Abstract
CISD2, the causative gene for Wolfram syndrome 2 (WFS2), is a previously uncharacterized novel gene. Significantly, the CISD2 gene is located on human chromosome 4q, where a genetic component for longevity maps. Here we show for the first time that CISD2 is involved in mammalian life-span control. Cisd2 deficiency in mice causes mitochondrial breakdown and dysfunction accompanied by autophagic cell death, and these events precede the two earliest manifestations of nerve and muscle degeneration; together, they lead to a panel of phenotypic features suggestive of premature aging. Our study also reveals that Cisd2 is primarily localized in the mitochondria and that mitochondrial degeneration appears to have a direct phenotypic consequence that triggers the accelerated aging process in Cisd2 knockout mice; furthermore, mitochondrial degeneration exacerbates with age, and the autophagy increases in parallel to the development of the premature aging phenotype. Additionally, our Cisd2 knockout mouse work provides strong evidence supporting an earlier clinical hypothesis that WFS is in part a mitochondria-mediated disorder; specifically, we propose that mutation of CISD2 causes the mitochondria-mediated disorder WFS2 in humans. Thus, this mutant mouse provides an animal model for mechanistic investigation of Cisd2 protein function and help with a pathophysiological understanding of WFS2.
Keywords: Cisd2, Wolfram syndrome 2, autophagy, knockout mice, mitochondria, premature aging
CISD2 is the second member of the gene family containing the CDGSH iron sulfur domain. There are currently three members in this gene family: CISD1 (synonyms ZCD1, mitoNEET), CISD2 (synonyms ZCD2, Noxp70, and Miner1) and CISD3 (synonym Miner2). CISD1 is an outer mitochondrial membrane protein that was originally identified as a target protein of the insulin sensitizer drug pioglitazone used to treat type 2 diabetes (Colca et al. 2004). CISD1 protein contains a transmembrane domain, a CDGSH domain, and a conserved amino acid sequence for iron binding; biochemical experiments suggest that CISD1 is involved in the control of respiratory rates and regulates oxidative capacity (Wiley et al. 2007). However, CISD2 and CISD3 are novel genes with previously uncharacterized functions. The only molecular documentation for CISD2 is that CISD2 was one of the markers for early neuronal differentiation in a cell culture study (Boucquey et al. 2006).
Recently, the CISD2 gene has been identified as the second causative gene (Amr et al. 2007) associated with Wolfram syndrome (WFS; MIM 222300), which is an autosomal recessive neurodegenerative disorder. WFS is highly variable in its clinical manifestations, which include diabetes insipidus, diabetes mellitus, optic atrophy, and deafness; thus, it is also known as the “DIDMOAD syndrome” (Barrett and Bundey 1997). Positional cloning and mutation studies have revealed that WFS is a genetically heterogeneous disease with a complex molecular basis involving more than one causative gene in humans (Domenech et al. 2006). A portion of WFS patients belonging to the WFS1 group (MIM 606201) carried loss-of-function mutations in the WFS1 (wolframin) gene, which encodes a transmembrane protein primarily localized in the endoplasmic reticulum (ER) (Inoue et al. 1998; Strom et al.1998; Takeda et al. 2001). In addition to this, a homozygous mutation of the CISD2 gene has been identified in three consanguineous families with WFS (Amr et al. 2007), and these patients have been classified as WFS2 (MIM 604928). However, the function of the CISD2 protein in these patients and in all other organisms remains unknown, and its physiological role has not been explored.
Significantly, the CISD2 gene is located within the region on human chromosome 4q where a genetic component for human longevity has been mapped. Previously, Puca et al. (2001) studied 137 sets of extremely old siblings (308 individuals in all) and conducted a genome-wide scan search for predisposing loci that might confer longevity; this linkage study revealed a single region on chromosome 4q and suggests that there may be at least one master gene contributing to life-span control; however, the responsible gene has not been identified.
In this study, we apply a mouse genetics approach and demonstrate that Cisd2 is involved in mammalian life-span control and plays an essential role in mitochondrial integrity. Cisd2 deficiency causes mitochondria-mediated phenotypic defects in mice. Furthermore, cell culture and biochemical investigations revealed that Cisd2 is a mitochondrial protein. Additionally, Cisd2 knockout mice exhibit many clinical manifestations of WFS patients including early-onset degeneration of central (e.g., optic) and peripheral (e.g., sciatic) nerves and premature death, as well as impaired glucose tolerance. This study therefore provides an animal model for mechanistic understanding of WFS, specifically WFS2, pathogenesis.
