Abstract
The nuclear exosome functions in a variety of pathways catalyzing formation of mature RNA 3′-ends or the destruction of aberrant RNA transcripts. The RNA 3′-end formation activity of the exosome appeared restricted to small noncoding RNAs. However, the nuclear exosome controls the level of the mRNA encoding the poly(A)-binding protein Nab2p in a manner requiring an A26 sequence in the mRNA 3′ untranslated regions (UTR), and the activities of Nab2p and the exosome-associated exoribonuclease Rrp6p. Here we show that the A26 sequence inhibits normal 3′-end processing of NAB2 mRNA in vivo and in vitro, and makes formation of the mature 3′-end dependent on trimming of the transcript by the core exosome and the Trf4p component of the TRAMP complex from a downstream site. The detection of mature, polyadenylated transcripts ending at, or within, the A26 sequence indicates that exosome trimming sometimes gives way to polyadenylation of the mRNA. Alternatively, Rrp6p and the TRAMP-associated Mtr4p degrade these transcripts thereby limiting the amount of Nab2p in the cell. These findings suggest that NAB2 mRNA 3′-end formation requires the exosome and TRAMP complex, and that competition between polyadenylation and Rrp6p-dependent degradation controls the level of this mRNA.
Keywords: RNA processing, TRAMP complex, exosome, mRNA 3′-end formation
INTRODUCTION
Most eukaryotic mRNAs require cleavage and polyadenylation to form their mature 3′-ends (Edmonds 2002). Large complexes of proteins recognize sequence elements in the 3′-untranslated (UTR) regions of nascent RNA polymerase II transcripts leading to cleavage of the precursor mRNAs (pre-mRNAs), which elicits transcription termination, at least in part as a result of the action of the 5′–3′ exonuclease Rat1p/Xrn2p on the portion of the transcript still bound to the paused RNA polymerase (Proudfoot and O'Sullivan 2002; Kim et al. 2004; West et al. 2004; Bentley 2005; Buratowski 2005). The released pre-mRNA undergoes polyadenylation by a process in which specific poly(A)-binding proteins regulate the activity of poly(A)-polymerase and influence the eventual length of the poly(A) tail (Wahle 1991, 1995; Bienroth et al. 1993; Hector et al. 2002; Dheur et al. 2005). The binding of these proteins, as well as others, forms a ribonucleoprotein (RNP) complex that profoundly affects the physical stability of the mRNA in the nucleus, as well as the efficiency of its export to the cytoplasm (Jensen et al. 2003; Rodriguez et al. 2004; Fasken and Corbett 2005).
Defects in mRNA 3′-end processing result in significant defects in pre-mRNA metabolism. Mutations that inhibit cleavage and polyadenylation, either by disrupting 3′-end processing sequences or by decreasing the activity of essential 3′-end formation factors, cause a dramatic decrease in mRNA levels (Patel and Butler 1992; Proweller and Butler 1994; Das et al. 2000; Hilleren et al. 2001; Libri et al. 2002). Partial reversal of this effect by depletion of components of the nuclear exosome revealed the existence of a surveillance system that degrades improperly processed pre-mRNAs (Burkard and Butler 2000; Torchet et al. 2002; Milligan et al. 2005). In this pathway, failure to cleave and/or polyadenylate pre-mRNAs produces an RNP recognized and degraded by the exoribonucleolytic activities of the exosome. In some cases, where defects produce 3′ extended transcripts, it appears that release of the transcript as the exosome nears the normal 3′-end processing site allows polyadenylation and the production of functional transcripts (Torchet et al. 2002). Recent evidence showed that this process plays a role in the 3′-end formation of the CTH2 mRNA in budding yeast (Ciais et al. 2008).
The exosome contains a nine-subunit core complex found in archael cells and in the nucleus and the cytoplasm of eukaryotic cells (Butler 2002; Houseley et al. 2006; Lorentzen and Conti 2006). This complex contains exonucleolytic activity in archaea, but not in humans or budding yeast. Instead, the ribonuclease activity of the core exosome resides in Dis3p/Rrp44p, which acts as an endonuclease and a hydrolytic exonuclease (Dziembowski et al. 2007; Greimann and Lima 2008; Schaeffer et al. 2009; Schneider et al. 2009). Two additional components, the putative helicase Dob1p/Mtr4p and the 3′–5′ exoribonuclease Rrp6p, function with the core exosome in the nucleus (Briggs et al. 1998; de la Cruz et al. 1998; Allmang et al. 1999). A variety of studies indicate that the nuclear exosome plays a role in an RNA surveillance pathway that degrades improperly processed mRNAs, rRNAs, and tRNAs, as well as normal mRNAs retained in the nucleus (Bousquet-Antonelli et al. 2000; Burkard and Butler 2000; Hilleren et al. 2001; Das et al. 2003; Fang et al. 2004; Galy et al. 2004; Kadaba et al. 2004; Kuai et al. 2004). The nuclear exosome, in cooperation with Lrp1p (Rrp47p), also plays a critical role in the 3′-end processing of 5.8S rRNA and many snRNAs and snoRNAs (Allmang et al. 1999; van Hoof et al. 2000; Mitchell et al. 2003; Peng et al. 2003; Phillips and Butler 2003).
In mammalian cells, the cytoplasmic exosome appears to function in the major mRNA degradation pathway, which contrasts with the predominant role of the 5′–3′ pathway of mRNA degradation in yeast (Wang and Kiledjian 2001; Mukherjee et al. 2002; Lejeune et al. 2003). Experiments in mammalian cells indicate that RNA-binding proteins may play a critical role in the degradation of certain normal mRNAs by the cytoplasmic exosome. Specifically, mRNAs carrying destabilizing AU-rich elements (AREs) in their 3′-UTRs require ARE-binding proteins to activate transcript degradation by the exosome (Chen et al. 2001). In some cases, the accumulation of truncated, polyadenylated mRNA intermediates after exosome depletion in mammalian cells implied a role for exosome-mediated surveillance of mRNAs (West et al. 2006).
Surveillance of mRNA 3′-end processing by the nuclear exosome implies that competition may occur between factors involved in mRNA production and degradation (Bousquet-Antonelli et al. 2000; Burkard and Butler 2000; Torchet et al. 2002; Milligan et al. 2005). Indeed, evidence suggested that this competition might act to regulate the concentration of the poly(A)-binding protein Nab2p of Saccharomyces cerevisiae (Roth et al. 2005). These experiments showed that the nuclear exosome limits the level of NAB2 mRNA in a manner dependent on a sequence of 26 consecutive adenosines (A26) in the NAB2 mRNA 3′-untranslated region. The Nab2 protein also limited the amount of its own mRNA in an A26- and Rrp6p-dependent manner, implying that Nab2p binds this sequence and recruits the exosome to degrade its own transcript. These experiments also provided evidence that the A26 sequence might inhibit normal mRNA 3′-end processing of NAB2 mRNA, thereby exposing the transcript to surveillance by the exosome. In this study, we present evidence that Nab2p acts to repress 3′-end formation of its own mRNA leading to 3′-end formation downstream from the mature 3′-end of the mRNA. The exosome and the TRAMP complex trim this 3′ extended transcript back to the A26 sequence at which time Rrp6p and the RNA helicase Mtr4p accelerate degradation of the mRNA, or polyadenylation occurs to produce a stable, mature transcript. We propose that the concentration of Nab2p determines whether this novel mechanism of 3′-end formation results in polyadenylation or degradation of NAB2 mRNA.
