Abstract
During B cell maturation, immunoglobulin (Ig) genes frequently acquire premature translation-termination codons (PTCs) as a result of the somatic rearrangement of V, D, and J gene segments. However, it is essential for a B lymphocyte to produce only one kind of antibody and therefore to ensure that the heavy and light chain polypeptides are expressed exclusively from the corresponding functional alleles, whereas no protein is made from the nonproductively rearranged alleles. At the post-transcriptional level, a well-studied mRNA quality control mechanism, termed nonsense-mediated mRNA decay (NMD), recognizes and degrades PTC-containing mRNAs in a translation-dependent manner. In addition, transcriptional silencing of PTC-containing Ig-μ and Ig-γ heavy chain reporter genes was observed in HeLa cells. To investigate the silencing of nonproductively rearranged Ig genes in a more physiological context, we analyzed a monoclonal line of immortalized murine pro-B cells harboring one productively (PTC−) and one nonproductively (PTC+) rearranged Ig-μ heavy chain allele. We show that the steady-state abundance of PTC+ mRNA was ∼40-fold lower when compared to that of the PTC− mRNA. However, both the PTC+ and PTC− allele seemed to be equally well transcribed since the abundances of PTC+ and PTC− pre-mRNA were very similar and chromatin immunoprecipitations revealed comparable occupancy of RNA polymerase II and acetylated histone H3 on both alleles. Altogether, we found no evidence for transcriptional silencing of the PTC+ allele in this pro-B cell line; hence, the efficient down-regulation of the PTC+ Ig-μ mRNA results entirely from NMD.
Keywords: nonsense-mediated transcriptional gene silencing (NMTGS), nonsense-mediated mRNA decay (NMD), VDJ rearrangement, immunoglobulin, pro-B cells, allelic exclusion
INTRODUCTION
To ensure accurate gene expression and to prevent the synthesis of faulty proteins, cells have evolved several quality control mechanisms. One of these surveillance mechanisms is termed nonsense-mediated mRNA decay (NMD) and protects eukaryotic cells from accumulation of truncated and potentially harmful proteins. Thereby, mRNAs harboring premature translation-termination codons (PTCs) are recognized in a translation-dependent manner and degraded rapidly (for reviews, see Chang et al. 2007; Isken and Maquat 2007; Mühlemann et al. 2008). The molecular mechanism is not fully understood, but the three NMD core factors UPF1, UPF2, and UPF3 are conserved in all eukaryotes analyzed so far (Hodgkin et al. 1989; Leeds et al. 1992; Applequist et al. 1997; Lykke-Andersen et al. 2000; Gatfield et al. 2003). PTCs arise from nonsense and frameshift mutations in the genome and from errors during RNA processing, mainly by alternative pre-mRNA splicing. During lymphoblast maturation, PTCs are frequently generated during the programmed recombination of the different variable (V), diversity (D), and joining (J) segments in the genes coding for immunoglobulins or T cell receptors, respectively.
In pro-B cells, VDJ recombination of the immunoglobulin heavy chain alleles (IgH) occurs in a sequential and regulated manner (Mostoslavsky et al. 2004; Jung et al. 2006): First, one D segment is combined with one J segment on both IgH alleles. After that, one V segment recombines with the DJ segment on one of the two alleles. If this rearrangement is productive, a pre-B cell receptor (pre-BCR) complex is expressed on the cell surface and a feedback mechanism inhibits the rearrangement of the second allele (Alt et al. 1984). If the rearrangement of the first allele is nonproductive, the second allele also recombines one V with its DJ region. If both rearrangements are nonproductive, cells undergo apoptosis. As a result of these recombination events, ∼60% of the mature, circulating B cells contain one productively rearranged allele and one nonrearranged allele (VDJ+/DJ), and ∼40% of the cells have one nonproductively and one productively rearranged allele (VDJ−/VDJ+).
