Abstract
Transfer RNA is highly modified. Nucleotide 37 of the anticodon loop is represented by various modified nucleotides. In Escherichia coli, the valine-specific tRNA (cmo5UAC) contains a unique modification, N6-methyladenosine, at position 37; however, the enzyme responsible for this modification is unknown. Here we demonstrate that the yfiC gene of E. coli encodes an enzyme responsible for the methylation of A37 in tRNA1Val. Inactivation of yfiC gene abolishes m6A formation in tRNA1Val, while expression of the yfiC gene from a plasmid restores the modification. Additionally, unmodified tRNA1Val can be methylated by recombinant YfiC protein in vitro. Although the methylation of m6A in tRNA1Val by YfiC has little influence on the cell growth under standard conditions, the yfiC gene confers a growth advantage under conditions of osmotic and oxidative stress.
Keywords: tRNA, modification, methylation, yfiC, N6-methyl adenosine
INTRODUCTION
Transfer RNA (tRNA) is extremely rich in modified nucleotides, constituting ∼10% of the entire tRNA sequence (Auffinger and Westhof 1998) . The role of many modified tRNA nucleotides has been studied; Position 37 is located adjacent to the anticodon triplet and is modified in 34 tRNA species in Escherichia coli, whereas unmodified adenosine is found at this position in only 13 tRNA species (Dunin-Horkawicz et al. 2006). The range of nucleotide 37 modifications include hypermodified t6A, m6t6A, i6A, and ms2i6A, as well as the simpler m2A, m1G, and m6A. Curiously, the latter m6A modification is found only in tRNA1Val (anticodon cmo5UAC) (Dunin-Horkawicz et al. 2006). During decoding, nucleotide 37 of the tRNA stacks over the 5′-codon 3′-anticodon base pair (Weixlbaumer et al. 2007). It has been suggested that some modifications at nucleotide 37 are necessary for stabilization of the anticodon loop structure (Stuart et al. 2003), binding of magnesium ions in the context of ribosomal decoding complex (Konevega et al. 2004), and/or maintenance of the reading frame (Urbonavicius et al. 2001, 2003).
The role of tRNA modifications in the cell can only be tested if the gene encoding the respective modification enzyme is identified and inactivated. However, this is no easy task since the proteins responsible for tRNA modifications are more numerous than the tRNA species themselves. Although the vast majority of genes involved in tRNA modification are known (Dunin-Horkawicz et al. 2006), several are still missing. Among them was the gene responsible for the m6A37 modification in tRNA1Val. In search of the “missing” methylation enzymes, we checked a set of eight open reading frames annotated as putative methyltransferases. Of these, the yfiC gene, which belongs to the COG4123 family, had already been suspected to encode a m6A37 methyltransferase in Mollicutes (de Crécy-Lagard et al. 2007). In this report, we demonstrate that the yfiC gene encodes the methyltransferase specific for tRNA1Val m6A37 formation.
RESULTS
YfiC is specific for modification of m6A37 in tRNA1Val
Recombinant protein YfiC, containing a His6 tag on its C-terminus, was expressed and purified from E. coli strain AG1 carrying the pCA24YfiC plasmid (Kitagawa et al. 2005). The resulting protein was pure according to the SDS-PAGE and was thus used to modify potential substrates. The substrates were prepared from the strain JW2559 lacking the yfiC gene on the chromosome (termed hereafter as ΔyfiC) (Baba et al. 2006). Cell lysate was fractionated into ribosome and S100 by ultracentrifugation. Both fractions were subsequently divided in half, and each of the ribosomal and S100 fractions were phenol deproteinized. Thus, four putative substrate groups were generated: Ribosomes, ribosomal RNA (rRNA), S100, and total small RNA. All substrate fractions were mixed with recombinant YfiC protein and radioactive S-adenosyl-L-[methyl-3H]methionine (3H-SAM). The reactions were then analyzed for tritium incorporation. The results showed that rRNA, either as a component of assembled ribosomal subunits or as pure RNA, could not be methylated by YfiC. In contrast, the total small RNA fraction, composed essentially of tRNA, was readily methylated as part of the S100 or in the deproteinized form.
