombinant tag-free γ-toxin C13A-C177A-C231A polypeptides were analyzed by SDS-PAGE. The Coomassie-blue-stained gel is shown. The amino acid at position 209 is indicated above each lane. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. (D) Anticodon nuclease reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 200 fmol 5′ 32P-labeled 17-mer stem–loop RNA (see Fig. 2D), and 200 fmol of the indicated γ-toxin preparations were incubated at 4°C for 30 min. Enzyme was omitted from the control reaction in lane “−.” The products were analyzed by denaturing PAGE. An autoradiograph of the gel is shown, with the sizes (in nucleotides) of the substrate and product indicated on the right.
RESULTS AND DISCUSSION
Mutagenesis strategy
The biological activity of γ-toxin was assayed using an S. cerevisiae strain containing a galactose-inducible γ-toxin expression cassette on a centromeric plasmid. In the presence of glucose, γ-toxin expression is suppressed and yeast cells grow normally. In the presence of galactose, γ-toxin is produced and growth is arrested (Fig. 1B). We tested the effects of 62 single alanine mutations on galactose-dependent toxicity. The residues targeted for alanine scanning (Fig. 1A) were ones we deemed most likely to be involved in catalysis of phosphoryl transfer (histidine, lysine, arginine, glutamate, cysteine) or RNA binding (lysine, arginine), based on general principles and the specific mechanisms of other well-studied ribonucleases that generate 2′,3′ cyclic phosphodiesters, such as RNase A, RNase T1, colicin E5, and tRNA splicing endonuclease (Steyaert 1997; Raines 1998; Xue et al. 2006; Yajima et al. 2006).
Forty of the alanine mutations did not affect toxicity in vivo (Table 1). The inessential functional groups are highlighted in yellow in the primary structure of the intracellular γ-toxin polypeptide (Fig. 1A). They include three cysteines, 18 glutamates, 13 lysines, and six arginines. By contrast, 22 of the alanine mutations inactivated γ-toxin and thereby allowed yeast cells to grow on medium containing galactose (Fig. 1; Table 1). The 22 essential amino acid side chains are highlighted in green in Figure 1A. They include the sole histidine in the γ-toxin polypeptide (His209), 10 lysines, seven arginines, three glutamates, and one cysteine. We proceeded to determine structure–activity relationships for each of the 22 essential residues by testing the effects of 44 conservative side chains substitutions: histidine was replaced by glutamine and asparagine; arginine was changed to lysine and glutamine; lysine was substituted by arginine and glutamine; glutamate was mutated to glutamine and aspartate; and cysteine was changed to serine and histidine. The results are summarized in Table 1 and are discussed below.
TABLE 1.
Mutational effects on γ-toxin activity in vivo
A lone essential histidine
The “classical” mechanism of RNA cleavage by transesterification exemplified in RNase A relies on two histidine side chains that serve, respectively, as: (1) a general base catalyst that abstracts a proton from the attacking ribose 2′-OH, and (2) a general acid catalyst that donates a proton to the ribose 5′-O leaving group (Raines 1998). K. lactis γ-toxin is instantly distinguished from RNase A in that it has only one histidine side chain (His209) in the active intracellular form of the enzyme (Fig. 1A). We find that His209 is essential for cytotoxicity in yeast. Whereas wild-type γ-toxin arrested growth in galactose, the H209A, H209Q, and H209N mutants did not (Fig. 1B). Asparagine and glutamine can, in principle, mimic the hydrogen-bonding properties of the histidine Nδ and Nε atoms, respectively, but cannot serve as proton donors or acceptors. These findings are consistent with His209 being functionally analogous to one of the histidine catalysts in RNase A. We note that Jablonowski et al. (2006) had previously isolated a nontoxic mutant of γ-toxin with three missense changes: Y152C, H209T, and L226I. Our results point to the H209T change as the likely cause of the loss of function.
Essential lysines
Structural and functional studies of RNase A highlight a single essential lysine that interacts with the scissile phosphodiester and stabilizes the pentacoordinate transition state (Raines 1998). (A glutamine side chain in RNase A also donates a hydrogen bond to the scissile phosphodiester.) By contrast, the RNase A-like Rana catesbeiana ribotoxin uses two lysines to coordinate the scissile phosphodiester (Leu et al. 2003). Lysine can also serve as a general acid to expel a 5′-OH polynucleotide leaving group, for example, during the DNA cleavage transesterification reaction catalyzed by topoisomerase IB (Krogh and Shuman 2000). A similar role for lysine in RNA transesterification is suggested by the crystal structure of substrate-bound colicin E5, a remarkable endoribonuclease that has no histidine side chain and instead relies on an active site composed of one arginine, one glutamine, and two lysines (one of which coordinates the O5′ leaving atom) (Yajima et al. 2006). The imputed active site of colicin D contains one histidine and two lysines (Graille et al. 2004; Yajima et al. 2004). The active site of RNA-bound tRNA splicing endonuclease is composed of one histidine and one lysine, plus a tyrosine (Xue et al. 2006). Lysines can also coordinate RNA phosphates flanking the scissile phosphodiester (Yajima et al. 2006).