Interestingly, nonproductively rearranged (PTC+) transcripts of the immunoglobulin superfamily appear to be exceptionally good NMD targets, as illustrated by the strong down-regulation of their mRNA levels (Baumann et al. 1985; Jäck et al. 1989; Li and Wilkinson 1998; Bühler et al. 2004). In addition to this post-transcriptional effect, transcriptional silencing of PTC+ Ig minigenes (Ig-μ and Ig-γ) stably expressed in HeLa cells was observed, but not with other reporter genes (β-globin, TCRβ or glutathione peroxidase 1) (Bühler et al. 2005). This so-called nonsense-mediated transcriptional gene silencing (NMTGS) is accompanied by a change from hyperacetylated histone H3 typically found in transcriptionally active chromatin to methylated lysine 9 of histone H3, a hallmark of transcriptionally inactive heterochromatic regions. Remarkably, NMTGS was inhibited by overexpression of the siRNAse 3′hEXO, suggesting that small RNAs might be involved in NMTGS. However, siRNA-like molecules complementary to the reporter transcript could not be detected until now (O. Mühlemann, unpubl.). Furthermore, translation of the mRNA and the NMD factor UPF1 are both required for NMTGS of the PTC+ Ig-μ reporter, pointing to a common mechanism of PTC recognition in NMD and NMTGS (Stalder and Mühlemann 2007). In contrast, no evidence for a reduced transcription rate of PTC+ Ig-μ genes was observed previously in mouse hybridoma cells (Baumann et al. 1985; Mühlemann et al. 2001) or in Abelson virus-transformed 18-81-derived pre-B and plasma cell lines (Jäck et al. 1989). These earlier studies, however, did not analyze a PTC+ and a PTC− Ig-μ allele in the same cell, and we therefore decided to generate and select a clonal line of immortalized murine pro-B cells containing one nonproductively (PTC+) and one productively (PTC−) rearranged Ig-μ heavy chain allele to investigate if in this context transcriptional silencing contributes to monoallelic expression of Ig-μ genes. We show here that the mRNA level of the PTC+ allele was efficiently reduced in these cells compared to the PTC− allele. In contrast, pre-mRNA levels, association with RNA polymerase II, and with acetylated histone H3 were very similar between the PTC+ and the PTC− Ig-μ allele, indicating that both alleles are equally well transcribed and that monoallelic Ig-μ expression at this early stage of B lymphocyte development relies entirely on a post-transcriptional surveillance mechanism, most likely NMD.
RESULTS
Identification of pro-B cell lines containing two rearranged Ig-μ alleles
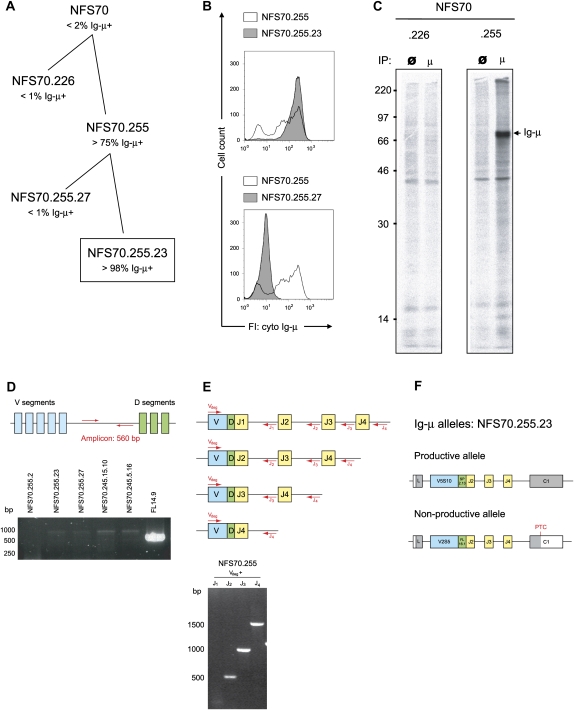
To establish clonally related pro-B cells with one productively and one nonproductively rearranged VDJ exon, we stepwise subcloned the pro-B line NFS70 by limiting dilution method. NFS70 was chosen for this analysis since it contains DJ rearrangements on both IgH alleles (Davidson et al. 1984) and since some cells that produce Ig-μ chains were detected by flow cytometry in a NFS70 culture, indicating that a productive V-DJ rearrangement had occurred in these cells (Fig. 1A). To isolate those, subclones were obtained from NFS70 cultures by limiting dilution and screened by flow cytometry for the presence of cytoplasmic Ig-μ chain (Fig. 1B). SDS-PAGE of metabolically labeled and immunoprecipitated Ig-μ chains from NFS70 subclones confirmed the presence of a full-length Ig-μ with an expected molecular mass of ∼72 kDa. A representative result is shown in Figure 1C for the Ig-μ-negative subclone NFS70.226 and the Ig-μ-positive clone NFS70.255. To assess the configuration at the second IgH allele, genomic DNA was isolated from different Ig-μ-positive NFS70 subclones. A first PCR with oligos annealing in the germline region between the VH segments and the D segments (Wasserman et al. 1998) confirmed the absence of intervening sequences between the VH and the D region, and thus the presence