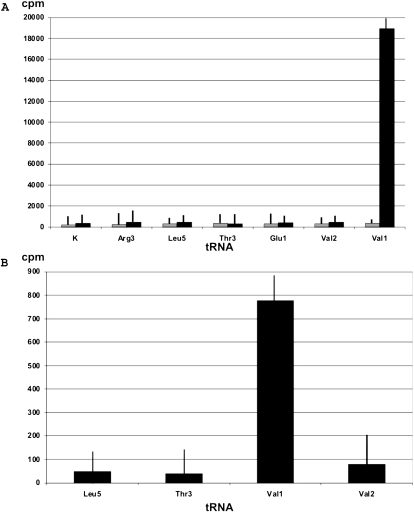
Very few tRNA methylation enzymes are left to be identified in E. coli, namely, for modification of Cm34 in tRNA5Leu, m6t6A37 in tRNA3,4Thr, and m6A in tRNA1Val. In order to determine if YfiC is responsible for one of those modification, we performed in vitro [3H]-methylation using recombinant YfiC protein of a total tRNA fraction purified from the ΔyfiC strain. Biotin-conjugated oligonucleotides, complementary to the divergent sequence regions in T- and acceptor stems of tRNA5Leu, tRNA3Thr, and tRNA1Val, were used to affinity purify the respective tRNA species. In parallel, the same method was used to prepare tRNA3Arg, tRNA1Glu, and tRNA2Val to serve as a control. As shown in Figure 1A, only the fraction purified using the oligonucleotide complementary to tRNA1Val was radiolabeled. This result indicates that the YfiC methyltransferase can methylate tRNA1Val, but none of other tRNAs tested.
FIGURE 1.
Methylation of tRNA species by recombinant YfiC protein in vitro. [3H]-SAM radioactivity introduced into various tRNA species by recombinant YfiC protein is indicated. (A) Methylation of tRNA species affinity purified from the cells. Gray bars correspond to methylation of tRNAs extracted from the wild-type strain, i.e., premethylated with nonradioactive SAM. Black bars correspond to methylation of tRNAs extracted from the yfiC knockout strain. K corresponds to mock tRNA purification with specific oligodeoxyribonucleotide omitted. (B) Methylation of in vitro transcribed tRNA species. Error bars indicate SD borders.
A similar result could also be obtained using in vitro transcribed tRNAs. Genes encoding tRNA5Leu, tRNA3Thr, tRNA1Val, and tRNA2Val were PCR-amplified from E. coli genomic DNA. The T7 promoter sequence was added to the 5′-end of each gene to allow in vitro transcription of the full-length tRNAs using T7 RNA polymerase. The transcripts were supplied as substrates for in vitro methylation using recombinant YfiC protein and 3H-SAM. Consistently with the in vivo purified tRNAs, methylation was only observed using the in vitro transcript of tRNA1Val (Fig. 1B).
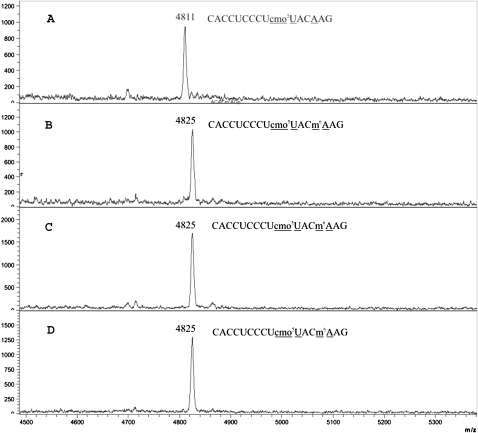
Since tRNA1Val contains only one methylated residue for which the respective enzyme has to be identified, it was logical to suggest that the function of YfiC is the formation of m6A37. To confirm this hypothesis, tRNA1Val from the wild-type and ΔyfiC strains was treated with RNase T1, and the products of the digestion were analyzed by MALDI-MS (Fig. 2A,B). The fragment of tRNA1Val carrying m6A37 from the wild-type strain (Fig. 2B) is 14 Da heavier than the equivalent fragment from the ΔyfiC strain (Fig. 2A), consistent with an additional methylation being present in this fragment. Indeed, the theoretical masses calculated for the methylated and unmethylated variants of the tRNA1Val RNase T1 fragments precisely match the masses observed by MALDI MS. Since A37 is the only methylated residue in this RNA fragment, YfiC is indeed responsible for the methylation of m6A37 in tRNA1Val.
FIGURE 2.
MALDI MS analysis of the T1 digest of the tRNA1Val. Heavy part of the spectrum is shown. MALDI spectrum corresponding to fragments of the tRNA1Val from the (A) ΔyfiC strain, (B) wild-type strain, (C) ΔyfiC strain transformed with pCA24YfiC, and (D) ΔyfiC strain treated with SAM and recombinant YfiC in vitro. Oligonucleotides corresponding to the peak are indicated.