The present alanine scan of γ-toxin pinpointed 10 of the 23 lysines as essential for activity in vivo. Conservative mutational effects highlighted four types of essential lysines (Table 1). In only one case, Lys21, was the lysine side chain strictly essential, i.e., neither arginine nor glutamine was active in lieu of Lys21. In seven instances (Lys57, Lys90, Lys98, Lys156, Lys168, Lys179, and Lys204), arginine substitutions restored activity in vivo, whereas glutamine did not, implicating positive charge as the key property of these eight residues. At Lys67, activity was restored by glutamine, but not by arginine, suggesting the importance of hydrogen bonding by Lys67. Only in the case of Lys89 was activity revived by either arginine or glutamine; we surmise that hydrogen bonding is the relevant feature at Lys89. Based on these results, we speculate that Lys21 is the best candidate to act as a “missing” nonhistidine general acid, assuming that His209 is a general base, because it alone cannot be replaced by arginine. (Arginine has a predicted pKa value of 12.5 versus a lysine pKa of 10.5, which might limit its efficacy as a general acid.) The other essential lysines that can be substituted by arginine include plausible candidates to either: (1) contact the scissile phosphodiester in tRNAGlu; (2) contact flanking or remote phosphodiesters in the folded tRNA structure; (3) contribute to recognition of the modified wobble base; or (4) stabilize the protein fold, for example, via salt bridges.
Essential arginines
Arginines classically play a role in ground-state binding and transition-state stabilization during phosphoryl transfer reactions by making bidentate contacts to the phosphate oxygens. Two arginines serve this role during DNA transesterification by topoisomerase IB (Tian et al. 2005). Several of the transesterifying ribonucleases with known structures assimilate a catalytic arginine in their active sites. For example, the colicin E5 active site includes a single arginine that coordinates both nonbridging oxygens of the scissile phosphodiester (Yajima et al. 2006). The active site of RNase T1 also has a single arginine that contacts the scissile phosphodiester (Zegers et al. 1998). By contrast, the active site of barnase includes two arginines (and a lysine) that contact the scissile phosphodiester (Buckle and Fersht 1994).
Here we found that seven of the 13 arginines in γ-toxin were essential for activity in vivo. Conservative mutational effects highlighted three classes of essential arginines (Table 1). Arg84, Arg148, and Arg151 were strictly essential and could not be functionally replaced by either lysine or glutamine. Thus, these three residues are candidates for an essential role by virtue of bidentate phosphate contacts, either at the cleavage site, or remotely on the tRNA backbone (Xue et al. 2006). Arginine is also a plausible candidate to recognize the modified wobble base, via bidentate hydrogen bonds to the uracil O4 and/or mcm5 oxygen atoms. Of course, the mutational findings at Arg84, Arg148, and Arg151 could also be explained if one or more of these arginines make bidentate salt bridges to glutamate or aspartate side chains that are essential for proper protein folding. We note that the earlier report of a nontoxic double-mutant with I45M and R151K changes (Jablonowski et al. 2006) can now be interpreted as the consequence of the R151K single change, which sufficed here to ablate activity in vivo.
Two of the arginines—Arg48 and Arg200—could be replaced functionally by lysine but not glutamine, thus testifying to the sufficiency of positive charge at these positions. These residues are good candidates to make electrostatic contacts to the tRNA backbone. In the case of Arg119, activity was restored by either lysine or glutamine, implying that neutral hydrogen bonding capacity is the relevant property of Arg119. Similar results pertained at Arg185, although the galactose-dependent growth arrest elicited by the R185K and R185Q mutants was not as strong as seen with wild-type γ-toxin.
Critical glutamates
RNase T1 and barnase exemplify the catalytic role of a glutamate in RNA transesterification enzymology. The active site of RNase T1 is composed of two histidines, one arginine, and one glutamate that contact the scissile phosphate and are critical for activity (Steyaert 1997). The prevailing model for the RNase T1 reaction posits that one of the histidines acts as a general acid to expel the leaving group. The role of general base is imputed to the glutamate, assisted by the second histidine residue (Steyaert 1997; Loverix and Steyaert 2001). Barnase relies on a single histidine general acid; a single glutamate is proposed to act as the general base (Buckle and Fersht 1994).