of a V to DJ rearrangement on both Ig-μ alleles in all tested subclones (Fig. 1D). Next, PCR analysis with a degenerated forward primer, which binds to framework sequences of most VH segments and four different reverse primers, each binding in the intron downstream from one of the four different JH segments (ten Boekel et al. 1997), was performed to identify the JH segments used in the pro-B cell clones. This analysis revealed that the JH2 segment was used on both alleles in subclone NFS70.255 (Fig. 1E). Subsequently, VDJ rearrangements were amplified from the clone NFS70.255.23, since almost all cells in this subclone produce full-length Ig-μ chain, and should therefore be derived from a single cell (Fig. 1A–C). The amplified VDJ fragments were then cloned, and the sequences of both VDJ alleles were determined. This analysis confirmed that both alleles utilized the JH2 segment and revealed that the productively rearranged VDJ allele (PTC−, i.e., without a premature translational stop codon) is composed of the VH segment V5S10 and the D segment SP2.13 (nomenclature according to http://imgt.cines.fr). The nonproductively rearranged allele (PTC+) comprises the VH segment V2S5 and the D segment FL16.1 and harbors the PTC at amino acid position 178 (Fig. 1F).
FIGURE 1.
Analysis of subclones from the pro-B line NFS70. (A) Pedigree illustrating subcloning of the cell line NFS70.255.23 from the parental NFS70 line by the limited dilution method. (B) Flow cytometric analysis of cytoplasmic Ig-μ production in NFS70 subclones. Cells were fixed, permeabilized, stained with FITC-conjugated goat antibodies against Ig-μ and analyzed by FACS. (C) Analysis of immunoprecipated and 35S-labeled Ig-μ chains from NFS70 subclones. Metabolically labeled cell lysates from the Ig-μ-negative subclone NFS70.226 and the Ig-μ-positive subclone NFS70.255 were precipitated with anti-Ig-μ (lanes μ). Precipitation with protein G sepharose only served as negative controls (lanes ø). Cell lysates were separated by 10% SDS-PAGE and radioactivity was detected in a PhosphoImager. (D) PCR-based analysis of IgH rearrangement. Schematic illustration of VH and D segments in the germline Ig locus before rearrangment and corresponding PCR primers (upper part). When VDJ rearrangement has occurred on both alleles, no 560 bp PCR amplicon can be generated. Pro-B cell line FL14.9 with one DJ-rearranged allele was used as control (lower part). (E) PCR-based analysis of JH segment usage. Schematic illustration of VH, D, and JH segments in the germline IgH locus after rearrangement and corresponding PCR primers (upper part). Depending on the JH segment combined to the D segment, different amplicon patterns can be detected on an agarose gels. JH2 is used on both alleles in subclone NFS70.255 (lower part). (F) Schematic illustration of the productively and nonproductively rearranged allele of the Ig-μ-positive subclone NFS70.255.23. Gray and colored boxes indicate protein coding sequences, white boxes untranslated exonic sequences.
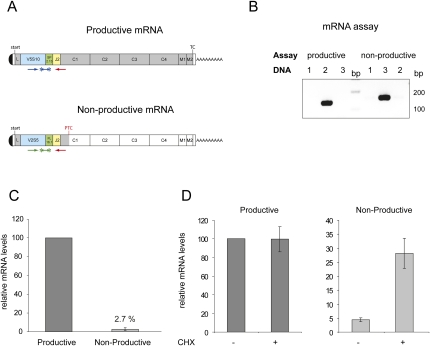
Based on the sequences, two allele-specific TaqMan assays for reverse transcription-coupled quantitative PCR (RT-qPCR) were designed: The forward primers and the TaqMan probes of both assays specifically bind to the VDJ region of the respective allele, whereas the common reverse primer is spanning over the JH2-Cμ1 junction to discriminate mRNA from pre-mRNA and genomic DNA (Fig. 2A). To test allele specificity of the assays, qPCR analysis was performed with plasmids containing either the nonproductively or the productively rearranged allele (Fig. 2B; data not shown). To allow quantitative comparison of the data obtained with the two TaqMan assays, the amplification rates of both assays were determined (data not shown) and found to be comparable: amplification rates for PTC− and PTC+ were 1.889 and 1.867, respectively.