RESULTS
Nab2p inhibits mRNA 3′-end formation of its own mRNA
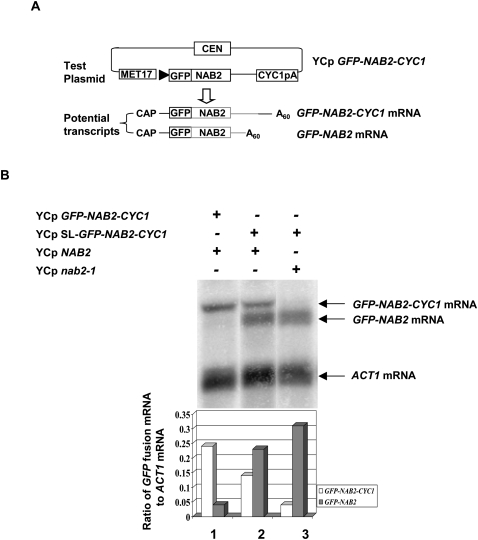
Previous experiments showed that overexpression of Nab2p caused a decrease in the level of its mature mRNA (Roth et al. 2005). This effect did not occur after replacement of the transcript's 3′-UTR-encoded A26 with U26, suggesting that Nab2p binds the A26 sequence and destabilizes its own mRNA. Moreover, a loss-of-function mutation (nab2-1) in Nab2p caused increased levels of NAB2 mRNA. In an effort to understand the relative contributions of mRNA degradation and 3′-end formation to NAB2 mRNA levels, we analyzed the effect of altering the gene dosage of NAB2 on its own expression. These experiments used a strain with a chromosomal deletion of NAB2 and a single-copy plasmid bearing either the normal or the nab2-1 allele of NAB2. The strain also contained a reporter plasmid expressing a GFP-NAB2 fusion with the strong CYC1 polyadenylation signal 250 nucleotides (nt) 3′ to the A26 sequence (Fig. 1A). This plasmid expresses two transcripts—one ending in the NAB2 3′-UTR at the A26 and the second ending at the CYC1 poly(A) site. Previous experiments showed that loss of Rrp6p increased the level of the shorter NAB2 3′-UTR transcript without changing the level of the longer transcript, suggesting that Rrp6p affects the degradation, but not 3′-end formation of this mRNA (Roth et al. 2005). Here, we analyzed the effect of changing Nab2p levels on production of these two reporter mRNAs. In the presence of a normal copy of NAB2, this plasmid produces predominantly transcripts ending at the CYC1 site (Fig. 1B, lane 1). We placed a stem–loop designed to block translation (Oliveira et al. 1993) upstream of the GFP-NAB2 start codon and found that this mutation abolished the presence of Gfp-Nab2p in the nucleus of cells bearing the plasmid (data not shown). The stem–loop mutation resulted in a sixfold increase in the level of the transcript ending at the NAB2 site and twofold decrease in the level of the transcript ending at the CYC1 site (Fig. 1B, lane 2). Thus, the decrease in the level of Gfp-Nab2p caused by blocking translation of the plasmid-borne gene fusion results in a significant increase in the level of stable transcript produced from the reporter plasmid and, consistent with published results (Roth et al. 2005), indicates that the normal plasmid produces Gfp-Nab2p, which functions as a negative regulator of its own expression. Expression of the stem–loop GFP-CYC1 plasmid in a strain carrying the nab2-1 mutation caused a 1.3-fold increase in the level of the transcript ending at the NAB2 site, and a fourfold decrease in production of the transcript ending at the CYC1 site (Fig. 1B, lane 3). Thus, a further decrease in Nab2p levels results in nearly all of the 3′-end formation occurring at the NAB2 site. These findings indicate that as the activity of Nab2p increases in the cell, it inhibits 3′-end formation at its own site and destabilizes its own mRNA.
FIGURE 1.
Nab2 protein inhibits 3′-end formation of its own mRNA. (A) Diagram of the test plasmid, which allows transcription from the MET17 promoter of the inserted test gene fused to green fluorescent protein (GFP). CYC1pA designates the polyadenylation site of CYC1. The potential transcripts, distinguished by their different 3′-ends, are shown below the diagram of the test plasmid. (B) Strain YSB501 (YCpNAB2) or YSB502 (YCpnab2-1) carrying the indicated plasmids was grown in SCD–URA-LEU at 30°C; total RNA was isolated and the levels of GFP-NAB2 transcripts determined by Northern blot analysis with an oligonucleotide probe complementary to the GFP portion of the mRNA. YCpSL-GFP-NAB2-CYC1 designates the use of the reporter plasmid with the stem–loop insertion in the 5′-UTR of the reporter mRNA. The bar graph below reports the ratio of the GFP-fusion transcripts to ACT1 mRNA and is based on two independent measurements.
The NAB2 mRNA 3′-end maps to the A26 sequence in the 3′-UTR
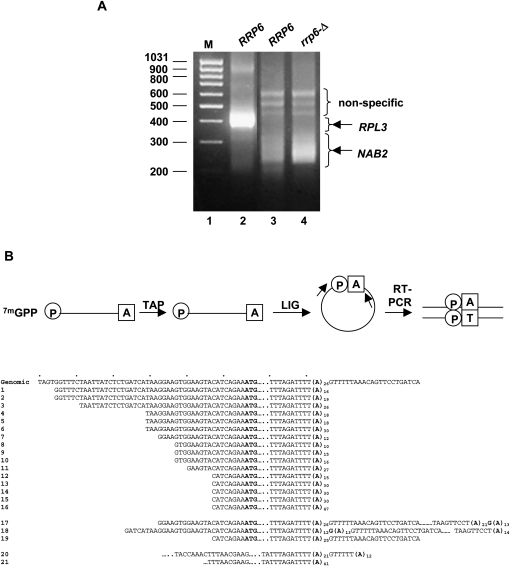
We analyzed the NAB2 mRNA 3′-ends using the ligation-mediated poly(A) tail assay (LM-PAT) (Salles et al. 1999). We carried out LM-PAT using mRNA-specific primers to RPL3 (previously called TCM1) mRNA (as a positive control) and NAB2 mRNA, and then separated the products by gel electrophoresis to compare the size of the NAB2 products with that of the RPL3 products (Fig. 2A). Previous experiments mapped the polyadenylation site of RPL3 mRNA (Proweller and Butler 1994), and the LM-PAT procedure yielded the expected ∼375–450-bp products (Fig. 2A, lane 2). The length heterogeneity of these products reflects the known heterogeneity of mRNA poly(A) tails in yeast and confirms that the LM-PAT procedure preserves their lengths. LM-PAT using a NAB2-specific primer (OSB217) (Fig. 2B) produced DNA products ∼250–350 bp in length, consistent with 3′-end formation of NAB2 mRNA near the A26 sequence within the 3′-UTR (Fig. 2A, lanes 3,4). The results also show increased amounts of NAB2 cDNAs in the rrp6-Δ strain compared with the RRP6 strain, consistent with the increase in NAB2 mRNA levels in rrp6 mutants (Fig. 2A, lane 4). LM-PAT of NAB2 also produced several longer DNA products, but subsequent DNA sequencing did not reveal any similarity of these products to the NAB2 locus.