Our alanine scan of γ-toxin identified three out of 21 glutamates as essential for toxicity. (Note that the “hit-rate” in the alanine scan was much lower for glutamate than for the basic residues arginine and lysine.) Toxicity was restored when Glu118 was replaced conservatively by either glutamine or aspartate (Table 1), signifying that hydrogen-bonding capacity is the pertinent property of this residue and effectively excluding Glu118 as a candidate general base. Glu9 was strictly essential; neither glutamine nor aspartate revived activity. Thus, Glu9 could be a candidate general base, assuming that His209 is the general acid of γ-toxin. Activity was restored when Glu24 was changed to aspartate but not glutamine, indicating that an acidic residue sufficed. Of course, our data do not discriminate between a catalytic role for Glu9 and Glu24 versus a structural role, for example, in forming a salt bridge to an arginine.
Our data do bear on the previous isolation of nontoxic mutants bearing single missense changes at either Glu49 (to lysine) or Glu145 (to phenylalanine). Given our findings that alanine mutations at Glu49 and Glu145 do not affect toxicity in the same assay employed by Jablonowski et al. (2006), we conclude that the loss of toxicity they observed reflects the drastic consequences of charge inversion or an acidic-to-hydrophobic transition, rather than any functional requirement for these two glutamates per se.
A lone essential cysteine
The tertiary structure and stability of RNase A are enforced by four intramolecular cysteine-disulfide bonds. The full-length secreted form of K. lactis γ-toxin contains six cysteines, two of which are located within the N-terminal signal peptide. Of the four cysteines present in the active intracellular version of γ-toxin, three were deemed nonessential (Cys13, Cys177, and Cys231), because single alanine substitutions did not affect toxicity in S. cerevisiae. These results might be rationalized if one putative intramolecular disulfide linkage sufficed for activity. To address this point, we simultaneously changed all three nonessential cysteines to alanine and found that the C13A-C177A-C231A mutant was active in vivo (Table 1). Because this mutant contains only one remaining cysteine (the essential Cys216), we conclude that formation of intramolecular disulfides (if it does occur) is not important for γ-toxin activity in this assay format. Conservative replacement of Cys216 with either serine or histidine failed to revive activity (Table 1), signifying that a sulfhydryl is specifically required at this position. Any of several roles can be envisioned for a cysteine, including interaction with the mcm5s2U wobble base or a role in proton transfer during the transesterification step.
Recombinant γ-toxin
Investigators in the Byström laboratory have produced K. lactis γ-toxin in bacteria as a GST-fusion, isolated the recombinant protein by glutathione-affinity chromatography, and demonstrated its anticodon nuclease activity in vitro (Lu et al. 2005). Our aim here was to produce a tag-free version of γ-toxin in order to avoid the issue of forced dimerization by the GST-domain. Thus, we produced γ-toxin in bacteria as a His10Smt3 fusion, a method that allows facile cleavage of the tag by the Smt3-specific protease Ulp1 (Mossessova and Lima 2000). Initial attempts to make the wild-type polypeptide in E. coli resulted in substantial protein accumulation, albeit exclusively in an insoluble form. Because γ-toxin is naturally a secreted protein and has multiple cysteines within the intracellular domain lacking the signal sequence used presently, we considered the prospect that the bacterial version might misfold or aggregate because of inappropriate disulfide formation, either intermolecularly or intramolecularly. To preclude intramolecular disulfides, and to try to lessen the risk of intermolecular aggregations, we attempted to express in bacteria the triple cysteine-to-alanine mutant, C13A-C177A-C231A, that we found to be active in vivo in yeast. This version of His10Smt3-(γ-toxin) was sufficiently soluble to allow its purification by Ni-affinity chromatography, followed by tag cleavage and separation of the tag-free γ-toxin from His10Smt3, yielding a predominant 27-kDa γ-toxin polypeptide (Fig. 1C).