FIGURE 2.
Releative mRNA levels of productively and nonproductively rearranged Ig-μ alleles. (A) Schematic illustration of productively and nonproductively rearranged Ig-μ mRNAs. Location of primers and TaqMan probes for the allele-specific TaqMan assays are shown below the mRNA. 5′ and 3′ UTRs are depicted as white boxes, VH segments are blue, D segments green, JH2 segments yellow, and the remaining parts of the ORFs are shown in gray. In the nonproductively rearranged mRNA, the ORF is truncated by a PTC at amino acid position 178. (B) For analysis of allele specificity of TaqMan assays, both assays were measured by qPCR with plasmids containing the sequence of the productively (DNA, lane 2) or the nonproductively rearranged allele (DNA, lane 3). qPCR products were resolved on an 2% agarose gel. Mouse cDNA from liver (DNA, lane 1) was used as negative control. (C) Relative mRNA abundances of the two Ig-μ alleles were measured by RT-qPCR using the allele-specific TaqMan assays. Values and standard deviations of three independent experiments with two runs each were calculated using the previously determined amplification rates. (D) Effect of cycloheximide (CHX) treatment on relative abundances of productive (left) and nonproductive Ig-μ mRNAs (right). Cells were incubated for four hours with 50 μg/mL CHX and relative mRNA levels were determined by RT-qPCR. Values and standard deviations of one representative experiment were calculated as in C.
NMD contributes to the low mRNA abundance of the PTC+ Ig-μ allele
To measure the steady-state abundance of mRNA from the nonproductively rearranged and the productively rearranged VDJ allele, RNA was isolated from pro-B cell clone NFS70.255.23 and analyzed by RT-qPCR with the allele-specific assays described above. The mRNA abundance of the PTC+ allele was ∼40-fold lower than that of the productively rearranged allele (Fig. 2C). This low PTC+ mRNA abundance could originate from reduced transcription, for example NMTGS, and/or represent a post-transcriptional effect, for example NMD. To test for NMD in this pro-B cell line, we added the translation inhibitor cycloheximide (CHX) to the cells four hours before harvesting. While this had no effect on the mRNA abundance of the productive allele, CHX treatment caused a sevenfold increase of the PTC+ mRNA (Fig. 2D), consistent with the mRNA from the nonproductively rearranged allele being a substrate for NMD. Although the amount of PTC+ mRNA increased upon CHX treatment, it did not reach the abundance of the PTC- mRNA. Therefore, we analyzed if, in addition to NMD, transcriptional silencing was also contributing to the low abundance of PTC+ mRNA.
No evidence for chromatin remodeling of the PTC-containing Ig-μ allele
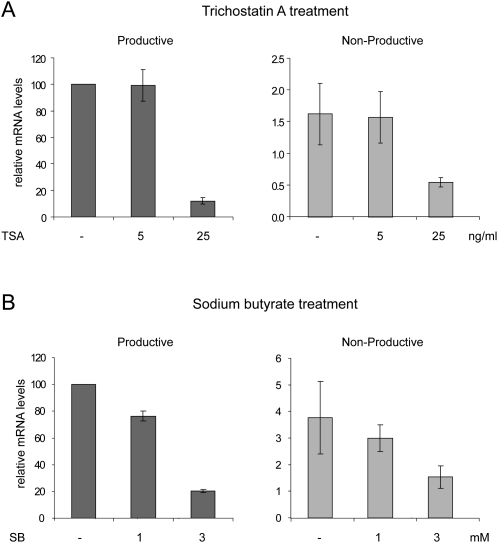
NMTGS in HeLa cells is accompanied with chromatin remodeling and can be reversed by treating the cells with histone deacetylase inhibitors like sodium butyrate (SB) or trichostatin A (TSA) (Bühler et al. 2005). Thus, if the nonproductively rearranged allele in the pro-B cell line was silenced at a transcriptional level by the same mechanism, the PTC+ mRNA level would be expected to increase upon treatment of the cells with SB or TSA. In contrast, the mRNA level of the PTC− allele should not be affected by histone deacetylase inhibitors, because it is already actively transcribed.