YfiC is sufficient for methylation of m6A37 of tRNA1Val
To prove that the YfiC protein is directly responsible for modification of A37 in tRNA1Val, rather than a product of another gene that is affected by inactivation of yfiC gene, we performed in vivo complementation of the ΔyfiC strain with the pCA24YfiC plasmid. tRNA1Val was therefore isolated from the wild-type, ΔyfiC strain, as well as the ΔyfiC strain complemented with pCA24YfiC that expresses YfiC exogenously. Following affinity purification, tRNA1Val was digested by RNase T1, and the resulting fragments were analyzed by MALDI MS (Fig. 2C). As expected, methylation of A37 in the ΔyfiC strain was restored only when YfiC was expressed from the plasmid. Thus, only the absence of YfiC protein, rather than any changes in the genomic context resulting from the genetic knockout, is the cause of the lack of m6A37 formation. To demonstrate that YfiC alone is necessary and sufficient for the tRNA1Val A37 methylation, we used SAM and the recombinant YfiC protein to modify tRNA1Val purified from the ΔyfiC strain. The product of modification was treated with RNase T1 and subjected to MALDI MS analysis (Fig. 2D). The results show clearly that the YfiC protein alone is sufficient for tRNA1Val methylation at A37.
Methylation of m6A37 of tRNA1Val is dispensable for growth of E. coli cells under standard growth conditions
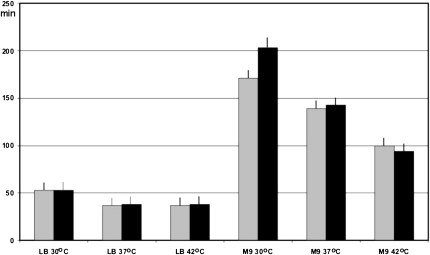
We next examined how the m6A37 modification of tRNA1Val by YfiC influences E. coli cell fitness under different growth conditions. E. coli doubling time was measured in rich LB and minimal M9 media at 30°C, 37°C, and 42°C (Fig. 3). Comparison of the growth of the wild-type and ΔyfiC strains in rich LB media revealed no significant difference at any of the different temperatures tested. In the minimal M9 media, very small growth retardation, resulting from yfiC inactivation was observable.
FIGURE 3.
Doubling time of the wild-type (gray bars) and ΔyfiC (black bars) strains. Growth conditions, such as temperature and medium, are indicated below the bars. Error bars indicate SD borders.
In addition, we checked the ability of the wild-type strain to compete for growth with ΔyfiC strain. Both rich LB and minimal M9 media were used for growth competition assays at 30°C, 37°C, and 42°C. Equal numbers of cells of the kanamycin-sensitive parental strain and kanamycin-resistant ΔyfiC strain (A600 0.001) were mixed and grown for 24 h. The growth cycles were repeated so that at the end of each cycle 1/1000th of the media containing the mixture of the cells was used to re-inoculate fresh media with 1000-fold dilution. The cells amplify 1000-fold with each growth cycle, which corresponds to ∼10 doublings. After each growth cycle, titers of the wild-type and ΔyfiC strains were measured. After 80 generations, no significant (more than one order of magnitude) drop in the ratio of ΔyfiC to wild-type cells was detected. We concluded that the modification of A37 in tRNA1Val by YfiC does not affect growth of E. coli cells under the standard growth conditions tested here.
Methylation of m6A37 of tRNA1Val influences survival of E. coli cells under conditions of osmotic and oxidative stress
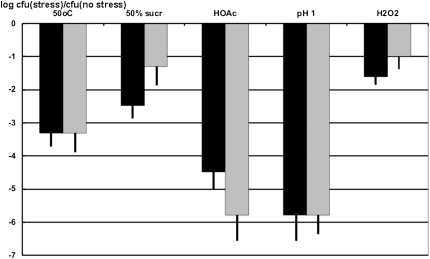
It is generally believed that if no readily detectable phenotype is observed for a knockout strain, the corresponding gene might be helpful for survival under stress conditions. Therefore, we tested if inactivation of yfiC gene would cause cell hypersensitivity to a number of different stress conditions, such as high temperature (50°C), hyperosmotic stress (50% sucrose) (Weber et al. 2006), acetic acid (50 mM at pH 4.5), phosphorus acid (100 mM at pH 1.5) (Blankenhorn et al. 1999, Kannan et al. 2008), and oxidative stress (5 mM hydrogen peroxide) (Almeida et al. 2006). Cells were kept under stress conditions for a period of 2 h, and then the titer of surviving cells was measured by serial dilutions and plating. The conditions were chosen to be at the border of E. coli cell survival, and therefore, as expected, all tested conditions led to a drop of several orders of magnitude in the wild-type cell titer (Fig. 4, gray boxes). Absence of yfiC gene causes hypersensitivity of E. coli to hyperosmotic and, to a lesser extent, oxidative stress (Fig. 4, black boxes). It could also be noted that the presence of modification at A37 decreases cell survival in the presence of acetic acid.