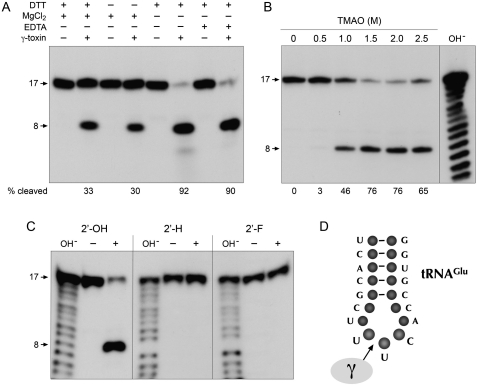
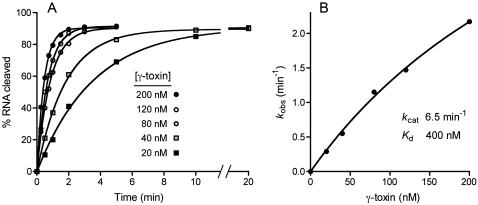
To monitor the nuclease activity of γ-toxin, we employed a minimized synthetic 17-mer RNA oligonucleotide substrate corresponding to the anticodon stem–loop of tRNAGlu (Fig. 2D). Lu et al. (2008) have shown that this RNA, although lacking the modified mcm5s2U wobble base, is nonetheless incised by γ-toxin at the correct phosphodiester 3′ of the first uridine of the UUC anticodon. Inspired by the prior report (Jiang et al. 2001) that bacterial PrrC gained activity in cleaving unmodified tRNA anticodons in the presence of high concentrations of trimethylamine oxide (TMAO), Lu et al. (2008) found that TMAO had a similar effect on γ-toxin, enabling it to cleave the unmodified version of the tRNAGlu anticodon stem–loop. The conditions employed by Lu et al. (2008) to study the model tRNase reaction were notable for the low reaction temperature and the inclusion of 10 mM MgCl2 in the reaction mixture. When we assayed our recombinant γ-toxin preparation under such conditions, we observed site-specific breakage of the 5′ 32P-labeled stem–loop RNA to yield a single radiolabeled product (Fig. 2A). We were immediately interested in whether a divalent cation was required for the RNA cleavage reaction catalyzed by γ-toxin, especially given that RNase A and other 2′,3′ cyclic phosphate-forming endoribonucleases typically do not rely on a catalytic metal ion. We found that not only was magnesium not required for tRNase activity, its omission significantly enhanced the extent of anticodon breakage (Fig. 2A). The increased activity was maintained in the presence of 10 mM EDTA, arguing against an essential role for a divalent metal in the anticodon nuclease activity of γ-toxin. We infer that γ-toxin employs an RNase A-like metal-independent transesterification mechanism reliant primarily on general acid base catalysis and transition-state stabilization by purely enzymic functional groups (Raines 1998). Further assays of model substrate cleavage were therefore performed in the absence of magnesium. In agreement with Lu et al. (2008), we found that cleavage of the synthetic 17-mer RNA was stimulated strongly by TMAO; whereas virtually no cleavage product was detected in the absence of TMAO or in the presence of 0.5 M TMAO, the 17-mer was efficiently converted to a single incised product at 1.5 to 2.5 M TMAO (Fig. 2B). Polyacrylamide gel electrophoresis (PAGE) analysis in parallel with a partial alkaline digest of the 5′ 32P-labeled 17-mer substrate verified that the cleavage product corresponds to an 8-mer formed by incision 3′ of the unmodified wobble U (Fig. 2B). The extent of RNA cleavage was proportional to input γ-toxin in the range of 0.1–5 nM concentration and attained a maximum of 90% substrate consumption at saturating enzyme (Fig. 3A). From the slope of the titration curve, we estimated that 3 fmol of RNA stem–loop was cleaved per fmol of input γ-toxin. The incised product accumulated steadily with reaction time at an RNA:enzyme ratio of 2:1, again attaining an endpoint of 91% cleavage (Fig. 3B).
FIGURE 2.
Requirements for anticodon nuclease activity in vitro. (A) Nuclease reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, and 200 fmol 5′ 32P-labeled 17-mer RNA (depicted in D) were supplemented with either 1 mM DTT, 10 mM MgCl2, 10 mM EDTA, or 5 pmol γ-toxin where indicated by “+” and then incubated at 4°C for 30 min. The products were analyzed by denaturing PAGE and visualized by autoradiography. The extents of RNA cleavage are indicated below the lanes. (B) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 200 fmol 5′ 32P-labeled 17-mer RNA, 50 fmol γ-toxin, and TMAO as specified were incubated at 4°C for 30 min. The products were analyzed by denaturing PAGE in parallel with a size marker ladder generated by partial alkaline hydrolysis of the 5′ 32P-labeled 17-mer RNA (lane OH−). An autoradiogram of the gel is shown. (C) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 200 fmol 5′ 32P-labeled 17-mer RNA with a ribose at the wobble nucleoside (2′-OH) (left), with a deoxyribose at the wobble nucleoside (2′-H) (middle), or with a 2′-fluoro sugar at the wobble nucleoside (2′-F) (right), and 100 fmol γ-toxin (lanes +) were incubated at 4°C for 30 min. Enzyme was omitted from control reactions in lanes –. The reaction products were analyzed by denaturing PAGE in parallel with size marker ladders generated by partial alkaline hydrolysis of the respective 5′ 32P-labeled 17-mer RNAs (lanes OH−). An autoradiogram of the gel is shown. (D) The 17-mer RNA oligonucleotide substrate corresponding to the anticodon stem–loop of tRNAGlu is shown. The site of cleavage by γ-toxin 3′ of the wobble base of the tRNAGlu anticodon nucleoside is indicated by the arrow.