To analyze the chromatin state of the two Ig-μ alleles in our pro-B cell line, cells were treated with different concentrations of SB or TSA for 24 h and RNA abundance was quantitated by RT-qPCR. The mRNA levels of both Ig-μ alleles were lower when treated with the relatively high concentration of histone deacetylase inhibitors (Fig. 3A,B), indicating an unspecific secondary effect of these drugs on pro-B cells. More importantly, however, no PTC-specific increase of mRNA was observed in any of the different tested concentrations and incubation times (Fig. 3A,B; data not shown). These data indicate that histone deacetylases do not contribute to the low abundance of PTC+ Ig-μ mRNA, suggesting that NMTGS as described previously (Bühler et al. 2005) might not occur on the nonproductive Ig-μ allele in our pro-B cell line.
FIGURE 3.
Effect of histone deacetylase inhibitors on relative mRNA abundances of the productively and the nonproductively rearranged Ig-μ alleles. (A) TSA at two different concentrations (5 and 25 ng/mL) was added to the cells for 24 h before RNA isolation. (B) Cells were incubated with 1 or 3 mM SB for 24 h before RNA isolation. Relative mRNA abundances were determined by RT-qPCR as in Figure 2D.
No evidence for transcriptional silencing of the nonproductively rearranged Ig-μ allele
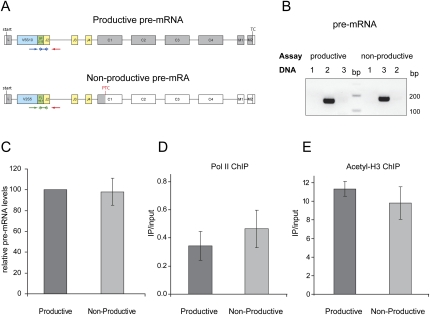
Because the sequences of the PTC− and the PTC+ Ig-μ alleles were not sufficiently different to determine the transcription rates of both alleles individually by nuclear run-on analysis, we decided to indirectly assess the transcription activity of both Ig-μ alleles by measuring the amount of PTC− and the PTC+ pre-mRNAs by RT-qPCR and by performing chromatin immunoprecipitation (ChIP) experiments using an antibody against RNA polymerase II (pol II).
To measure pre-mRNA levels of both alleles, RT-qPCR was performed with the allele-specific forward primers and TaqMan probes used for mRNA measurements combined with a reverse primer annealing in the intron downstream from the VDJ exon (Fig. 4A). Again, the amplification rates and the allele specificity of both assays were determined as above (Fig. 4B; data not shown). Control reactions using RNA samples that were not reverse-transcribed (−RT controls) confirmed that the background signal produced from possible contamination of the RNA samples with genomic DNA was <1% of the measured pre-mRNA values and therewith negligible (data not shown). Comparison of the PTC− and the PTC+ pre-mRNA levels revealed no significant difference between the two Ig-μ alleles (Fig. 4C), suggesting comparable transcription rates of these alleles.
FIGURE 4.
No evidence for transcriptional silencing of PTC-containing Ig-μ allele. (A) Schematic illustration of the pre-mRNA from the productively and nonproductively rearranged Ig-μ alleles as described in Figure 2A. (B) Analysis of the allele specificity of the two pre-mRNA TaqMan assays. Both assays were tested by qPCR as described in Figure 2B, and the amplicons were analyzed on a 2% agarose gel. (C) Relative pre-mRNA levels of productive and nonproductive allele were measured by RT-qPCR. Average values and standard deviations of four independent experiments with one run each are shown. For accurate calculation of the relative pre-mRNA abundances, the amplification rates were determined first. (D) Two independent pol II ChIPs were performed and the extracted DNA analyzed by qPCR using the two allele-specific TaqMan assays. The relative amount of DNA in the immunoprecipitated fraction was normalized to the input (input set to 1). (E) Two independent acetyl-H3 ChIPs were performed and analyzed as in D.