FIGURE 2.
The mature 3′-end of NAB2 mRNA maps to the A26 sequence in the 3′-UTR. (A) Agarose gel electrophoretic analysis of the product of LM-PAT analysis of the strains with the indicated genotypes. (M) The lane with molecular size standards. (B) Diagram of the cRT-PCR method for a model mRNA. TAP (tobacco acid pyrophosphatase) treatment removes the cap, leaving a 5′-phosphate and the 3′-poly(A) tail, which is ligated (LIG) with RNA ligase to produce a circular RNA that is amplified by RT-PCR with (arrows) specific primers. The sequences obtained using primers to NAB2 mRNA are shown below the genomic sequence with the start codon (ATG) and A26 sequence in bold.
The DNA sequence of several NAB2 LM-PAT products suggested that the majority end within the A26 sequence and sometimes carry more than the template-encoded adenosines (data not shown). However, the presence of the A26 sequence could cause artifacts in this procedure, so we determined the NAB2 mRNA 3′-end formation site by sequencing circular reverse transcription-polymerase chain reaction (cRT-PCR) products (Couttet et al. 1997; Mullen and Marzluff 2008). Total RNA samples from the RRP6 strain YSB3001 were decapped and then circularized with RNA ligase (Fig. 2B). RT-PCR was carried out with primers complementary to regions 3′ to the ATG start codon and 5′ to the TAG stop codon of NAB2 mRNA. The cDNAs were cloned and the sequence of the ligation junction determined to identify the 5′- and 3′-ends of the mRNAs. Of the 21 independent clones sequenced, 17 have 3′-ends corresponding to the A26 sequence in the NAB2 3′-UTR (Fig. 2B, clones 1–16,21). These clones contain eight different 5′-ends suggesting multiple transcription start sites for NAB2, although we do not discount the possibility that some of these represent intermediates in the 5′–3′ mRNA decay pathway. The length of the poly(A) tails in these clones ranged from 10 to 47 adenosines, suggesting the addition of some of the adenosines by a post-transcriptional mechanism.
Four clones end downstream from the A26 sequence: two of these end with poly(A) tails added 114 nt downstream from the beginning of the A26 sequence (Fig. 2B, clones 17,18). Remarkably, each of these clones clearly shows the presence of a single G within the poly(A) sequences: one within the A26 and one within the 3′ poly(A) tail. A third clone has an unadenylated 3′-end 21 nt downstream from the template-encoded adenosine stretch, which, in this case, is 25 adenosines long (Fig. 2B, clone 19).
Two clones have 5′-ends within the NAB2 coding sequence (Fig. 2B, clones 20,21). One has a 41-nt poly(A) tail at the A26 sequence, while the second has a 12-nt poly(A) tail added 6 nt beyond an A21, rather than an A26 sequence. The truncated 5′-ends of these transcripts suggest that they may represent intermediates in the 5′–3′ mRNA decay pathway.
Two of the sequenced clones (Fig. 2B, clones 17,21) ended downstream from the template-encoded A26 sequence, but contained fewer than the expected number of internal adenosines. We amplified this region by PCR from genomic DNA isolated from strain YSB3001, cloned the PCR products, and determined their sequence. Five clones contained 25 consecutive adenosines, four contained 26 adenosines, and one contained 27 adenosines. This distribution is consistent with the propensity of DNA polymerases to produce deletion mutations upon replication of repetitive sequences (Shinde et al. 2003; Garcia-Diaz and Kunkel 2006). These findings suggest that small variations in the length of the adenosine tract in the 3′-UTR of NAB2 cDNAs may result from slippage during replication or the process of RT-PCR. Overall, the data suggest that the major 3′-end of NAB2 mRNA maps in the A26 sequence, with a minor 3′-end occurring 114 nt downstream.
NAB2 mRNA 3′-end formation requires the core exosome
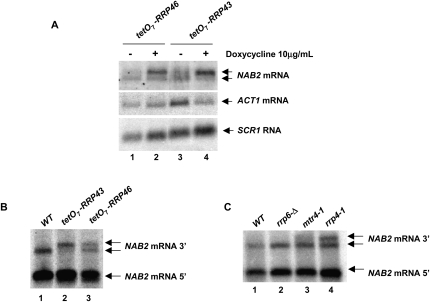
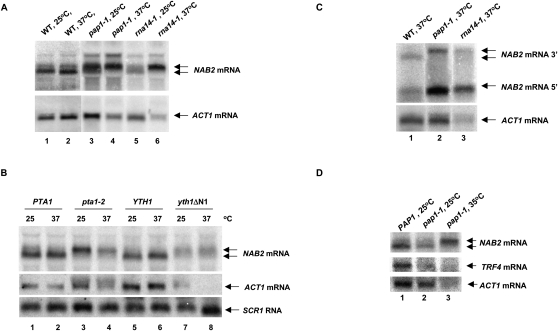
Previous experiments showed that the presence of 26 consecutive adenosines (A26) 140 nt 3′ to the NAB2 stop codon inhibited mRNA 3′-end formation at this site and rendered the transcript sensitive to degradation by the nuclear exosome (Roth et al. 2005). In support of the view that the A26 sequence inhibits normal mRNA 3′-end processing, its replacement with U26 resulted in efficient 3′-end formation at that site (Roth et al. 2005). Nevertheless, the presence of more than 26 adenosines at this site in cDNAs derived from mature NAB2 mRNAs implies that polyadenylation occurs at this site, perhaps by a mechanism different from classical cleavage and polyadenylation. Since the nuclear exosome plays an A26-dependent role in the regulation of NAB2 mRNA levels, we asked whether depletion of core exosome components might alter the pattern of 3′-end formation of NAB2 mRNA. We compared NAB2 mRNAs produced from the chromosomal allele in cells in which the endogenous promoters for RRP43 or RRP46 were replaced with the tetO7 promoter. Treatment of these cells with doxycyline represses expression of these genes, depletes the cells of the core exosome components Rrp43p and Rrp46p, and results in the expected defects in rRNA processing (Fang et al. 2004). Depletion of Rrp43p or Rrp46p causes a clear decrease in the mobility of NAB2 mRNA but has no detectable effect on the mobility of ACT1 mRNAs or SCR1 RNA, a stable RNA polymerase III transcript (Fig. 3A). Moreover, depletion of Rrp43p or Rrp46p increases the amount of NAB2 mRNA twofold relative to the control transcripts.