FIGURE 3.
γ-toxin catalyzes multiple turnovers and sediments as a monomer. (A) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 200 fmol 32P-labeled 17-mer RNA, and increasing amounts of γ-toxin as specified were incubated at 4°C for 30 min. The extents of RNA cleavage are plotted as a function of input γ-toxin. Each datum is an average of three separate titration experiments ± SEM. (B) A reaction mixture (90 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, and 20 nM 32P-labeled 17-mer RNA, and 10 nM γ-toxin was incubated at 4°C. Aliquots (10 μL) were withdrawn at the times specified and quenched with formamide-EDTA. The extents of RNA cleavage are plotted as a function of time. Each datum is an average of three separate experiments ± SEM. (C) Aliquots (15 μL) of even-numbered glycerol gradient fractions were analyzed by SDS-PAGE (top). The Coomassie-blue stained gel is shown. The γ-toxin, catalase, BSA, and cytochrome c polypeptides are indicated. Molecular weight calibration for the PAGE analysis (kDa) is indicated at right. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 20 nM 32P-labeled 17-mer RNA, and 1 μL of the even-numbered glycerol gradient fractions were incubated at 4°C for 10 min (bottom). The extents of RNA cleavage are plotted.
The quaternary structure of tag-free γ-toxin was gauged by zonal velocity sedimentation in a 15%–30% glycerol gradient. Marker proteins catalase (native size 248 kDa), bovine serum albumin (BSA) (66 kDa), and cytochrome c (12 kDa) were included as internal standards in the gradient. The anticodon RNase activity profile paralleled the abundance of the γ-toxin polypeptide (calculated mass 27 kDa), which sedimented as a discrete peak (fractions 26–30) between cytochrome c and BSA, albeit closer to cytochrome c (Fig. 3C). An S value of 2.4 was calculated for γ-toxin by interpolation to a linear plot of the S values of the internal standards versus peak fraction number (not shown). We surmise from these results that γ-toxin is a monomer in solution.
Effect of ribose 2′ modifications at the scissile phosphodiester
Lu et al. (2008) showed that cleavage by γ-toxin of the minimized 17-mer anticodon stem–loop substrate is stringently dependent on the anticodon loop sequence U34U35C36A37 derived from wild-type tRNAGlu. Any single base change within the UUCA element abolished tRNase activity. Here we extended the analysis of γ-toxin substrate specificity by testing the effects of pentose sugar modifications at the scissile U34 phosphodiester. Synthetic 17-mer oligonucleotides (20 nM) containing either 2′-deoxy (2′-H) or 2′-fluoro (2′-F) nucleosides in lieu of the 2′-OH ribonucleoside at the wobble U position were reacted with γ-toxin (10 nM) (Fig. 2C). The salient findings were that (1) the 2′-H and 2′-F substrates were refractory to cleavage at the phosphodiester flanking the wobble base under conditions where 90% of the unmodified RNA was cleaved; and (2) prevention of RNA scission at the “normal” site did not precipitate the use of alternative cleavage sites within the anticodon loop. There was no detectable cleavage of the 2′-H and 2′-F substrates even when the concentration of γ-toxin was increased to 200 nM (i.e., a 10-fold excess over the RNA substrate; not shown). These results underscore the essential role of the ribose O2′ atom as the nucleophile for the transesterification reaction.
Single-turnover RNA cleavage
We measured the kinetics of cleavage of 20 nM RNA stem–loop at stoichiometric or superstoichiometric levels of enzyme, varying the concentration of γ-toxin from 20 to 200 nM. The kinetic profiles fit well to a single exponential with a common endpoint of 91% RNA cleavage (Fig. 4A). The cleavage rate increased steadily with enzyme concentration, signifying that initial binding of γ-toxin to the RNA substrate was rate limiting. A plot of the observed cleavage rate constants versus enzyme concentration fit well to a hyperbolic one-site binding model (Fig. 4B), from which we derived a Kd value of 400 nM and a kcat of 6.5 min−1.
FIGURE 4.