The pol II ChIP experiments confirmed our interpretation of the pre-mRNA analysis. The same relative amounts of both Ig-μ alleles were precipitated with the pol II antibody (Fig. 4D). Hence, we conclude that the transcription of the productive and the nonproductive Ig-μ allele is similar in this pro-B cell line.
To assess the chromatin state at the two Ig-μ alleles, we performed additional ChIP experiments using antibodies against specific posttranslational modifications of histone tails that are markers for either transcriptionally active or silent chromatin. ChIP analysis with an antibody against acetylated histone H3 (H3ac), a hallmark of actively transcribed euchromatin, showed no significant difference between the productive and the nonproductive Ig-μ allele (Fig. 4E). In fact, both alleles seemed to be highly enriched with acetylated histone H3. When the ChIP experiments were performed with an antibody against dimethylated histone H3 (H3K9me2), a marker for transcriptionally silent heterochromatin, the signal in the qPCR was almost undetectable for both alleles (data not shown), corroborating that both alleles are actively transcribed. This is in sharp contrast to our previous observations of NMTGS in HeLa cells, where the PTC+ Ig-μ minigene was associated with less H3ac but more H3K9me1, H3K9me2, and H3K9me3 compared to the PTC− Ig-μ minigene (Bühler et al. 2005; L. Stalder and O. Mühlemann, unpubl.).
DISCUSSION
In this study, transcriptional and post-transcriptional regulation of the productively and the nonproductively rearranged Ig-μ allele in the same pro-B cell line were analyzed and compared. In summary, we showed that the mRNA level of the nonproductive Ig-μ allele in a clone of the murine pro-B cell line NFS70 was silenced efficiently by a post-transcriptional mechanism. The sensitivity of this PTC-specific post-transcriptional response to the translation inhibitor CHX strongly suggests NMD to be responsible for this mRNA reduction. On the other hand, we found no evidence for transcriptional silencing of the PTC+ allele using a variety of different methods (pre-mRNA measurements, pol II ChIP, histone deacetylase inhibitors, and ChIP against histone modifications). Consequently, we conclude that transcriptional silencing, in particular NMTGS, is not involved in keeping nonproductively rearranged Ig-μ alleles silent in pro-B cells. Our finding of an entirely post-transcriptional down-regulation of PTC+ Ig-μ expression confirms earlier reports, where productive and nonproductive Ig-μ alleles were analyzed separately in different cells (Baumann et al. 1985; Jäck et al. 1989; Mühlemann et al. 2001). These results are further corroborated by recent RNA fluorescence in situ hybridization (FISH) data demonstrating transcription of both productively and nonproductively rearranged VDJ alleles, even after allelic exclusion of the nonproductive heavy chain locus (Daly et al. 2007). On the other hand, however, FISH experiments by Skok and colleagues showed that in primary splenic B cells, but not in transformed B cell lines, nonfunctional Ig alleles are recruited to centromeric heterochromatin after activation and exhibit differential expression patterns, suggesting that epigenetic factors and regulated nuclear localization contribute to maintain the monoallelic expression of Ig genes (Skok et al. 2001). Furthermore, signals from pre-BCR can also lead to rapid repositioning of one Ig-μ chain allele to repressive centromeric domains (Roldan et al. 2005).
Based on the NMTGS of Ig-μ and Ig-γ minigenes observed in HeLa and U2OS cells (Bühler et al. 2005), we hypothesized that this mechanism might contribute to ensure monoallelic IgH expression in B cells. However, the results reported here do not provide any evidence for this hypothesis, and the physiological role of NMTGS still remains elusive. While we cannot exclude that our hypothesis concerning the physiological role of NMTGS is simply wrong, it is also possible that transcriptional silencing of the nonproductive heavy chain allele is not yet established in pro-B cells, but occurs only at a later stage during B lymphocyte differentiation. To investigate this possibility, B lymphoid cells in different developmental stages, including late pre-B and mature B cells, should be analyzed.
MATERIALS AND METHODS
Cell culture
The pro-B line NFS70 (Davidson et al. 1984), purchased from ATCC (American type culture collection), was grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), supplemented with 10% heat-inactivated fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Amimed), 1 mM sodium pyruvate (Gibco), and 50 μM β-mercaptoethanol (Gibco) at 37°C in a humidified atmosphere containing 5% CO2.