FIGURE 4.
Cell titer drop (cfu indicates colony forming units) in the logarithmic scale after exposure of the culture to various stress conditions. Stress type is indicated above the bars. Gray bars correspond to the wild-type strain; black bars correspond to the ΔyfiC strain. Error bars indicate SD borders.
Modifications in tRNA are known to increase stability of its structure (Stuart et al. 2003, Vendeix et al. 2008); however, no decrease in temperature sensitivity was detected with the ΔyfiC strain. E. coli cells counteract hyperosmotic stress by accumulation of extreme, up to 1 M, concentrations of potassium in the cytoplasm (Dinnbier et al. 1988). At such ionic conditions, subtle advantage of tRNA1Val modified at m6A37 could be essential. Addition of acetic acid to the media, in contrast to addition of phosphorus acid, leads to acidification of internal cytoplasm (Blankenhorn et al. 1999). It might be possible that presence of methyl group at A37 could pose some disadvantage at intracellular pH lower than neutral. Oxidative stress causes the majority of modification mutants to be compromised (P.V. Sergiev, unpubl.), among them the yfiC deficiency displays a modest phenotype. The reason for this could be more a stringent requirement for the efficiency of the protein synthesis apparatus, which should replace cellular proteins damaged by oxidation. Increase of the survival of bacteria under such harsh conditions could be one function of the tRNA modifications.
Proteome analysis of the yfiC knockout strain
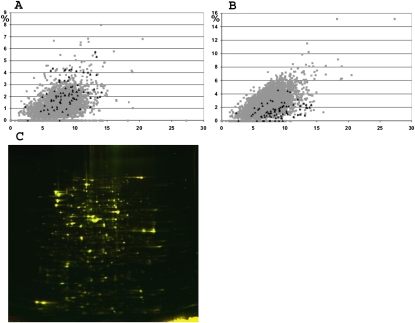
Valine codon distribution among variously expressed genes of E. coli was analyzed (Fig. 5A,B): While GUA codons are equally used in highly expressed genes and total proteins (Fig. 5A), GUG codons are underrepresented in highly expressed genes (Karlin et al. 1998, 2001) (Fig. 5B). Obviously, the GUA, but not the GUG, codon forms perfect Watson–Crick base-pairing with the cmo5UAC anticodon. To test the significance of A37 methylation for maintaining the correct balance of protein expression in the cell, we utilized a proteomic approach. Total proteins prepared from the ΔyfiC strain were compared with those from the parental wild-type strain by two-dimensional protein gel electrophoresis (Fig. 5C). However, no significant differences in protein expression profile were observed.
FIGURE 5.
Highly expressed gene codon bias for tRNA1Val GUA (A) and GUG (B) codons. Plotted is the particular codon frequency (in percent relative to total codons) against total number of valine codons (in percent relative to total codons). Gray boxes correspond to each of E. coli genes. Black triangles correspond to highly expressed genes (Karlin et al. 1998, 2001). (C) Two-dimensional protein gel, showing a comparison of protein composition of wild-type strain (labeled red) and ΔyfiC strain (labeled green). Yellow color corresponds to equal amount of protein in both strains.