Kinetics of single-turnover RNA cleavage. (A) Reaction mixtures (100 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 20 nM 32P-labeled 17-mer RNA, and 20, 40, 80, 120, or 200 nM γ-toxin were incubated at 4°C. Aliquots (10 μL) were withdrawn at the times specified and quenched with formamide-EDTA. The extent of RNA cleavage is plotted as a function of time for each enzyme concentration. Nonlinear regression curve fitting to a single exponential was performed in Prism. The apparent rate constants derived thereby are plotted in B as a function of γ-toxin concentration. kcat and Kd values were obtained by nonlinear regression curve fitting to a hyperbolic one-site binding model in Prism.
His209 is required for anticodon RNase activity in vitro
Missense mutations of His209 were introduced into the vector used to express the active C13A-C177A-C231A variant of γ-toxin in E. coli. The H209A, H209Q, and H209N proteins were purified from soluble bacterial extracts in parallel with the “wild-type” H209 protein (Fig. 1C). The salient findings were that the alanine, glutamine, and asparagine mutations abolished anticodon nuclease activity with the synthetic stem–loop substrate (Fig. 1D), suggesting that His209 is required for transesterification per se, rather than recognition of the modified wobble base or the tRNA fold.
Concluding remarks
Here we conducted a systematic in vivo structure–function analysis of a eukaryal tRNA anticodon nuclease. The identification of 22 essential side chains by alanine-scanning, the clarification of structure–activity relations by conservative substitutions, and the analysis of the requirements for RNA incision in vitro engender plausible ideas about the catalytic mechanism, especially the role of His209. The solution to the puzzle of how γ-toxin assembles its active site and specifically recognizes its tRNA target, and whether it has any structural similarities to other RNases, will ultimately hinge on crystallizing the protein, alone and in complex with RNA. The capacity to preclude catalysis by single-atom modifications of the RNA or single conservative mutations of the toxin may facilitate such structural studies.
MATERIALS AND METHODS
Inducible expression of γ-toxin in budding yeast
The S. cerevisiae strain used in this study is a haploid derivative of W303 transformed with plasmid pLF16 (CEN LEU2 UASGAL-γ-toxin), which allows for galactose-inducible, glucose-repressible expression of an intracellular 232-amino acid version of K. lactis γ-toxin that lacks the N-terminal 17- amino acid signal peptide (Jablonowski et al. 2006). The “wild-type” γ-toxin open reading frame in pLF16 encodes a polypeptide that differs from the original NCBI database entry (accession YP_001648056) at two positions, such that residues 127 and 141 in pLF16 are valine and glutamate, respectively, rather than isoleucine and lysine. These polymorphisms are functionally benign, insofar as the γ-toxin encoded on pLF16 is active in vivo and in vitro (vide supra). Missense mutations were introduced into the γ-toxin gene of pLF16 via two-stage overlap extension PCR. The inserts were sequenced completely to verify the absence of unwanted coding changes. The mutated pLF16 plasmids were transformed into yeast in parallel with the wild-type pLF16. Leu+ transformants were selected on glucose-containing medium lacking leucine. Cultures were grown in selective liquid media at 30°C, adjusted to A600 of 0.1, and then diluted serially in 10-fold increments. Aliquots (3 μL) of each dilution were spotted on glucose-containing and galactose-containing agar plates, which were incubated at 30°C.