Subcloning
Single-cell clones from the pro-B line NFS70 were established by the limiting dilution method. Cytoplasmic Ig-μ was detected by flow cytometry with affinity-purified FITC-conjugated goat antibodies against murine Ig-μ (Southern Biotechnology) as described before (Schuh et al. 2003). Data analyses were performed using a FACS-Calibur (BD Biosciences) and CellQuestTM software (v4.0.2).
Analysis of immunoprecipitated and metabolically labeled cell lysates
5 × 106 cells were washed twice in ice-cold PPS, resuspended in 2 mL methionin/cysteine-free minimal essential medium (MEM) medium (Bradl and Jäck 2001) and starved for 1 h at 37°C and 5% CO2. 3.7 × 106 Bq 35S-labeled methionin/cystein (MP Biochemicals) were added, and cells were incubated for 3 h at 37°C and 5% CO2. 35S-labeled cells were lysed for 30 min on ice with NET buffer (150 mM NaCl, 0.5% Nonidet P-40, 1 mM sodium vanadates, 50 mM NaF, 1 mM PMSF, 5 mM EDTA, 25 mM Tris-HCL at pH 7.5). Five micrograms of affinity-purified unconjugated goat antibodies against murine Ig-μ (Southern Biotechnology) and 30 μL equilibrated protein G-Sepharose (Pierce) were added to cell lysates. The mixtures were rotated for 2 h at RT. Sepharose was pelleted and washed three times in 1 mL PBS and once in 1 mL low salt buffer (50 mM Tris-HCl at pH 8.5). Immunoprecipitated proteins were eluted by boiling the pellets for 5 min in 50 μL SDS sample buffer (62.5 mM Tris-HCL at pH 6.8, 2% SDS, 50 mM DTT, 10% glycerol, 0.002% bromphenol blue) and analyzed by SDS-PAGE according to Laemmli (1970). Radioactive signals were detected in a PhosphoImager.
Isolation of genomic DNA, PCR, and sequencing
Genomic DNA was isolated using the “Wizard Genomic DNA Purification Kit” (Promega). PCR was performed with “PCR Core Kit” (Qiagen) using 200 ng DNA, 20 μM forward primer 5′-CAGTGCCATCAGACACCACAG-3′ and reverse primer 5′-GTGTGGAAAGCTGTGTATCCCC-3′ for analysis whether both alleles were rearranged (Wasserman et al. 1998). For analyzing which J segment is present, a degenerated forward primer 5′-AGGTSMARCTGCAGSAGTCWGG-3′ with four different reverse primers, 5′-GTCGTAGCAGAGTGTGGCAGATGGCC-3′, 5′-AGGTGTCCCTAGTCCTTCATGACCTG-3′, 5′-CCCAGACCCATGTCTCAACTTTGGGAC-3′, or 5′-CCCAACTTCTCTCAGCCGGCTCCCTCA-3′, was used (ten Boekel et al. 1997). The PCR-amplified fragments were cloned into pCRII-TOPO vector (Invitrogen) and sequenced with M13 primers.
Quantitative reverse transcription-coupled real-time PCR (RT-qPCR)
Total cellular RNA was isolated using “Absolutely RNA RT-PCR Miniprep Kit” (Stratagene), 1 μg RNA was reverse-transcribed, and RT-qPCR was performed as described by Eberle et al. (2008). The forward oligo and the TaqMan probe for the nonproductively rearranged Ig-μ allele were 5′-GGACAATTCCAAGAGCCAAGTTT-3′ and 5′-TTTAAAATGAACAGTCTGCAAGCTGATGACACA-3′; for the productively rearranged allele they were 5′-CAATGCCAAGAACACCCTGTA-3′ and 5′-TTGTATTACTGTGCAAGACTTTATTACCCAAGGG-3′. As a reverse primer, 5′-AAGGACTGACTCTCTGAGGAGACTGT-3′ was used for mRNA measurement, 5′-GAATAGAAGAGAGAGGTTGTAAGGACTCA-3′ for pre-mRNA measurements, and 5′-CCCCAACAAATGCAGTAAAATCTA-3′ for ChIP analysis. The amplification rate was determined for each assay by dilution series. To check allele specificity for each TaqMan assay, qPCR with plasmids containing the productively or nonproductively rearranged gDNA/pre-mRNA or mRNA sequences were performed. These plasmids were generated by introducing PCR fragments into pCRII-TOPO vector (Invitrogen) and confirmed by sequencing.