Results
Shortened life span in Cisd2−/− mice
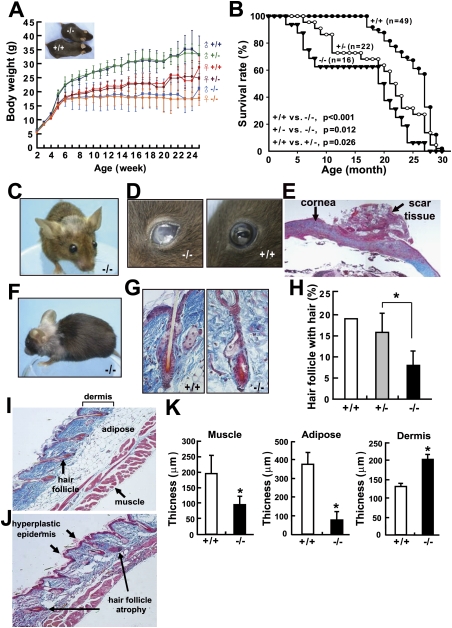
CISD2 is an evolutionarily conserved gene localized on human chromosome 4q24 (Supplemental Table 1; Supplemental Figs. 1, 2); the mouse syntenic region is on chromosome 3G3. Northern blot analysis showed that Cisd2 is a widely expressed gene in mice. Interestingly, quantitative real-time RT–PCR revealed that expression levels of Cisd2 decrease in an age-dependent manner in naturally aged mice (Supplemental Fig. 3). To study the role of Cisd2 involvement in development and pathophysiology, we generated Cisd2 knockout mice. Southern and Northern blot analyses demonstrated that the Cisd2 gene was disrupted and that there was undetectable mRNA expression in the homozygous knockout (Cisd2−/−) mice (Supplemental Fig. 4). Growth retardation and a smaller somatotype are clearly evident; it appears that there is almost no growth after 5 wk old in the Cisd2−/− mice (Fig. 1A). Early senescence is accompanied by a shortened life span when survival of the various genotypes is examined and there appears to be signs of haploinsufficiency for Cisd2 in view of the slightly lower survival rate for the heterozygous (Cisd2+/–) mice (Fig. 1B).
Figure 1.
The decreased body weight, the shortened life span, and the ocular and cutaneous symptoms of aging in Cisd2−/− mice. (A) Growth curves of the different genotypes. (B) Decreased survival rate of the Cisd2−/− mice. (C) The prominent eyes and protruding ears of the Cisd2−/− mice. (D) The Cisd2−/− mice go blind at ~6 mo old. (E) The opacity of cornea was analyzed by histological examination. Masson's trichrome staining indicated debris deposition in the scar tissue outside the cornea. (F) Early depigmentation and gray hair are present on top of the head and on the shoulders. The representative photo was taken from a 12-mo-old Cisd2−/− female. (G) Hair follicle atrophy in the Cisd2−/− mice was demonstrated by Masson's trichrome staining. (H) A decreased density of hair follicles containing hair in the Cisd2−/− mice was detected compared with wild-type skin. (I,J) Histological analyses of the skin from 12-mo-old wild-type and Cisd2−/− mice, respectively. The Cisd2−/− skin exhibits a phenotype involving a hyperplastic epidermis, hair follicle atrophy, a decrease in subcutaneous fat and muscle, and an increased thickness of the dermis layer. (K) Quantification of the subcutaneous muscle tissue, adipose tissue, and dermis for the histological sections of the wild-type and Cisd2−/− skins. (*) P < 0.05 was considered statistically significant.
Premature aging phenotype
Starting at 8 wk old, Cisd2−/− mice begin to acquire a set of aged appearance phenomena remarkably similar to those of premature aging syndrome (Hasty et al. 2003; Kipling et al. 2004). These include prominent eyes and protruding ears (Fig. 1C). Ocular abnormalities were observed as the Cisd2−/− mice developed opaque eyes and blindness, which was accompanied by cornea damage at 20 wk old (Fig. 1D). Histopathological examination revealed that the opacity of the cornea was due to debris deposition in the scar tissue outside the cornea (Fig. 1E). In addition, corneal neovascularization was observed in the Cisd2−/− mice; this can impair vision and is usually associated with pathogenesis due to eye trauma or the presence of a degenerative disorder (Supplemental Fig. 5). There was also early depigmentation in the fur at ~48 wk old (Fig. 1F; Supplemental Fig. 6); furthermore, hair follicle atrophy and a decreased hair density could be detected in Cisd2−/− mice (Fig. 1G,H). A decrease in the hair regrowth rate was also observed in the Cisd2−/− mice (Supplemental Fig. 7A,B). Additionally, the skin of 48-wk-old Cisd2−/− mice exhibits a phenotype with a noticeably thickened dermis, an expanded surface, and a significant decrease in subcutaneous adipose tissue and muscle (Fig. 1I–K).