DISCUSSION
tRNAs are particularly enriched in modified nucleotides (Auffinger and Westhof 1998). They play a role in stabilization of tRNA structure (Stuart et al. 2003, Vendeix et al. 2008) and enhancement of the correct codon–anticodon duplex formation (Konevega et al. 2004, Kothe and Rodnina 2007). Nucleotide 37 is located next to the codon–anticodon duplex, stacking over the base pair between the 5′-nucleotide of a codon and the 3′-nucleotide of anticodon (Weixlbaumer et al. 2007). This position in tRNA is represented exclusively by purines, in particular by modified purines. In E. coli, m1G is located at position 37 of tRNA2Arg and all species of tRNAPro, while an unknown derivative of guanosine is located at this position in tRNA1−3Leu. All other tRNAs contain either unmodified adenosine (tRNA1,2Ala, tRNA1−3Gly, tRNA4Ser, tRNA1,2Thr, tRNA2,3Val, and tRNAfMet) or modified adenosine residues. Methylation of the C2 atom of A37 is found for tRNA1Arg, tRNA1Asp, tRNA1,2Gln, tRNA1Glu, and tRNA1His. N6-threonylcarbamoyladenosine occupies position 37 of tRNA3,4Arg, tRNA1Asn, tRNA1−3Ile, tRNA1Lys, tRNA1Met, and tRNA3Ser, while N6-threonylcarbamoyl, N6-methyl adenosine is located in tRNA3Thr. N6-isopentenyl adenosine could be found exclusively in tRNASec, while the more complex 2-methylthio, N6-isopentenyl adenosine is located in tRNA1Cys, tRNA4,5Leu, tRNA1Phe, tRNA1,2Ser, tRNA1Trp, and tRNA1,2Tyr. The monomethylation of the N6 position is found exclusively at nucleotide 37 of tRNA1Val (cmo5UAC) (Dunin-Horkawicz et al. 2006).
tRNA1Val (cmo5UAC) belongs to a group of tRNA species carrying a uridine 5-oxyacetic acid residue at the 5′-position of the anticodon. These tRNA species are characteristic for reading four-codon families and can pair with all nucleotides at the wobble position of a codon (Nasvall et al. 2004; Kothe and Rodnina 2007). While additional tRNA species, specific for reading of pyrimidines at wobble position, are usually present, their function can be assumed by cmo5U-containing tRNAs. Evidence for in vivo and in vitro translation of the codons ending with pyrimidines by cmo5U-containing tRNAs was previously reported by the Björk (Nasvall et al. 2004), Agris et al. (2007) and Rodnina (Kothe and Rodnina 2007) laboratories. Although modification of U is dispensable for reading of 3′-adenine codons, it is required for reading codons ending with a pyrimidine (Nasvall et al. 2004). The structural basis for this phenomenon was studied by Ramakrishnan and co-workers (Weixlbaumer et al. 2007), revealing that the modification allows tautomerization of the uridine derivative to an enol form, enabling more efficient pairing with guanosine. The unusual geometry of cmo5U-U and cmo5U-C pairs is also facilitated by 5-oxyacetylation.
While modification of U in tRNA1Val (cmo5UAC) is important for decoding of some codons, modification of A37 is less so. However, it is known that like other modified bases at position 37, m6A stacks over the codon–anticodon duplex (Weixlbaumer et al. 2007), facilitating codon–anticodon interaction. Specifically, the methyl group causes distortion in the structure of the anticodon loop, preventing intraloop pairing (Vendeix et al. 2008). However, the exact function of the methylated adenosine at position 37 of tRNA1Val(cmo5UAC) in the cell remained unknown. It is also curious that this particular modification is specific for only one tRNA. To test the importance of this modification, it is necessary to identify the enzyme responsible for the modification.
In our study we have identified yfiC gene as encoding the methyltransferase responsible for A37 methylation of tRNA1Val. YfiC is a 27-kDa protein, classified as a putative methyltransferase. It contains the conserved N-methyltransferase motifs (Jeltsch 2002) 50DxGxGxG56 and 122NPPY125. The protein belongs to the COG4123 (Altschul et al. 1997), which has representatives among several phylum of bacteria, such as Aquificae, Thermotogae, Fusobacteria, Firmicutes, and Proteobacteria, exemplified by E. coli. YfiC has no orthologs in archaea and eukarya. Phylogenetic distribution of the yfiC gene is in agreement with the distribution of tRNAVal species, carrying m6A at position 37. The nonuniversal distribution of this particular modification goes in concert with nonessential nature of this modification in E. coli.
One possible function for the A37 methylation is that it is necessary for maintaining a balance in terms of the translational decoding efficiencies of GUA and GUG codons. Genes that have different expression levels (Karlin et al. 2001) also display a different preference for the codons (Karlin et al. 1998) recognized by tRNA1Val(cmo5UAC) (Fig. 5A,B), such that highly expressed genes are biased against GUG codons (Fig. 5B). Thus, balancing the intracellular protein levels might be dependent on fine-tuning the decoding efficiency. However, at least under the conditions tested here, we find no evidence for any difference in E. coli proteome (Fig. 5C) caused by loss of A37 methylation.