FIGURE 3.
Depletion of core exosome components results in longer NAB2 mRNAs. (A) YSB3002 (tetO7-RRP43) or YSB3003 (tetO7-RRP46) was grown in YPD at 30°C and treated (+) or not (−) with doxycycline for 8 h after which total RNA was prepared and RNA levels determined by Northern blotting. (B,C) Northern blot analysis of total RNA after RNase H treatment as described in Materials and Methods using OSB313 and total RNA from strains (B) YSB3001 (wt), YSB3002 (tetO7-RRP43), or YSB3003 (tetO7-RRP46); and (C) CWO4 (WT), YSB214 (rrp6-Δ), MS157-1A (mtr4-1), or YRP1222 (rrp4-1).
Next, we cleaved the NAB2 transcripts with RNase H and an oligonucleotide complementary to a site approximately one-third of the way from the start to the stop codon of NAB2 mRNA (Fig. 3B). Northern blot analysis of the products revealed that the distal product of RNase H cleavage increases by ∼100 nt after depletion of Rrp43p and Rrp46p, indicating that the increase in length results from a defect in 3′-end formation (Fig. 3B). Previous experiments showed that a temperature-sensitive mutation (rrp4-1) in the core exosome component Rrp4p caused an increase in NAB2 mRNA levels, but the resolution of the transcript did not clearly indicate a change in its length (Roth et al. 2005). RNase H cleavage shows that the rrp4-1 mutation clearly causes a defect in 3′-end formation of NAB2 mRNA (Fig. 3C, lane 4). Interestingly, deletion of RRP6 does not result in 3′ extended transcripts (Fig. 3C, lane 2). These findings indicate that normal 3′-end formation of NAB2 mRNA requires the activity of the core exosome, but not the nucleus-specific component Rrp6p. The fact that we detected no change in the length of ACT1 mRNA indicates that this effect is specific and does not result from depletion of components of the canonical mRNA cleavage and polyadenylation system.
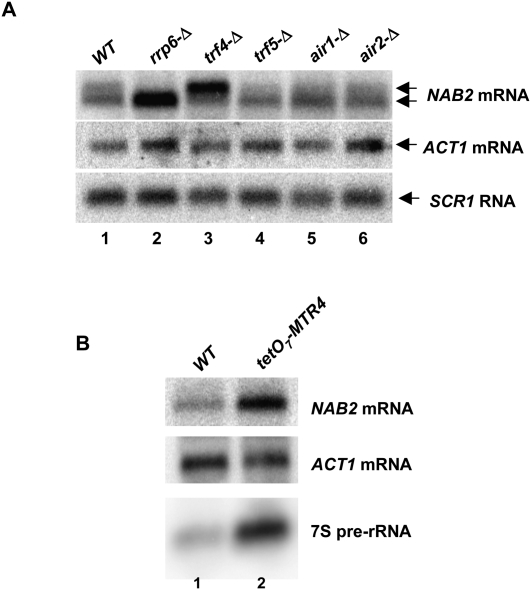
NAB2 mRNA 3′-end processing requires the TRAMP component Trf4p
The TRAMP complex composed of Mtr4p, Air1p or Air2p, and Trf4p or Trf5p enhances the rate of RNA degradation by the nuclear exosome (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005). Accordingly, we asked whether strains lacking TRAMP components cause defects in NAB2 mRNA 3′-end formation. Indeed, strains lacking Trf4p produce increased amounts of the longer form of NAB2 mRNA, without changing the amount or size of ACT1 mRNA or SCR1 mRNA (Fig. 4A, lane 3). In contrast, strains devoid of Trf5p, Air1p, or Air2p produce NAB2 mRNA in amounts and lengths similar to the normal control (Fig. 4A, lanes 4–6). The mtr4-1 mutation in the gene (DOB1/MTR4) encoding the putative helicase component of the TRAMP complex caused a marginal increase in the level of NAB2 mRNA (Roth et al. 2005). RNase H cleavage of NAB2 mRNA from an mtr4-1 mutant showed no significant change in the length of the transcript (Fig. 3C, lane 3). To avoid potential allele-specific effects of the mtr4-1 mutation, we revisited the role of Mtr4p in NAB2 expression by depleting the protein from cells in which the chromosomal gene promoter was replaced with the tetO7 promoter. After 5 h of treatment with doxycycline, Northern blot analysis revealed a sixfold increase in the level of 7S pre-rRNA, as expected since conversion of 7S to 5.8S rRNA requires Mtr4p and the core exosome (Fig. 4B, lane 2; de la Cruz et al. 1998). Likewise, depletion of Mtr4p results in a fourfold increase in the amount of NAB2 mRNA. However, loss of Mtr4p activity has no detectable effect on the length of the transcript. We conclude that the Trf4p component of the TRAMP complex plays a specific role in the 3′-end formation of NAB2 mRNA and that Mtr4p plays a role in controlling the ultimate level of the transcript.
FIGURE 4.
The TRAMP component Trf4p is required for normal 3′-end formation of NAB2 mRNA. (A) Northern blot analysis of total RNA from strains YSB1001 (RRP6), YSB1005 (rrp6-Δ), YSB1017 (trf4-Δ), YSB1020 (trf4-Δ), YSB1060 (air1-Δ), and YSB1061 (air2-Δ). (B) Northern blot analysis of total RNA from strains YSB3001 (WT) or YSB3017 (tetO7-RRP46) grown for 5 h in the presence of 10 μg/mL doxycycline.
Defects in the mRNA cleavage and polyadenylation system cause a 3′-end formation defect for NAB2 mRNA
The requirement for TRAMP and the core exosome in NAB2 mRNA 3′-end processing implies that defects in the classical cleavage and polyadenylation components should have no effect on the length of NAB2 mRNA. We tested this assumption by analyzing the length of NAB2 mRNAs in strains carrying temperature-sensitive mutations in the cleavage and polyadenylation factor (CPF) or cleavage factor IA (CFIA) complexes of the mRNA 3′-end processing apparatus. Northern blot analysis of mRNAs from strains with temperature-sensitive mutations in PAP1, RNA14, PTA1, and YTH1 reveal that these defects lead to the accumulation of 3′ extended NAB2 transcripts (Fig. 5A,B). Significantly, 3′ extended NAB2 transcripts accumulate in the pap1-1 mutant even at the permissive temperature (Fig. 5A, lane 3). Oligonucleotide-directed RNase H cleavage of transcripts from the pap1-1 and rna14-1 strains confirms that the increase in length results from the distal portion of the mRNA (Fig. 5C). These findings suggest that the CPF and CFIA mutations have effects similar to defects in TRAMP and the core exosome on NAB2 3′-end formation.