Microcomputer tomography (micro-CT) imaging showed that the trabeculae of the femur are noticeably thinner in Cisd2−/− mice (Fig. 2A). Dual energy X-ray absorpitometer (DEXA) detected a decrease in femur density after 8 wk old; interestingly, the decrease of femur density also started to emerge in heterozygous Cisd2+/− mice, but at 24 wk old, while a progressively more severe phenotype was observed at the same age with Cisd2−/− mice (Fig. 2B). This shows, in addition to what was observed in terms of life-span evaluation, that there is also an apparent Cisd2 haploinsufficiency with respect to femur density. The results from the gross anatomy viewpoint, from the X-ray radiography, and using micro-CT reveal a significant lordokyphosis phenotype after 12 wk old (Fig. 2C,D; Supplemental Fig. 7C,D); consequently, this seems to lead to a decrease in mean thoracic volume (Fig. 2E) and thence pulmonary function abnormalities. Indeed, we observed decreases in various respiratory parameters as measured by plethysmography after 20 wk old in the Cisd2−/− mice (Supplemental Fig. 8). Muscle degeneration was detectable at 3 wk old in the Cisd2−/− mice. There was a progressive degeneration of muscle fibers and the magnitude of the degeneration exacerbated with age (Fig. 2F–J); muscle degeneration was further confirmed by transmission electron microscopy (TEM) (Supplemental Fig. 9). In addition, angular fibers, which are an indicator of muscle atrophy caused by neuron degeneration, could be observed in the Cisd2−/− mice (Fig. 2H).
Figure 2.
Abnormalities of skeleton and muscle and decrease of thoracic volume in the Cisd2−/− mice. (A) Micro-CT imaging of the femur trabecula at 12 wk old. Thinner trabecular thickness was observed in the Cisd2−/− mouse. (B) Osteopenia was analyzed by DEXA. A decrease in femur density was detected with the 8-wk-old Cisd2−/− mice. At 24 wk old, a decrease in femur density also was detected with the Cisd2+/− mice and, at 24 wk old, the phenotype had become more severe in the Cisd2−/− mice. (C) Radiographs of a wild-type mouse and a Cisd2−/− mouse at 16 wk old. The Cisd2−/− mouse displays a lordokyphosis (curvature of the spinal column) phenotype. (D) Micro-CT scanning allowed three-dimensional reconstruction of the thoracic and spinal column. (E) Lordokyphosis was evident and had led to a decrease in thoracic volume in the Cisd2−/− mice compared with their wild-type littermates. (F,G) H&E staining of transverse sections of skeletal muscle of 4-wk-old and 28-mo-old wild-type mice. (H,I) Muscle degeneration of 4-wk-old and 8-wk-old Cisd2−/− mice was examined by H&E staining of transverse sections of the skeletal muscle. Black arrows indicate degenerated transverse fibers that are present in the Cisd2−/− and also in spontaneously aged mice. The blue arrow indicates an angular fiber, which is an indicator of muscle atrophy caused by neuron degeneration. (J) Quantification of the degenerating fibers in the skeletal muscles. (*) P < 0.05; (**) P < 0.005.
One possible mechanism for the accelerated aging phenotypes is a defect in cellular proliferation in the Cisd2−/− mice. To test this possibility, we created several primary mouse embryonic fibroblast (MEF) cell lines from individual embryos with different genotypes. Our results revealed no significant difference in the doubling time and MEF cell growth (Supplemental Fig. 10), suggesting that accelerated aging in the Cisd2−/− mice is not due to an intrinsic defect in cellular proliferation.
A summary of the aging-related phenotypes in Cisd2−/− mice is provided in Supplemental Table 2. These mutant mice exhibit a premature aging phenotype with 100% penetrance for both sexes using either a C57BL/6 (B6) or a 129Sv/B6 mixed background.