Recombinant γ-toxin from bacteria
The gene encoding the intracellular γ-toxin C13A-C177A-C231A protein was excised from pLF16 with endonucleases NdeI and SalI and inserted between the NdeI and SalI sites in pET23a. This plasmid was used as the template to PCR-amplify the open reading frame with primers that introduced a BamHI site 5′ of the start codon and a XhoI site 3′ of the stop codon. The PCR product was inserted into pET28-His10Smt3 to generate a phage T7 RNA polymerase-driven expression vector encoding the γ-toxin C13A-C177A-C231A polypeptide fused to an N-terminal His10Smt3 tag. Missense mutations H209A, H209Q, and H209N were introduced into γ-toxin-(C13A-C177A-C231A) gene by two-stage overlap extension PCR. The pET28 plasmid inserts were sequenced completely to verify the intended coding sequence. The pET28-His10Smt3-(γ-toxin) plasmids were transformed into E. coli BL21(DE3)-RIL. Cultures (1 L) amplified from single transformants were grown at 37°C in Luria-Bertani medium containing 0.05 mg/mL kanamycin until the A600 reached ∼0.6. The cultures were adjusted to 0.2 mM isopropyl-β-D-thiogalactopyranoside and 2% (v/v) ethanol and then incubated at 17°C for 6 h with constant shaking. Cells were harvested by centrifugation, and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 50 mL of buffer A (50 mM Tris-HCl at pH 7.5, 0.5 M NaCl, 10% sucrose). Cell lysis was achieved by the addition of lysozyme, phenylmethanesulphonylfluoride, and Triton X-100 to final concentrations of 1 mg/mL, 0.2 mM, and 0.1%, respectively. The lysates were sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extracts were applied to 4-mL columns of Ni-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with buffer A containing 0.1% Triton X-100. The columns were washed with buffer B (50 mM Tris-HCl at pH 8.0, 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol) containing 50 mM imidazole and eluted with buffer B containing 500 mM imidazole. The polypeptide compositions of the column fractions were monitored by SDS-PAGE. The peak fractions containing the His10Smt3-(γ-toxin) polypeptide were pooled and supplemented with the Smt3-specific protease Ulp1 to attain a His10Smt3-(γ-toxin):Ulp1 ratio of 200:1. The mixtures were dialyzed overnight against buffer C (50 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol). The dialysates were applied to 4-mL nickel-agarose columns equilibrated with buffer C. Tag-free γ-toxin was recovered in the flow-through fractions. The γ-toxin preparations were dialyzed against buffer D (50 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 2 mM DTT, 5 mM EDTA, 0.05% Triton X-100, 10% glycerol) and stored at −80°C. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The yield of γ-toxin was ∼2 mg from a 1 L bacterial culture.
Anticodon nuclease assay
Reaction mixtures containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 20 nM 5′ 32P-labeled 17-mer stem–loop RNA (see Fig. 2D), and γ-toxin and other components as specified were incubated at 4°C. The reactions were quenched by adding 10 μL of 90% formamide, 50 mM EDTA. The samples were heated at 95°C for 1 min and then analyzed by electrophoresis through a 15-cm 24% polyacrylamide gel containing 8 M urea in 0.5× TBE (45 mM Tris-borate, 1.2 mM EDTA) at 12 W for ∼90 min. The radiolabeled products were visualized by autoradiography, and the extents of RNA cleavage were quantified by scanning the gel with a Fujix BAS2500 imager. The 5′ 32P-labeled RNA ladders used to size the cleavage product were prepared by incubating aliquots of the 5′ 32P-labeled 17-mer RNAs in a solution of 40 mM NaHCO3, 60 mM Na2CO3 (pH 10.2) for 12 min at 95°C. The partially cleaved RNAs were then stored at −20°C.
Materials
RNA oligonucleotides used as substrates in anticodon nuclease assays were purchased from Dharmacon. The RNAs were deprotected according to the vendor's instructions and 5′ 32P-labeled by reaction with T4 polynucleotide kinase and [γ32P]ATP (Amersham). The labeled RNA was purified by preparative gel electrophoresis and stored at −20°C. TMAO was purchased from Sigma.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1637809.
REFERENCES
- Amitsur M., Benjamin S., Rosner R., Chapman-Shimshoni D., Meidler R., Blanga S., Kaufmann G. Bacteriophage T4-encoded Stp can be replaced as activator of anticodon nuclease by a normal host cell metabolite. Mol. Microbiol. 2003;50:129–143. doi: 10.1046/j.1365-2958.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S., Amitsur M., Azem A., Kaufmann G. PrrC-anticodon nuclease: Functional organization of a prototypal bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle A.M., Fersht A.R. Subsite binding in an RNase: Structure of a barnase-tetranucleotide complex at 1.76-Å resolution. Biochemistry. 1994;33:1644–1653. doi: 10.1021/bi00173a005. [DOI] [PubMed] [Google Scholar]
- Butler A.R., Porter M., Stark M.J. Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast. 1991;7:617–625. doi: 10.1002/yea.320070610. [DOI] [PubMed] [Google Scholar]