FIGURE 5.
Mutations in CPF and CFIA components cause NAB2 mRNA 3′-end formation defects. (A,B) Northern blot analysis of total RNA isolated from strains (A) YSB213 (WT), YSB215 (pap1-1), or YSB202 (rna14-1); and (B) FY41 (PTA1), FY1284 (pta1-2), YT2 (YTH1), and YT3 (yth1ΔN1) before and after shift for 1 h from 25°C to 35°C. All strains were grown in YPD, except YT2 and YT3, which were grown in synthetic complete dextrose without uracil (Fink 1991). (C) Northern blot analysis of the products of RNase H cleavage of total RNA from strains with the indicated genotypes after a 1-h shift to 35°C. (D) Northern blot analysis of total RNA prepared from strains with the indicated genotypes before (25°C) and after shift for 1 h to 35°C.
Three observations suggest that the effects of these mutations may affect NAB2 3′-end processing indirectly. First, while shift of the pap1-1 and rna14-1 strains to the nonpermissive temperature causes the expected decrease in ACT1 mRNA levels, it does not lower the level of NAB2 mRNA in the pap1-1 strain and appears to increase the level in the rna14-1 strain (Fig. 5A). Second, while defects in CPF and CFIA affect 3′-end processing of many mRNAs, the defects in TRAMP and the core exosome appear specific to NAB2 mRNA (Figs. 3, 4A). Third, the pap1-1 defect causes the accumulation of a 3′ extended NAB2 mRNA. Since the cleavage step of mRNA 3′-end processing in yeast does not require Pap1p, 3′ extended transcripts do not accumulate in pap1-1 mutants (Patel and Butler 1992; Proweller and Butler 1994). Instead, the levels of most mRNAs decrease upon inactivation of Pap1p owing to the inhibition of polyadenylation and subsequent degradation of the transcripts by the nuclear exosome (Burkard and Butler 2000). However, some poly(A)− mRNAs accumulate after inactivation of Pap1p in a pap1-1 strain, and they end at their normal 3′-end cleavage sites (Proweller and Butler 1994).
We reasoned that if the expression of a key component of the TRAMP or core exosome complex was unusually sensitive to Pap1p activity, then a defect in Pap1p would indirectly result in defective 3′-end formation of NAB2 mRNA. Accordingly, we analyzed the level of TRF4 mRNA in a pap1-1 strain since this transcript, as well as six of the 10 components of the core exosome, exhibits a relatively short half-life (5–8 min) that should result in its relatively rapid depletion upon inhibition of polyadenylation (Wang et al. 2002). Indeed, Northern blot analysis of TRF4 mRNA levels shows that the pap1-1 mutation results in a significant loss of TRF4 mRNA even at the permissive temperature (Fig. 5D). We conclude that the 3′ extension of NAB2 mRNA seen in the CPF and CFIA mutants likely reflects depletion Trf4p.
Nab2p defect bypasses the requirement for Trf4p in NAB2 mRNA 3′-end formation
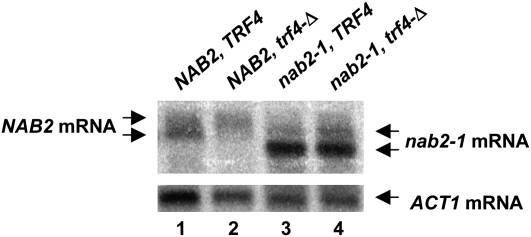
The results presented in Figure 1 indicated that decreasing Nab2p activity results in increased 3′-end formation of the NAB2 transcript at the site within the A26 sequence. This suggests that Nab2p may act to repress 3′-end formation at this site, leaving the processing of the transcript dependent on the exosome and TRAMP complex. This view implies that a defect in Nab2p activity should bypass the requirement for the exosome and TRAMP in 3′-end formation of the mRNA. We tested this by asking where the 3′-end of NAB2 mRNA lies in a strain carrying the nab2-1 and trf4-Δ mutations. The results show that while the majority of NAB2 transcripts end at the distal site in the NAB2, trf4-Δ strain, the majority of the transcripts end at the proximal site in the nab2-1, trf4-Δ strain (Fig. 6). These latter transcripts appear shorter and more abundant than normal since the nab2-1 mutation deletes a small portion of the N terminus of Nab2p and increases the level of the transcript fourfold owing to the inability of the defective Nab2p to enhance mRNA degradation by Rrp6p (Roth et al. 2005). These findings support the idea that Nab2p represses 3′-end formation at the A26 sequence, thereby rendering 3′-end processing dependent on the exosome and TRAMP complex.
FIGURE 6.
The nab2-1 mutation bypasses the requirement for Trf4p in NAB2 mRNA 3′-end processing. Northern blot analysis of total RNA from strains YSB501 [nab2∷HIS3 pACY717 (NAB2 LEU2 CEN)], YSB503 [nab2∷HIS3 trf4∷KANMX pACY717 (NAB2 LEU2 CEN)], YSB502 [nab2∷HIS3 pACY1152 (nab2-1 LEU2 CEN)], and YSB504 [nab2∷HIS3 trf4∷KANMX pACY1152 (nab2-1 LEU2 CEN)] grown in YPD at 30°C.