Mitochondrial degeneration and autophagy
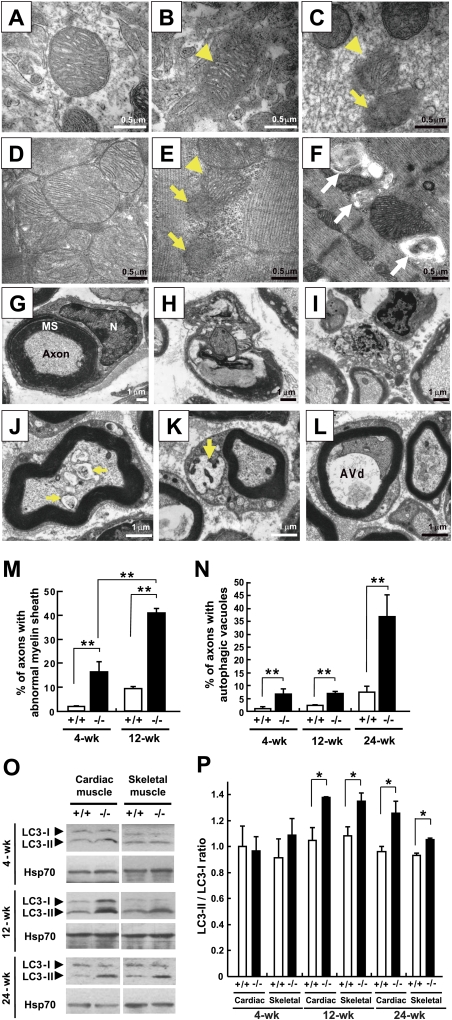
The observation of premature aging phenotypes involving muscle degeneration prompted a detailed examination of the tissue ultrastructure of the homozygous knockout mice. A TEM study revealed that mitochondrial degeneration occurs in the axons of sciatic nerves, brain cells (Fig. 3A–C), cardiac muscle cells, and skeletal muscle cells (Fig. 3D–F) in the Cisd2−/− mice. Notably, the mitochondrial outer membrane (OM) appeared to have broken down prior to the destruction of the inner cristae (Fig. 3B,E). In wild-type mice, the myelinated axons are enveloped with a myelin sheath formed by the fusion of many layers of plasma membrane from Schwann cells (Fig. 3G). However, considerable disintegration of the myelin sheath and degeneration of axon was detected in the Cisd2−/− sciatic nerves (Fig. 3H,I). Importantly, these mitochondrial abnormalities, involving destruction of mitochondria, myelin sheath disintegration, and axonal lesions, are already present to a certain extent in 2-wk-old Cisd2−/− mice (Fig. 3J–L; Supplemental Fig. 11), a stage prior to the first premature aging phenotype of muscle and nerve degeneration in these mice. Interestingly, the damaged mitochondria appear to induce autophagy to eliminate the dysfunctional organelles (Kim et al. 2007) because we identified morphologically distinct autophagic vacuoles (Eskelinen 2008) in muscle, sciatic nerve, optic nerve, and brain tissue (Fig. 3J–L; Supplemental Fig. 12). The general term autophagic vacuole refers to an autophagosome, amphisome, or autolysosome. Morphologically, autophagic vacuoles can be classified into two categories: (1) early or initial autophagic vacuoles (AVis)—i.e., autophagosomes, which are double-membraned structures containing undigested cytoplasmic material or organelles; (2) late or degradative autophagic vacuoles (AVds), including amphisomes and autolysosomes, which contain partially degraded cytoplasmic material (Eskelinen 2008; Fader and Colombo 2009). Remarkably, mitochondrial degeneration exacerbates with age, and the magnitude of the autophagy increases in parallel to the development of premature aging phenotype (Fig. 3M,N). We measured the thickness of myelin sheaths and counted the numbers of myelinated axons in sciatic nerve; our results revealed no significant differences between different genotypes (Supplemental Fig. 13), indicating that these two factors are not involved in the nerve degeneration of Cisd2−/− mice. We also examined the autophagosome marker LC3-II (Kabeya et al. 2000) in skeletal and cardiac muscles, which are the most sensitive tissues to in vivo autophagic degradation (Mizushima et al. 2004); indeed, the ratio of LC3-II/LC3-I was significantly higher in Cisd2−/− mice than in their wild-type littermates. This biochemical evidence confirms the TEM results and provides a quantitative basis for the autophagy induction (Fig. 3O,P).
Figure 3.
Mitochondrial degeneration and autophagy induction in the muscles and neurons of the Cisd2−/− mice. (A) Wild-type mitochondria in the brain (hippocampus). (B) A Cisd2−/− mitochondrion in the brain (hippocampus). Note that the outer mitochondrial membrane has broken down (arrowhead), while the inner cristae appear to be intact. (C) Cisd2−/− mitochondria in sciatic nerve. One mitochondrion (arrowhead) has a destroyed OM, but with cristae still visible; the other mitochondrion (arrow) has destroyed OMs and IMs. (D) Wild-type mitochondria in cardiac muscle. (E) Cisd2−/− mitochondria in cardiac muscle. This micrograph shows one mitochondrion (arrowhead) with a destroyed OM and two degenerated mitochondria consisting of debris (arrows). (F) A cluster of autophagic vacuoles and abnormal mitochondria was observed between the myofibrils of Cisd2−/− skeletal muscle (white arrows). (G) A wild-type myelinated axon of the sciatic nerve. (N) Nucleus of Schwann cell; (MS) myelin sheath. (H) A myelinated axon of sciatic nerve from a Cisd2−/− mouse. An ovoid with a disintegrating myelin sheath and a degenerating axonal component are shown. (I) Debris from an axon undergoing degeneration in the Cisd2−/− sciatic nerve. (J–L) Early or AVis enclosing mitochondria (arrows) and late or AVds were detected in the axonal component and cytoplasm of a Schwann cell from a 2-wk-old Cisd2−/− sciatic nerve. (M,N) Percentage of myelinated axons present in the sciatic nerves showing disintegration of their myelin sheaths and autophagic vacuoles, including AVi and AVd, in their axonal component. There were three mice for each group. (O) Western blotting to detected the presence of the proteins LC3-I and LC3-II. (P) Ratios of the LC3-II to LC3-I. There were three mice for each group. (*) P < 0.05; (**) P < 0.005. Mouse age in A–I is 4 wk old.