Drug treatment
50 μg/mL CHX (Merck) was added to the cells four hours before harvesting. Cells were incubated with 1 mM and 3 mM SB (Upstate) or 5 ng/mL and 25 ng/mL TSA (Sigma) for 24 h.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described by Bühler et al. (2005) with modifications in sonication conditions (pulse: 1 sec; pause: 5 sec, 35-fold with an intensity of 35%). Five micrograms of antibody against pol II (Santa Cruz, H-224X) or acetyl-histone H3 (Upstate, 17-245) was used for pulldown; 60–75 ng DNA was analyzed by qPCR.
ACKNOWLEDGMENTS
We thank Edith Roth for excellent technical assistance and all members of the Jäck laboratory for their fruitful discussions. The work was financed in part by the Interdisciplinary Center for Clinical Research (IZKF) and research grant FOR832 (JA968/4) from the Deutsche Forschungsgemeinschaft (DFG) to H.-M.J., and in part by research grants to O.M. from the Swiss National Science Foundation (SNSF), the European Research Council (ERC-StG), the Roche Research Foundation, and the Kanton Bern. The stipend of K.H. was supported by an intramural ELAN grant and the DFG training grant GK592. O.M. is supported by a fellowship from the Max Cloëtta Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1516409.
REFERENCES
- Alt F.W., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist S.E., Selg M., Raman C., Jack H.M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash M.J., Kohler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradl H., Jäck H.M. Surrogate light chain-mediated interaction of a soluble pre-B cell receptor with adherent cell lines. J. Immunol. 2001;167:6403–6411. doi: 10.4049/jimmunol.167.11.6403. [DOI] [PubMed] [Google Scholar]
- Bühler M., Paillusson A., Mühlemann O. Efficient downregulation of immunoglobulin μ mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 2004;32:3304–3315. doi: 10.1093/nar/gkh651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M., Mohn F., Stalder L., Mühlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol. Cell. 2005;18:307–317. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Daly J., Licence S., Nanou A., Morgan G., Martensson I.L. Transcription of productive and nonproductive VDJ-recombined alleles after IgH allelic exclusion. EMBO J. 2007;26:4273–4282. doi: 10.1038/sj.emboj.7601846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson W.F., Fredrickson T.N., Rudikoff E.K., Coffman R.L., Hartley J.W., Morse H.C., III A unique series of lymphomas related to the Ly-1+ lineage of B lymphocyte differentiation. J. Immunol. 1984;133:744–753. [PubMed] [Google Scholar]
- Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D., Unterholzner L., Ciccarelli F.D., Bork P., Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans . Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Jäck H.M., Berg J., Wabl M. Translation affects immunoglobulin mRNA stability. Eur. J. Immunol. 1989;19:843–847. doi: 10.1002/eji.1830190510. [DOI] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leeds P., Wood J.M., Lee B.S., Culbertson M.R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae . Mol. Cell. Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wilkinson M.F. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Alt F.W., Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Mühlemann O., Mock-Casagrande C.S., Wang J., Li S., Custodio N., Carmo-Fonseca M., Wilkinson M.F., Moore M.J. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell. 2001;8:33–43. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- Mühlemann O., Eberle A.B., Stalder L., Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim. Biophys. Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Roldan E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A. Locus “decontraction” and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh W., Meister S., Roth E., Jäck H.M. Cutting edge: Signaling and cell surface expression of a mu H chain in the absence of λ 5: A paradigm revisited. J. Immunol. 2003;171:3343–3347. doi: 10.4049/jimmunol.171.7.3343. [DOI] [PubMed] [Google Scholar]
- Skok J.A., Brown K.E., Azuara V., Caparros M.L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- Stalder L., Mühlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin micro genes requires translation of its mRNA. J. Biol. Chem. 2007;282:16079–16085. doi: 10.1074/jbc.M610595200. [DOI] [PubMed] [Google Scholar]
- ten Boekel E., Melchers F., Rolink A.G. Changes in the V(H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- Wasserman R., Zeng X.X., Hardy R.R. The evolution of B precursor leukemia in the Emu-ret mouse. Blood. 1998;92:273–282. [PubMed] [Google Scholar]