- Davidoff E., Kaufmann G. RloC: A wobble nucleotide-excising and zinc-responsive bacterial tRNase. Mol. Microbiol. 2008;69:1560–1574. doi: 10.1111/j.1365-2958.2008.06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M., Mora L., Buckingham R.H., van Tilbeurgh H., de Zamaroczy M. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 2004;23:1474–1482. doi: 10.1038/sj.emboj.7600162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Johansson M.J.O., Byström A.S. An early step in wobble tRNA modification requires the elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D., Schaffrath R. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 2007;35:1533–1537. doi: 10.1042/BST0351533. [DOI] [PubMed] [Google Scholar]
- Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Meidler R., Amitsur M., Kaufmann G. Specific interactions between anticodon nuclease and the tRNALys wobble base. J. Mol. Biol. 2001;305:377–388. doi: 10.1006/jmbi.2000.4282. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Blanga S., Amitsur M., Meidler R., Krivosheyev E., Sundaram M., Bajii A., Davis D.R., Kaufmann G. Structural features of tRNALys favored by anticodon nuclease as inferred from reactivities of anticodon stem and loop substrate analogs. J. Biol. Chem. 2002;277:3826–3841. doi: 10.1074/jbc.M110072200. [DOI] [PubMed] [Google Scholar]
- Klassen R., Teichert S., Meinhardt F. Novel yeast killer toxins provoke S-phase arrest and DNA damage checkpoint activation. Mol. Microbiol. 2004;53:263–273. doi: 10.1111/j.1365-2958.2004.04119.x. [DOI] [PubMed] [Google Scholar]
- Klassen R., Paluszynski J.P., Emhoff S., Pfeiffer A., Fricke J., Meinhardt F. The primary target of the killer toxin from Pichia acaciae is tRNAGln. Mol. Microbiol. 2008;69:681–697. doi: 10.1111/j.1365-2958.2008.06319.x. [DOI] [PubMed] [Google Scholar]
- Krogh B.O., Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- Lacadena J., Alvarez-Garcia E., Carreras-Sangrà N., Herrero-Galán E., Alegre-Cebollada J., García-Ortega L., Oñaderra M., Gavilanes J.G., Martínez del Pozo A. Fungal ribotoxins: Molecular dissection of a family of natural killers. FEMS Microbiol. Rev. 2007;31:212–237. doi: 10.1111/j.1574-6976.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- Leu Y.J., Chern S.S., Wang S.C., Hsiao Y.Y., Amiraslanov I., Liaw Y.C., Liao Y.D. Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and x-ray crystallography. J. Biol. Chem. 2003;278:7300–7309. doi: 10.1074/jbc.M206701200. [DOI] [PubMed] [Google Scholar]
- Lin Y.L., Elias Y., Huang R.H. Structural and mutational studies of the catalytic domain of colicin E5: A tRNA-specific ribonuclease. Biochemistry. 2005;44:10494–10500. doi: 10.1021/bi050749s. [DOI] [PubMed] [Google Scholar]
- Loverix S., Steyaert J. Deciphering the mechanism of RNase T1. Methods Enzymol. 2001;341:305–323. doi: 10.1016/s0076-6879(01)41160-8. [DOI] [PubMed] [Google Scholar]
- Lu J., Huang B., Esberg A., Johanson M., Byström A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Esberg A., Huang B., Byström A.S. The Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem–loop of tRNA. Nucleic Acids Res. 2008;36:1072–1080. doi: 10.1093/nar/gkm1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H., Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie. 2002;84:433–438. doi: 10.1016/s0300-9084(02)01425-6. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Lima C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Nandakumar J., Schwer B., Schaffrath R., Shuman S. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol. Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H., Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Tomita K., Ueda T., Watanabe K., Uozumi T., Masaki H. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- Raines R.T. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Stark M.J.R., Boyd A. The killer toxin of Kluyveromyces lactis: Characterization of the toxin subunits and identification of the genes which encode them. EMBO J. 1986;5:1995–2002. doi: 10.1002/j.1460-2075.1986.tb04455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert J. A decade of protein engineering on ribonuclease T1: Atomic dissection of the enzyme–substrate interactions. Eur. J. Biochem. 1997;247:1–11. doi: 10.1111/j.1432-1033.1997.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- Tian L., Claeboe C.D., Hecht S.M., Shuman S. Mechanistic plasticity of DNA topoisomerase IB: Phosphate electrostatics dictate the need for a catalytic arginine. Structure. 2005;13:513–520. doi: 10.1016/j.str.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Calvin K., Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- Yajima S., Nakanishi K., Takahashi K., Ogawa T., Hidaka M., Kezuka Y., Nonaka T., Ohsawa K., Masaki H. Relation between tRNase activity and the structure of colicin D according to X-ray crystallography. Biochem. Biophys. Res. Commun. 2004;322:966–973. doi: 10.1016/j.bbrc.2004.07.206. [DOI] [PubMed] [Google Scholar]
- Yajima S., Inoue S., Ogawa T., Nonaka T., Ohsawa K., Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA–tRNA interaction. Nucleic Acids Res. 2006;34:6074–6082. doi: 10.1093/nar/gkl729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers I., Loris R., Dehollander G., Haikal A.F., Poortmans F., Steyaert J., Wyns L. Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography. Nat. Struct. Biol. 1998;5:280–283. doi: 10.1038/nsb0498-280. [DOI] [PubMed] [Google Scholar]