The NAB2 A26 sequence inhibits mRNA 3′-end formation in vitro
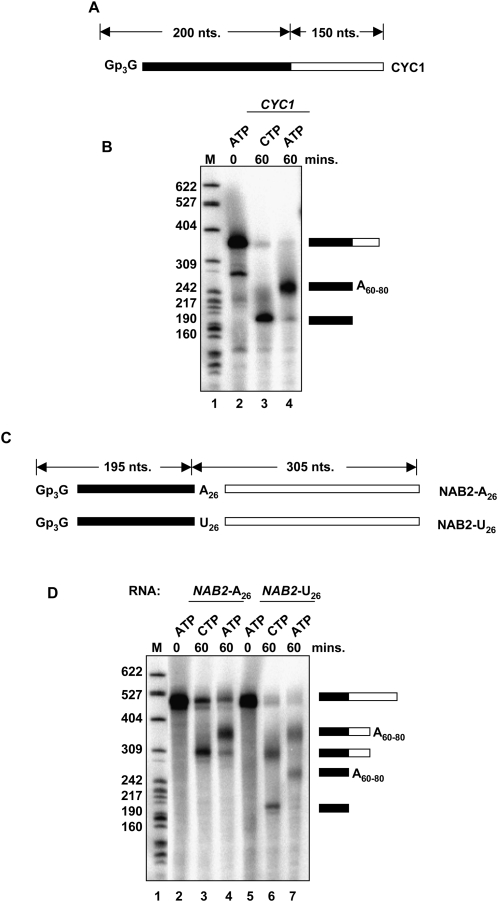
The dependence of NAB2 3′-end formation on the Trf4p and the exosome and the apparent repressive effect of the A26 sequence on normal 3′-end formation prompted us to analyze NAB2 3′-end processing in vitro. In this assay, synthetic radiolabeled RNAs are incubated in a yeast extract fraction capable of carrying out accurate 3′-end cleavage and polyadenylation (Butler et al. 1990). Figure 7 shows a diagram of the control CYC1 substrate and the results of incubating the 32P-labeled RNA in the extract. In the presence of CTP, the substrate undergoes cleavage at the polyadenylation site, but polyadenylation fails because of the lack of ATP (Fig. 7B, lane 3). In the presence of ATP, the cleavage product is polyadenylated, as expected (Fig. 7B, lane 4). A synthetic 32P-labeled NAB2 RNA containing its A26 sequence and 305 nt of sequence downstream undergoes cleavage and polyadenylation in the extract, but cleavage occurs ∼110 nt downstream from the A26 sequence. In contrast, a substrate in which U26 replaces A26 undergoes cleavage and polyadenylation at the downstream site, as well as at the U26 sequence. This result parallels that seen when U26 replaces A26 in vivo causing all 3′-end formation to occur at the U26 site and making the transcript insensitive to Rrp6p (Roth et al. 2005). These findings support the conclusions that formation of the mature 3′-end of NAB2 mRNA does not occur by canonical cleavage and polyadenylation and that the A26 sequence inhibits cleavage and polyadenylation at this site.
FIGURE 7.
The NAB2 A26 sequence inhibits mRNA 3′-end processing in vitro. Processing of synthetic 32P-labeled RNAs in vitro. (A) Diagram of the synthetic CYC1 RNA showing the portion (white) 5′ to the cleavage site and (black) 3′ to the cleavage site. (B) Processing of the CYC1 RNA in vitro. (Right) The diagrams show the positions of the substrate and products of the reaction. (Left) The numbers refer to the positions of the length markers (M). (C) Diagram of the synthetic NAB2 RNAs showing the portion (white) 5′ and (black) 3′ to the A26 or U26. (D) Processing of the CYC1 RNA in vitro. (Right) The diagrams show the positions of the substrate and products of the reaction. (Left) The numbers refer to the positions of the length markers (M).
DISCUSSION
This study presents evidence indicating that normal 3′-end formation of the NAB2 mRNA occurs by a novel mechanism requiring the action of the core exosome and the Trf4p component of the TRAMP complex. The major 3′-end of NAB2 mRNA maps to a site at or within an A26 sequence in its 3′-UTR (Fig. 2). However, several observations suggest that, in the presence of excess Nab2p, this end does not form by the normal cleavage and polyadenylation process. First, Nab2p represses the usage of this site in addition to destabilizing the transcript (Fig. 1). This conclusion refines the previous model that only featured destabilization of NAB2 mRNA by Nab2p (Roth et al. 2005). Second, a defect in Nab2p bypasses the requirement for Trf4p in 3′-end formation (Fig. 6). Third, our findings suggest that the mature 3′-end of NAB2 mRNA results from exosome trimming of a 3′ extended pre-mRNA, followed by polyadenylation at, or within, the A26 sequence. Depletion or mutation of core exosome components or Trf4p resulted in the accumulation of NAB2 transcripts ending 3′ of the normal site (Figs. 3–5). In all cases in which the 3′ extended transcript exists, its level exceeds that of the mature transcript found in normal cells. The fact that only mature transcripts accumulate upon depletion of Rrp6p or Mtr4p indicates that they do not play a necessary role in formation of the NAB2 mRNA 3′-end. However, the elevated level of mature NAB2 mRNA observed under these conditions suggests a role for Rrp6p and Mtr4p in controlling the ultimate amount of the transcript (Roth et al. 2005).
Inactivation of components of CPF and CFIA, required for cleavage and polyadenylation of mRNAs, results in 3′ extended NAB2 mRNAs (Fig. 5). Three observations suggest that this effect does not reflect inhibition of processing at the A26 site of the pre-mRNA. First, inactivation of these components results in stable 3′ extended NAB2 mRNAs, while other mRNAs are degraded by the exosome under these conditions (Fig. 5; Burkard and Butler 2000; Torchet et al. 2002; Milligan et al. 2005). Second, defects in CPF and CFIA may abrogate 3′-end trimming by the exosome indirectly by depleting Trf4p (Fig. 5). The apparent loss of Trf4p activity upon inactivation of Pap1p in a pap1-1 strain may also explain the reported requirement for Pap1p in the polyadenylation of some noncoding RNAs (van Hoof et al. 2000; Kuai et al. 2004; Carneiro et al. 2007; Grzechnik and Kufel 2008). Third, the A26 site is not cleaved and polyadenylated in vitro, but is activated for this process by replacement of U26 for A26 (Fig. 7).
Based on these and other observations, we propose the following model for NAB2 mRNA 3′-end formation. The region just 5′ of the A26 contains sequence elements similar to those necessary for formation of CFIA and CPF complexes (Guo and Sherman 1995; Dichtl and Keller 2001), so we suggest that the canonical 3′-end processing system recognizes the mature site, but fails to cleave because of the inhibitory effect of the A26 sequence, most likely because it binds Nab2p. Instead, a NAB2 mRNA end forms downstream from the mature 3′-end of the transcript, probably at the secondary 3′-end site observed in vivo and in vitro ∼110 nt downstream from the mature 3′-end (Figs. 2, 7). Next, the core exosome and TRAMP degrade the transcript in cycles of adenylation by Trf4p and degradation by the core. When the core exosome reaches the A26 sequence, it dissociates and two alternative events may occur: (1) If Nab2p levels are insufficient for it to remain bound to the A26 sequence, polyadenylation occurs, forming the mature mRNA; (2) if Nab2p levels are high, the protein remains bound to the A26 sequence and recruits or activates Rrp6p to degrade the transcript with the aid of Mtr4p, and possibly the core exosome. Since core exosome and Trf4p defects trap the transcript in the longer form, our experiments cannot tell if degradation requires their activity. Nevertheless, since core exosome and Trf4p depletion elevate NAB2 mRNA levels, we conclude that degradation of the transcript by Rrp6p and Mtr4p requires, at the very least, prior 3′-end processing by the core exosome. Clarification of the details of this model, including how the initial 3′-end of the pre-mRNA forms, requires further experimental work.