Autophagy can lead to cell death by directly activating autophagic (type II programmed) cell death that produces self-degradation of the dying cells (Shimizu et al. 2004; Mizushima et al. 2008); alternatively, autophagy might cause cell death through activation of apoptosis (Scott et al. 2007). To determine whether there is an increased level of apoptosis in the Cisd2−/− mice, we performed TUNEL assays on various mouse tissues to detect apoptotic cells in situ and found no evidence of increased apoptosis in Cisd2−/− mice (Supplemental Fig. 14). In addition, it has been reported that starvation can induce muscle autophagy (Mizushima et al. 2004). To test this possibility, we measured the metabolic indices including intake of food and water and generation of urine and stool. Our results revealed no significant difference in these metabolic indices between Cisd2−/− and wild-type mice at 6 wk old (Supplemental Fig. 15A); this is 4 wk after the detection of autophagic activation at 2 wk old. This excludes starvation/malnutrition as the cause of autophagic induction in Cisd2−/− mice. A decrease in the metabolic index becomes evident after 12 wk old (Supplemental Fig. 15B), and this is likely to be a consequence of the aging phenotype.
Cisd2 is probably a mitochondrial OM protein
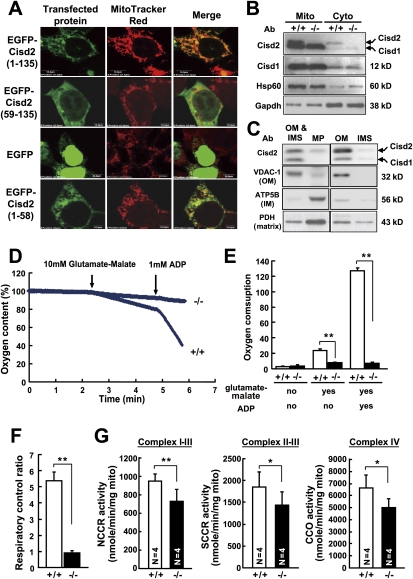
The annotated characteristics of Cisd2 protein are very similar to Cisd1, which is an outer mitochondrial membrane protein (Supplemental Fig. 16A; Wiley et al. 2007). To address the subcellular localization, we expressed the EGFP-tagged Cisd2 protein in NIH3T3 cells. Our result indicated that Cisd2 was colocalized with the mitochondrial marker (Supplemental Fig. 16B). However, deletion of the N-terminal 58 amino acids completely abolished the mitochondrial localization; furthermore, when the N-terminal 58 amino acids were fused to EGFP, this construct was able to redirect EGFP from a nuclear and cytoplasmic localization to the mitochondria (Fig. 4A), suggesting that Cisd2 is a nucleus-encoded mitochondrial protein and its N-terminal 58 amino acids are both necessary and sufficient to direct mitochondrial localization. To confirm the subcellular localization of the Cisd2 protein, the cytosolic and mitochondrial fractions were prepared from skeletal muscle of wild-type mice. Antibodies against Cisd1 and Cisd2 were generated. Western blot analysis revealed that Cisd2 protein, like the mitochondrial proteins Cisd1 and Hsp60 (Samali et al. 1999; Colca et al. 2004; Wiley et al. 2007), is primarily localized in the mitochondrial fraction (Fig. 4B). To further define the submitochondrial localization of Cisd2, we separated mouse liver mitochondria into the following fractions: OM, mitoplasts (MP, inner membrane [IM] and matrix), and intermembrane space (IMS, soluble material between the IM and OM). Immunoblotting each fraction with antibodies against Cisd2 and known markers revealed that Cisd2 was highly enriched in the OM fraction, as was the OM marker VDAC-1; this result strongly suggests that Cisd2 is a mitochondrial OM protein (Fig. 4C).
Figure 4.