It became a common point to speculate that nonessential modifications in tRNA could increase tRNA stability and favor uniform codon–anticodon recognition, which could be essential at extreme conditions. We have determined how inactivation of the yfiC gene affects cell survival under conditions of heat, osmotic, acidic, and oxidative stress. It turned out that methylation of tRNAVal at A37 enhances cell survival during osmotic and oxidative stress. Increasing osmolarity should decrease the value of ionic interactions and increase the value of hydrophobic interactions. Thus, necessity for A37 methylation at high osmotic conditions could be explained by hydrophobic interactions of the methyl group that shields the codon–anticodon minihelix from the solution. The dependence of cell survival on m6A37 formation during oxidative stress is harder to explain. One possible explanation might be the necessity to increase the overall efficiency of protein synthesis, which would be necessary for replacement of damaged protein following oxidative stress. A role for which the nonessential tRNA1Val modification m6A37 plays in cell fitness under extreme conditions, must encourage a search for the functional value of other nonessential RNA modifications in processes of cell survival, adaptation, and regulation.
MATERIALS AND METHODS
Strains and plasmids
BW25141 parental strain (Datsenko and Wanner 2000) and JW2559 strain, carrying kanR cassette, inserted at the place of yfiC gene (Baba et al. 2006), as well as pCA24yfiC plasmid (Kitagawa et al. 2005) were kindly provided by Dr. H. Mori (Keio University).
Protein preparation
Recombinant N-terminal hexahistidine tagged YfiC protein was prepared from AG1 E. coli cells, harboring pCA24N plasmid with yfiC gene cloned under control of T5lac promoter (Kitagawa et al. 2005). Cells were grown in LB media at 37°C until A600 0.5 and induced by IPTG, 0.5 mM final concentration. After induction, cells were grown for an additional 3 h and lysed by sonication in a buffer of 20 mM Hepes K at pH 7.5, 1 mM Mg(OAc) 2, and 200 mM NH4OAc. Protein purification on Ni-NTA agarose (Qiagen) was done in the same buffer, containing 10 mM imidazole. After washing three times with the same buffer containing 30 mM imidazole, the protein was eluted by increasing imidazol concentration up to 200 mM. Eluted YfiC protein was dialyzed, and its purity was proven by SDS-PAGE.
Substrate preparation
To prepare YfiC putative substrates, wild-type cells and cells of JW2559 strain were grown to A600 0.8 in LB media at 37°C. Cells were harvested by centrifugation and lysed by grinding with 2× cell mass of aluminum oxide at 4°C. Cell debris and aluminum oxide was pelleted by 30-min centrifugation at 16000 rpm in JA20 rotor. Lysates were subjected to 18-h ultracentrifugation in Ti70 rotor at 35000 rpm to pellet ribosomes. Ribosomes were resuspended in 20 mM Hepes K (pH 7.5), 1 mM Mg(OAc)2, 200 mM NH4OAc, and 4 mM DTT buffer. Ribosomes and S100 were used to prepare rRNA and total tRNA by phenol extraction in buffer 300 mM NaOAc, 0.5% SDS, and 5 mM EDTA, followed by ethanol precipitation.
In vitro methylation
For in vitro methylation, 100 pmol of ribosomes, rRNA, or 2 A600 units of S100 or total tRNA were mixed with 2 mkCi S-adenosyl-L-[methyl-3H]methionine (GE Healthcare) and 10 pmol of recombinant YfiC protein in 200 μL volume. The ionic conditions for in vitro methylation were kept to 20 mM Hepes K (pH 7.5), 1 mM Mg(OAc) 2, 200 mM NH4OAc, and 4 mM DTT. After 30-min incubation at 37°C, reaction was quenched by equal volume of 300 mM NaOAc, 0.5% SDS, and 5 mM EDTA, and RNA was extracted by phenol. After ethanol precipitation, the pellet was washed five times vigorously with 70% ethanol to remove residual S-adenosyl-L-[methyl-3H]methionine. Radioactivity incorporated into RNA was measured by scintillation counting.
Bulk tRNA, prepared from the wild type, JW2559, JW2559 transformed with pCA24YfiC, and those modified with YfiC in vitro was used to affinity purify tRNA5Leu, tRNA3Thr, tRNA1Glu, tRNA3Arg, tRNA1Val, and tRNA2Val. Purification was done by a procedure, resembling one published by Miyauchi et al. (2007). Biotinylated oligonucleotides, complementary to the most divergent part of T and acceptor stems, were annealed to samples of total tRNA by heating to 75°C and slow cooling to 37°C. We used following primers:
BIO-GGGTGATGACGGGAT for tRNA1Val;
BIO-GCGTCCGAGTGGACT for tRNA2Val;
BIO-GCTGATAGGCAGATT for tRNA3Thr;
BIO-GCCGAAGGCCGGACT for tRNA5Leu;
BIO-GTCCCCTAGGGGATT for tRNA1Glu; and
BIO-GCGCCCTGCAGGATT for tRNA3Arg.