We assume that Trf4p carries out its role in the initial trimming of NAB2 mRNA in the context of the TRAMP complex, but we have no direct evidence to support this claim. The lack of an apparent requirement for the Mtr4p in the initial trimming of the pre-mRNA might reflect the absence of this putative helicase in the complex, or a lack of a requirement for it to remove secondary structures or proteins bound to the RNA in this region. Likewise, our experiments do not indicate a requirement for Air1p or Air2p, the putative RNA binding components of the TRAMP complex. We could not isolate a viable air1, air2 mutant, making it impossible to test if these proteins play a redundant role in formation of the NAB2 3′-end. Nevertheless, considering the facts that (1) the role of Trf4p in RNA degradation appears to occur in the context of the TRAMP complex; (2) the activity of Trf4p requires the activity of at least one of the Air proteins; and (3) Mtr4p plays a key role in the subsequent degradation of the NAB2 transcript, it seems likely that TRAMP works with the core exosome to form the 3′-end of NAB2 mRNA (Kadaba et al. 2004, 2006; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006).
These experiments suggest that the TRAMP complex and the core exosome participate in the 3′-end formation of a normal mRNA. Previous studies showed that defects in Pap1p, the mRNA poly(A) polymerase, resulted in the Rrp6p-dependent degradation of pre-mRNAs, indicating that Rrp6p plays a surveillance role that carries out the degradation of slowly polyadenylated transcripts (Burkard and Butler 2000; Milligan et al. 2005). Likewise, defects in mRNA 3′-end processing factors Rna14p and Rna15p resulted in the production of 3′ extended transcripts that underwent degradation by the exosome (Torchet et al. 2002). In some cases, it appeared that exosome trimming could stop at, or near, the poly(A) site, whereupon the transcript underwent polyadenylation (Torchet et al. 2002). The cell appears to have adopted this surveillance mechanism to form the mature 3′-end of the budding yeast CTH2 mRNA (Ciais et al. 2008) and, in the present case, of NAB2 mRNA. In the case of NAB2, the A26 and, perhaps, a nonproductive cleavage and polyadenylation complex just upstream, serve to elicit a transition in exosome degradation such that either polyadenylation occurs, or degradation resumes using Rrp6p and Mtr4p. In this view, trimming of the transcript into the A26 sequence would allow it to act as a primer for poly(A) polymerase in the stalled polyadenylation complex. In the case of CTH2 mRNA, specific sequence elements in the pre-RNA may bind Nrd1p and Nab3p, which then recruit the exosome to the transcript in a manner similar to that which serves to regulate the levels of NRD1 mRNA (Arigo et al. 2006; Ciais et al. 2008). Previous studies showed that a defect in Nrd1p results in increased levels of NAB2 mRNA, suggesting a role for Nrd1p in regulating 3′-end formation (Vasiljeva and Buratowski 2006). The NAB2 mRNA contains several potential Nrd1p and Nab3p binding sites, but the role of these factors in NAB2 mRNA processing remains to be addressed.
The novel mechanism of mRNA 3′-end formation proposed here for NAB2 mRNA serves to regulate the level of Nab2p in the cell. Nab2p plays a key role in this regulation since increases in its activity decrease its mRNA levels, and loss of its function results in more NAB2 mRNA (Roth et al. 2005). This autoregulatory circuit requires the presence of the A26 in the transcript's 3′-UTR, which almost certainly reflects the poly(A)-binding specificity of Nab2p (Anderson et al. 1993). Nab2p plays important roles in mRNA export and the control of poly(A) tail lengths during mRNA 3′-end processing (Green et al. 2002; Hector et al. 2002). Moreover, changes in Nab2p activity by mutation or alteration of Nab2p levels cause defects in mRNA metabolism that lead to significant growth defects in the affected cells. These observations provide a rationale for understanding the evolution of the autoregulatory mechanism that controls NAB2 mRNA levels.
MATERIALS AND METHODS
Strains and reagents
Yeast media and experimental reagents were prepared by standard protocols (Fink 1991). Yeast strains are described in Table 1. Oligonucleotides are listed in Table 2. Chemicals were obtained from Sigma, US Biological, or Fisher.
TABLE 1.
Yeast strains
TABLE 2.
Oligonucleotides
LM-PAT and cRT-PCR analysis
The protocol for the LM-PAT was followed directly as specified in Salles and Strickland (1999). The oligonucleotide used as phosphorylated oligo(dT)13 was OSB349. The oligonucleotide used as the oligo(dT)12-anchor was OSB290. The 5′ RPL3 and NAB2 primers used to amplify the resulting LM-PAT cDNAs by PCR were OSB5 and OSB217, respectively. PCR products were cloned into the pGEM-T-Easy blue/white-screening vector (Promega) and transformed to XL1 Escherichia coli, and appropriate clones were selected for subsequent sequencing.
cRT-PCR was performed as described by Mullen and Marzluff (2008). Fifty micrograms of total yeast RNA was treated with 25 units of RQ1 DNase (Promega) in 50 μL of the manufacturer's buffer containing 40 units of Ribolock (Fermentas) for 20 min at 37°C. The products were extracted once with phenol:chloroform:isoamyl alcohol (50:48:2), and the RNA in the aqueous layer was precipitated with ethanol. The RNA was collected by centrifugation, dried, and resuspended in 25 μL of H2O. Ten micrograms of the RNA was treated with 5 units of tobacco acid pyrophosphatase (Epicentre) in 20 μL of the manufacturer's buffer containing 20 units of Ribolock (Fermentas) for 1 h at 37°C. Four micrograms of the decapped RNA was treated with 20 units of RNA ligase (New England Biolabs) in 400 μL of the manufacturer's buffer containing 20 units of Ribolock for 12 h at 15°C. The products were precipitated with ethanol, collected by centrifugation, washed with 70% ethanol, dried, and resuspended in 12 μL of H2O. cDNA synthesis was carried out on 6 μL of the ligated RNA with 200 units of M-MLV reverse transcriptase (Invitrogen) in 20 μL of the manufacturer's first-strand synthesis buffer containing 20 units of Ribolock, 0.5 mM dNTP, and 25 pmol of OSB769 for 50 min at 37°C, followed by incubation for 15 min at 70°C. PCR was carried out on 1 μL of the cDNA using OSB316 and OSB772. The products were purified using a Qiaquick PCR Purification Kit (QIAGEN) and cloned directly into pGEM-T-easy (Promega). DNA sequence analysis was carried out on individual clones.