Cisd2 is primarily localized in the outer mitochondrial membrane, and Cisd2 deficiency leads to mitochondrial dysfunction. (A) EGFP-tagged Cisd2 protein is directed to the mitochondria by an N-terminal signal sequence. The EGFP-Cisd2 proteins were expressed in NIH3T3 cells. EGFP-tagged full-length Cisd2 protein was colocalized with MitoTracker Red, whereas deletion of the N-terminal 58 amino acids completely abolished mitochondria localization. When the N-terminal 58-amino-acid sequence was fused to EGFP, this construct was able to redirect EGFP from a nuclear and cytoplasmic localization to the mitochondria. (B) Subcellular localization of the Cisd2 and Cisd1 proteins analyzed by Western blotting using protein extracts of the mitochondrial (Mito) and cytosolic (Cyto) fractions prepared from skeletal muscles of 12-wk-old mice. Polyclonal antibody (Ab) against Cisd2 protein (15 kDa) was generated; this antibody cross-reacts with Cisd1 protein (12 kDa). Antibodies against mitochondrial proteins Cisd1 and Hsp60 were used as controls. (C) Ten micrograms of each submitochondrial fraction prepared from the livers of 4-wk-old mice were analyzed by Western blot using antibodies against Cisd2 and known mitochondrial marker proteins. OM marker: (VDAC-1) voltage-dependent anion channel-1; IM marker: (ATP5B) complex V β subunit; matrix marker: (PDH) pyruvate dehydrogenase. (MP) Microplast (IM and matrix); (IMS) intermembrane space. (D) Impaired mitochondrial respiration in the skeletal muscle of 4-wk-old Cisd2−/− mice. Representative oxygraphs of the mitochondria after adding first glutamate-malate and then ADP into the closed chamber of the oxygen meter. (E) Respiratory activity was expressed as oxygen consumption rate (nanomoles of O2 per minute per milligram of mitochondria) in the resting state, for glutamate-malate supported respiration, and for ADP activated respiration. A significant decrease in oxygen consumption was detected in the Cisd2−/− mitochondrial samples (n = 4) compared with wild-type samples (n = 3). (F) The RCR (O2 consumption rate after ADP addition/O2 consumption rate after glutamate-malate addition) was significantly lower in the Cisd2−/− mitochondria. (G) Comparison of electron transport activities of the respiratory enzyme complexes of mitochondria prepared from the skeletal muscles of 4-wk-old Cisd2−/− (n = 4) and wild-type mice (n = 4). (NCCR activity) Measurement of NCCR activity, which represents complexes I–III; (SCCR activity) measurement of SCCR activity, which represents complexes II and III; (CCO activity) cCCO activity, which represents complex IV. (*) P < 0.05; (**) P < 0.005.
Previously, Amr et al. (2007) reported that the Flag-tagged CISD2 protein colocalized with the ER marker calnexin in the transfected mouse P19 and human HEK293 cells. We sought to determine if there is a small portion of the Cisd2 protein sorted into the ER/sarcoplasmic reticulum (SR) using subcellular fractions prepared from skeletal muscles of wild-type mice. Our data indeed revealed a weak signal indicating the presence of Cisd2 protein in the post-mitochondrial supernatant, and this colocalized with the ER markers in the microsomal fractions. The ratio of the Cisd2 protein present in the mitochondria versus ER was estimated to be about 5.8:1 (Supplemental Fig. 17).
Mitochondria are the cellular energy factories that generate ATP via oxidative phosphorylation. To investigate whether the mitochondrial degeneration detected in this study has a direct functional consequence leading to a respiratory dysfunction, we assessed mitochondrial aerobic respiration using isolated mitochondria prepared from skeletal muscle. This was done by measuring the oxygen consumption after stimulating the mitochondria with glutamate-malate and ADP to activate the respiratory chain reactions. Our results revealed a significant decrease in the oxygen consumption and the respiratory control ratio (RCR) in the Cisd2−/− mitochondria (Fig. 4D–F). To further expand this investigation, we explored the iron-sulfur proteins, which are essential electron carriers in the mitochondrial respiratory chain; there are up to 12 different iron-sulfur clusters that shuttle electrons through complex I–III (Rouault and Tong 2008). We measured the activities of the various iron-sulfur proteins of complex I–III (NADH cytochrome c reductase, NCCR) and complex II–III (succinate cytochrome c reductase, SCCR). In addition, we also measured the activity of complex IV (cytochrome c oxidase, CCO), which contains hemes and copper centers for electron transport (Rouault and Tong 2008). Our results showed that there was an average 30% decrease in the electron transport activities of complex I–III, complex II–III, and complex IV in the Cisd2−/− mitochondria compared with wild-type mitochondria (Fig. 4G). Together with the oxygen consumption experiment, these results reveal a respiratory dysfunction in the Cisd2−/− mitochondria.
To test whether an increased level of reactive oxygen species (ROS), which is a by-product of mitochondrial oxidative phosphorylation, may contribute to the phenotypes of Cisd2−/− mice, we monitored the intracellular ROS, mainly H2O2, in MEF cells and primary cells obtained from the brains and livers of different genotypes of mice. There was no significant difference in the ROS levels in these primary cells between the different genotypes (Supplemental Fig. 18A). In addition, the mRNA levels of the enzymes that scavenge ROS were unaffected in brain, heart, liver, and skeletal muscle, suggesting that there was no ROS-induced stress response present in the Cisd2−/− mice (Supplemental Fig. 18B).