After annealing, hybridized tRNA species were separated by streptavidin resin and washed 10 times with the buffer of 20 mM Hepes K (pH 7.5), 1 mM Mg(OAc) 2, 200 mM NH4OAc, and 4 mM DTT. For scintillation counting, the resin was resuspended in scintillation cocktail without tRNA elution.
Genes coding for tRNA species were amplified from the total DNA of E. coli strain BW25141 using the following primers:
VAL1_5 TAATACGACTCACTATAGGGGTGATTAGCTCAGCTG;
VAL1_3 TmGmGTGGGTGATGACGGGA;
VAL2_5 TAATACGACTCACTATAGGCGTCCGTAGCTCAGTTGG;
VAL2_3 TmGmGTGCGTCCGAGTGGAC;
THR3_5 TAATACGACTCACTATAGGCTGATATAGCTCAGTTGG;
THR3_3 TmGmGTGCTGATAGGCAGATTC;
LEU5_5 TAATACGACTCACTATAGGCCGAAGTGGCGAAATCG; and
LEU5_3 TmGmGTGCCGAAGGCCGGAC.
PCR products were agarose gel-purified and used for in vitro transcription by T7 RNA polymerase. Transcription products were purified by electrophoresis in 10% polyacrylamide gel.
For MALDI MS analysis, tRNA was eluted by heating to 75°C in 6 M Urea, purified further by electrophoresis in denaturing 10% acrylamide gel, eluted from the gel, and dissolved in 50 mM ammonium citrate buffer. RNase T1 digestion and MALDI MS analysis were carried out as described for YbiN methyltransferase (Sergiev et al. 2008).
Examination of cell growth and survival characteristics
Growth rate measurements and growth competition experiments were carried as was previously described for YcbY (Lesnyak et al. 2006) and YgjO (Sergiev et al. 2006) methyltransferases. Survival of wild-type and JW2559 cells at stress conditions was checked by growing cell in LBK media (1% bacto tryptone, 0.5% yeast extract, 0.64% KCl) to A600 0.5 followed by application of a stress factor. A sample of the prestressed culture was used to determine initial cell titer. For heat-shock, the cultures were transferred to a 50°C water bath and incubated for 2 h. Hyperosmotic shock was introduced by addition of equal weight of sucrose. Acetic or phosphorus acid stress was made by addition of 1/10 a volume of 0.5 M KOAc-HOAc buffer (pH 4.5) or 1 M KH2PO4-H3PO4 buffer (pH 1.5). Oxidative stress was applied by addition of 1/10 of a volume of 50 mM hydrogen peroxide. In all cases, cells were incubated at a particular stress condition for 2 h in a cell culture shaker at 37°C. Following that, the survived cell titer was measured by serial dilutions and plating. A drop in cell titer was calculated, relative to cell titer before the stress.
Proteomics
Two-dimensional gel electrophoresis of total proteins was done as follows. Cell pellets were disrupted by sonication for 5 min at 4°C in ice water and centrifuged (10 min at 15,000g) to discard insoluble particles. Samples (10 μL) were dissolved in buffer: 8 M urea, 2 M thiourea, 4% CHAPS, and 30 mM TrisHCl (pH 8.5). Protein concentration was determined using Quick start Bradford dye reagent (Bio-Rad). In 50 μg of the sample, proteins was labeled with 400 pmol of either Cy3 or Cy5 CyDye DIGE Fluor minimal dyes (Amersham Biosciences) according to the manufacturer's instruction. Since protein equalization, Cy3 and Cy5 labeled samples were mixed, and 100 mM DTT (Bio-Rad) and 2% (v/v) Ampholine 3-10 (Bio-Rad) was added. IEF was performed using tube gels (20 cm × 1.5 mm) containing carrier ampholytes and applying a voltage gradient in an IEF-chamber produced in house (Dansenko and Wanner 2000). After IEF, the ejected tube gels were incubated in equilibration buffer (125 mM TrisHCl, 40% [w/v] glycerol, 3% [w/v] SDS, 65 mM DTT at pH 6.8) for 10 min. The tube gels were placed onto the polyacrylamide gels (9%–16%) of 1.5-mm thickness, 20 × 18 cm (Protean II Multi-Cell, Bio-Rad) and fixed using 1.0% (w/v) agarose containing 0.01% (w/v) bromphenol blue. The electrophoresis was carried out for 12–14 h, and then the gels were scanned using a Typhoon Trio Imager at 200-dpi resolution (Amersham Biosciences). Just afterward, the gels were fixed and silver stained according to the method described by Shevchenko et al. (1996). Image analysis was performed using PDQest 8.0 software (Bio-Rad). Two independent experiments were performed for each experimental setup. Spot quantities were normalized to remove nonexpression-related variations in spot intensity. The criterion of differential expression of a certain protein between the two subsets was posed as at least a twofold change in spot optical density between the two matched sets in duplicates.