Messenger RNA 3′-end processing of mRNA in vitro
The protocol for 3′-end processing of synthetic radiolabeled pre-mRNA substrates was performed as described in Butler et al. (1990). Oligonucleotides OSB317, which encodes the SP6 promoter, and OSB218 (3′ NAB2 primer) were used in PCR with plasmid DNA templates pGFPNAB2 and pNAB2-U26 to generate DNA templates for in vitro mRNA transcription. Templates for transcription to produce CYC1 RNAs were produced in two steps. First, for production of the CYC1-A26 substrate, separate PCR reactions were performed using oligonucleotides OSB491 and OSB498 or OSB493 and OSB497 using plasmid pRP497 (Muhlrad and Parker 1999) as a template. These products were mixed together and used as a template for PCR with OSB497 and OSB498. Likewise, for production of the CYC1-U26 substrate, separate PCR reactions were performed with oligonucleotides OSB493 and OSB498 or OSB494 and OSB497 using plasmid pRP497 as a template. These products were mixed together and used as a template for PCR with OSB497 and OSB498. The CYC1-normal substrate was made by PCR using OSB497 and OSB498 and pRP497 as a template. The resulting PCR products were cloned into HindIII-digested YCplac22 by gap repair. Isolated plasmid clones were recovered, and the DNA sequence of the CYC1 region was determined. Next, the CYC1 portions of the CYC1-normal, CYC1-A26, and CYC1-U26 substrates were PCR-amplified with OSB544, which encodes the SP6 RNA polymerase promoter, and OSB545. The products were used in SP6 RNA polymerase transcription reactions in vitro to produce 32P-labeled RNA transcripts (Butler et al. 1990).
RNA analysis
Total RNA was isolated from yeast strains grown to an A600 of 0.8–1.2 as described (Patel and Butler 1992), and Northern Blot analysis was carried out as described (Briggs et al. 1998). Oligonucleotides used for detection of RNA in Northern blot analyses are described in Table 2. The procedure for generating [γ-32P]ATP-labeled DNA oligonucleotide probes was described in Briggs et al. (1998). 7S rRNA, SCR1 RNA, and ACT1 mRNA were detected by 32P-N-terminal-labeled DNA oligonucleotides OSB157, OSB151, and OSB184, respectively. 5′ [α-32P]CTP-labeled DNA random primed hexamer probes were generated using the Random Primers DNA Labeling System (Invitrogen). GFP-NAB2 fusion transcripts were detected with 32P-labeled random-primed labeled probes to GFP generated from a PCR template generated with primers OSB272 and OSB273 using pGFP-N-FUS as a template. NAB2 mRNA was detected with 32P-labeled random-primed probes generated from a BamHI, EcoRI NAB2-containing fragment of pGAD424-NAB2 (Roth et al. 2005). TRF4 mRNA was detected with 32P-labeled random-primed labeled probes generated from a PCR template with primers OSB574 and OSB575 using YSB1001 genomic DNA as a template.
ACKNOWLEDGMENTS
We are grateful to Clair Moore (Tufts University) and Anita Corbett (Emory University) for providing yeast strains. We thank Jason Hoskins and Kevin Callahan for discussions and critical reading of the manuscript and Guangze Jin for help with DNA sequence analysis. This work was supported by grants from the National Institutes of Health (GM-59898) and the National Science Foundation (MCB-0817324).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.709609.
REFERENCES
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet M., Wyers F., Dufour M.E., Aggerbeck L.P., Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.S.J., Parker R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.T., Wilson S.M., Datar K.V., Swanson M.S. NAB2: A yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo J.T., Carroll K.L., Ames J.M., Corden J.L. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bienroth S., Keller W., Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti C., Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Butler J.S. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics. 1996;143:1149–1161. doi: 10.1093/genetics/143.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M.W., Burkard K.T., Butler J.S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Burkard K.T., Butler J.S. A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.S. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Butler J.S., Sadhale P.P., Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol. Cell. Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T., Carvalho C., Braga J., Rino J., Milligan L., Tollervey D., Carmo-Fonseca M. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Ciais D., Bohnsack M.T., Tollervey D. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 2008;36:3075–3084. doi: 10.1093/nar/gkn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P., Fromont-Racine M., Steel D., Pictet R., Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Guo Z., Russo P., Chartrand P., Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Butler J.S., Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;16:5502–5512. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Tollervey D., Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S., Nykamp K.R., Viphakone N., Swanson M.S., Minvielle-Sebastia L. Yeast mRNA poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent poly(A) nuclease activity. J. Biol. Chem. 2005;280:24532–24538. doi: 10.1074/jbc.M504720200. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Edmonds M. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- Egecioglu D.E., Henras A.K., Chanfreau G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Hoskins J., Butler J.S. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken M.B., Corbett A.H. Process or perish: Quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 2005;12:482–488. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- Fink C. Guide to yeast genetics and molecular biology. Academic Press; San Diego, CA: 1991. [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M., Kunkel T.A. Mechanism of a genetic glissando: Structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Green D.M., Marfatia K.A., Crafton E.B., Zhang X., Cheng X., Corbett A.H. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- Greimann J.C., Lima C.D. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P., Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Sherman F. 3′-End-forming signals of yeast mRNA. Mol. Cell. Biol. 1995;15:5983–5990. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.E., Nykamp K.R., Dheur S., Anderson J.T., Non P.J., Urbinati C.R., Wilson S.M., Minvielle-Sebastia L., Swanson M.S. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T.H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Dower K., Libri D., Rosbash M. Early formation of mRNP. License for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A.M., Hinnebusch A.G., Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Wang X., Anderson J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kuai L., Fang F., Butler J.S., Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lejeune F., Li X., Maquat L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E., Conti E. The exosome and the proteasome: Nano-compartments for degradation. Cell. 2006;125:651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Marfatia K.A., Crafton E.B., Green D.M., Corbett A.H. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- Milligan L., Torchet C., Allmang C., Shipman T., Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Gao M., O'Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T.E., Marzluff W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C.C., van den Heuvel J.J., McCarthy J.E. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: The roles of the stability and position of stem–loops in the mRNA leader. Mol. Microbiol. 1993;9:521–532. doi: 10.1111/j.1365-2958.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- Patel D., Butler J.S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.T., Robinson M.D., Mnaimneh S., Krogan N.J., Cagney G., Morris Q., Davierwala A.P., Grigull J., Yang X., Zhang W., et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Phillips S., Butler J.S. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N., O'Sullivan J. Polyadenylation: A tail of two complexes. Curr. Biol. 2002;12:R855–R857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- Proweller A., Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes & Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., Stutz F. Nuclear export of RNA. Biol. Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Roth K.M., Wolf M.K., Rossi M.L., Butler J.S. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol. 2005;25:1577–1585. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles F.J., Strickland S. Analysis of poly(A) tail lengths by PCR: The PAT assay. Methods Mol. Biol. 1999;118:441–448. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
- Salles F.J., Richards W.G., Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- Schaeffer D., Tsanova B., Barbas A., Reis F.P., Dastidar E.G., Sanchez-Rotunno M., Arraiano C.M., van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Leung E., Brown J., Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde D., Lai Y., Sun F., Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 2003;31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacahashi Y., Helmling S., Moore C.L. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 2003;31:1744–1752. doi: 10.1093/nar/gkg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C., Bousquet-Antonelli C., Milligan L., Thompson E., Kufel J., Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Lennertz P., Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- Wang Z., Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Norbury C.J., Proudfoot N.J. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kessler M., Helmling S., O'Connor J.P., Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]