WFS and Cisd2−/− mice
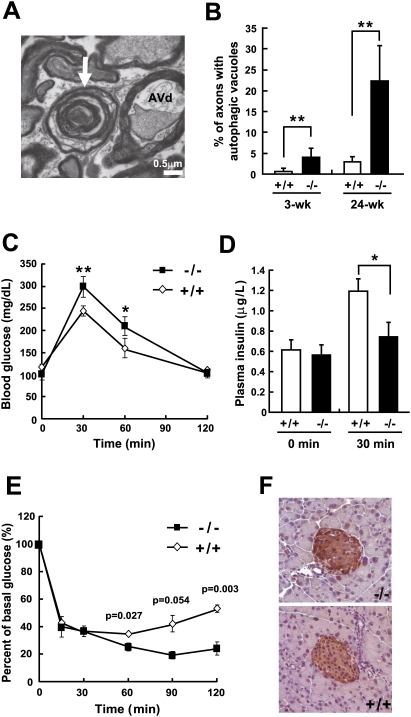
In order to evaluate the usefulness of Cisd2−/− mice as an animal model for WFS2 and gain insight into the mechanistic basis of WFS pathogenesis, we compared the clinical manifestations of this disease and the phenotype of Cisd2−/− mice. WFS is a clinically heterogeneous disease; only juvenile-onset diabetes mellitus and optic atrophy are necessary criteria for WFS diagnosis. Importantly, Cisd2−/− mice exhibit a progressive neurodegenerative phenotype that includes optic nerve defects (Fig. 5A,B; Supplemental Fig. 12). Regarding glucose homeostasis, we found that Cisd2−/− mice display a milder phenotype, namely, impaired glucose tolerance and decreased insulin secretion, which was revealed by the oral glucose tolerance test (Fig. 5C,D). In addition, insulin tolerance tests did not show insulin resistance in the Cisd2−/− mice; in fact, these mutant mice were somewhat more sensitive to insulin (Fig. 5E). Furthermore, immunohistochemistry (IHC) staining of the pancreatic islets revealed no obvious difference in insulin expression within the β cells between Cisd2−/− and wild-type mice (Fig. 5F). Taken together, these results indicate impaired glucose homeostasis in the Cisd2−/− mice, which seems to have an insulin secretory defect rather than insulin resistance. The importance of mitochondrial dysfunction in β-cell insulin secretion defects has been previously confirmed in other mouse models, which demonstrated that mitochondrial ATP production is a critical part of the β-cell signaling system and allows insulin release (Wallace 2001; Torraco et al. 2009). However, there was no overt diabetes observed in the Cisd2−/− mice with the C57BL/6 congenic background. This is consistent with a previous observation that C57BL/6 background confers a more diabetes-resistant phenotype (Coleman 1992); a similar finding of a genetic background effect also had been reported for WFS1 (wolframin) knockout mice (Ishihara et al. 2004). In addition to optic atrophy and glucose intolerance, the phenotypic features of Cisd2−/− mice reflect other aspects of the clinical manifestations of WFS patients including early (juvenile) onset and premature death (Supplemental Table 3). Thus, this mutant mouse may also provide an animal model for mechanistic investigation of Cisd2 protein function and help with the pathophysiological understanding of WFS2.
Figure 5.
Optic nerve degeneration and impaired glucose tolerance in Cisd2−/− mice. (A) A representative TEM micrograph showing a late or AVd detected in the axonal component of a myelinated axon of the optic nerve in 24-wk-old Cisd2−/− mice. The white arrow indicates a disintegrating myelinated axon. (B) Percentage of myelinated axons of the optic nerves containing autophagic vacuoles, including AVi and AVd, in the axonal component. There were three mice for each group; (wk) week. (C,D) Blood glucose levels and plasma insulin levels, respectively, before (0 min) and after the glucose load at the indicated time points. Oral glucose (1.5 g/kg body weight) tolerance tests were performed on 12-wk-old Cisd2−/− and wild-type mice, all of which had a C57BL/6 genetic background. Blood samples were collected to determine the mice's blood glucose levels and plasma insulin levels. (E) Insulin (0.75 U/kg body weight) tolerance tests were performed on 12-wk-old Cisd2−/− and wild-type mice. There were three mice in each group, and three independent measurements were carried out on each mouse. (*) P < 0.05; (**) P < 0.005. (F) IHC staining of insulin in the β cells of pancreatic islets using tissue sections prepared from 12-wk-old Cisd2−/− and wild-type mice.