ACKNOWLEDGMENTS
We thank Dr. H. Mori for providing us with the knockout strain JW2559 and a plasmid, encoding yfiC. We thank A. Bogdanov for fruitful discussions and wise advice. We thank Dr. D. Wilson for valuable comments and his help in translation of the manuscript to English. This work was supported by grants from the DFG-RFBR program, International Research Training Groups (no. 08-04-91972), Human Frontiers Science Program RGY0088/2008, Howard Hughes Medical Institute 55005605, Russian Foundation for Basic Research 07-04-00457, Leading Scientific Schools, and the DFG-RFBR program, International Research Training Groups (grants GRK 1384, 08-04-91972).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1494409.
REFERENCES
- Agris P.F., Vendeix F.A., Graham W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Almeida C.C., Romão C.V., Lindley P.F., Teixeira M., Saraiva L.M. The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 2006;281:32445–32450. doi: 10.1074/jbc.M605888200. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P., Westhof E. Location and distribution of modified nucleotides in tRNA. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington: 1998. pp. 569–576. [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants: The Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn D., Phillips J., Slonczewski J.L. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 1999;18:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-Lagard V., Marck C., Brochier-Armanet C., Grosjean H. Comparative RNomics and modomics in Mollicutes: Prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- Dinnbier U., Limpinsel E., Schmid R., Bakker E.P. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 1988;150:348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- Dunin-Horkawicz S., Czerwoniec A., Gajda M.J., Feder M., Grosjean H., Bujnicki J.M. MODOMICS: A database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Kannan G., Wilks J.C., Fitzgerald D.M., Jones B.D., Bondurant S.S., Slonczewski J.L. Rapid acid treatment of Escherichia coli: Transcriptomic response and recovery. BMC Microbiol. 2008;8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Mrázek J., Campbell A.M. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 1998;29:1341–1355. doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- Karlin S., Mrázek J., Campbell A.M., Kaiser D. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 2001;183:5025–5040. doi: 10.1128/JB.183.17.5025-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Konevega A.L., Soboleva N.G., Makhno V.I., Semenkov Y.P., Wintermeyer W., Rodnina M.V., Katunin V.I. Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U., Rodnina M.V. Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell. 2007;25:167–174. doi: 10.1016/j.molcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Lesnyak D.V., Sergiev P.V., Bogdanov A.A., Dontsova O.A. Identification of Escherichia coli m(2)G methyltransferases: I. The ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol. 2006;364:20–25. doi: 10.1016/j.jmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Miyauchi K., Ohara T., Suzuki T. Automated parallel isolation of multiple species of noncoding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasvall S.J., Chen P., Björk G.R. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev P.V., Lesnyak D.V., Bogdanov A.A., Dontsova O.A. Identification of Escherichia coli m(2)G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J. Mol. Biol. 2006;364:26–31. doi: 10.1016/j.jmb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Sergiev P.V., Serebryakova M.V., Bogdanov A.A., Dontsova O.A. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 2008;375:291–300. doi: 10.1016/j.jmb.2007.10.051. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stuart J.W., Koshlap K.M., Guenther R., Agris P.F. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J. Mol. Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J., Qian Q., Durand J.M., Hagervall T.G., Björk G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Stahl G., Durand J.M., Ben Salem S.N., Qian Q., Farabaugh P.J., Björk G.R. Transfer RNA modifications that alter +1 frameshifting in general fail to affect-1 frameshifting. RNA. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix F.A., Dziergowska A., Gustilo E.M., Graham W.D., Sproat B., Malkiewicz A., Agris P.F. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry. 2008;47:6117–6129. doi: 10.1021/bi702356j. [DOI] [PubMed] [Google Scholar]
- Weber A., Kögl S.A., Jung K. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 2006;188:7165–7175. doi: 10.1128/JB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A., Murphy F.V., 4th, Dziergowska A., Malkiewicz A., Vendeix F.A., Agris P.F., Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]