Discussion
For more than a decade, physicians and researchers have fiercely debated as to whether WFS is associated with mitochondria and a defect in ATP supply. Most WFS patients die prematurely with severe neurological disabilities involving the central nervous system and peripheral nerves (Barrett and Bundey 1997; Domenech et al. 2006). In 1993, Bu and Rotter proposed a dual genome defect model and hypothesized that mitochondrial DNA mutation and nuclear genetic defects that interfere with the normal function of mitochondria can independently lead to WFS (Bu and Rotter 1993). This hypothesis was based on the clinical observations that the affected tissues and organs in WFS patients have a high metabolic demand and most of the clinical manifestations of WFS are consistent with an ATP supply defect, which is often seen in mitochondria-mediated disorders. There were several studies supporting this hypothesis (Rötig et al. 1993; Vora and Lilleyman 1993; Barrientos et al. 1996). Notably, Bundey et al. (1992) described a WFS patient having morphologically and biochemical abnormal mitochondria in the muscle biopsy; this finding indicated that a mitochondrial defect may be involved in the pathogenesis of WFS. However, other clinical studies revealed no evidence supporting the hypothesis of mitochondrial deficiency (Hofmann et al. 1997; Barrett et al. 2000). This controversy seems to have been resolved by the identification of different causative genes for WFS, and this hypothesis is supported by the mouse works carried out in this study.
WFS1 is associated with an ER defect
Previous studies in patients had identified WFS1 (wolframin) as the causative gene for WFS1 (Inoue et al. 1998; Strom et al. 1998). Biochemical and cell culture investigations revealed that wolframin is a transmembrane protein primarily localized in the ER and may be involved in the regulation of ER stress and calcium homeostasis (Takeda et al. 2001; Fonseca et al. 2005; Zatyka et al. 2008). In animal studies, a pancreatic phenotype related to glucose intolerance and impaired insulin secretion, but not the neurodegenerative phenotype, has been reported in wolframin knockout mice. Wolframin deficiency in mice leads to progressive loss of β cells and impaired glucose homeostasis (Ishihara et al. 2004), which appears to be caused by increased ER stress and apoptosis in the pancreatic β cells (Riggs et al. 2005; Yamada et al. 2006). Obviously, the pathogenesis of WFS1 patients with wolframin mutations is mechanistically related to an ER rather than a mitochondrial defect. This provides an explanation for the discrepancy as to why there were contradictory observations in some WFS (specifically WFS1) patients who do not have any detectable abnormality in their mitochondria.
WFS2 is a mitochondria-mediated disorder
Recently, Amr et al. (2007) identified CISD2 homozygous mutations in WFS patients and suggested that CISD2 is the causative gene responsible for WFS2. Our Cisd2 gene knockout mouse work provides strong evidence supporting the hypothesis that WFS is a mitochondria-mediated disorder; thus, specifically, WFS2, which is caused by a CISD2 mutation, is a mitochondria-mediated disorder. Previous clinical studies in WFS patients suggested that optic atrophy probably represents a degeneration of the optic nerve (Mtanda et al. 1986; Barrett et al. 1997). Indeed, our mouse work has revealed that progressive degeneration of the optic nerve is one of the earliest phenotypic features detected at 2–3 wk of age, which is before weaning; this phenotype exacerbates with age in the Cisd2−/− offspring (Fig. 5; Supplemental Fig. 12). Regarding glucose homeostasis, although the phenotype is relatively milder and only glucose intolerance was observed in the diabetes-resistant C57BL/6 background, in the future, it will be of great interest to introduce the Cisd2 mutant allele into C57BLKS/J (Mao et al. 2006), 129/Sv (Terauchi et al. 2003), or other diabetes-prone strains of mice, which may contain genetic modifier(s) that increase susceptibility to diabetes. This will allow the effect of the genetic background on the severity of diabetes to be examined.
Our present study reveals that Cisd2 is primarily localized in the mitochondria. Cisd2 deficiency causes mitochondrial dysfunction accompanied by autophagic cell death, and these events precede neuron and muscle degeneration; together, they lead to a panel of phenotypic features suggestive of premature aging. Since muscles and nerves have the highest energy needs and are therefore the most dependent on mitochondrial function, this explains why neuronal lesions and muscle abnormalities are the two earliest manifestations and why they precede the gross premature aging phenotype. Accordingly, mitochondrial degeneration appears to have a direct phenotypic consequence that triggers the accelerated aging process in Cisd2−/− mice (Fig. 6). Our results thus provide strong evidence for the causal involvement of mitochondrial dysfunction in driving mammalian aging as suggested previously by other studies in mitochondrial DNA mutator mice (Trifunovic et al. 2004; Kujoth et al. 2005; Vermulst et al. 2008). There are many genetic factors that have the potential to shorten life span (Kuro-o et al. 1997; Hasty et al. 2003; Mounkes et al. 2003; Niedernhofer et al. 2006). However, the human genetic factor that has been specifically identified as present on human chromosome 4q is present in the same region as Cisd2, and this is highly suggestive. Nonetheless, experiments that shorten life span might be less informative than those that prolong a healthy life span (Kurosu et al. 2005; Schriner et al. 2005; Pinton et al. 2007). Accordingly, it will be of great interest to evaluate the life history of transgenic mice expressing elevated levels of Cisd2 protein to see whether Cisd2 is the genetic determinant on human chromosome 4 that may regulate mammalian longevity and allow an unusually long life span to be achieved.
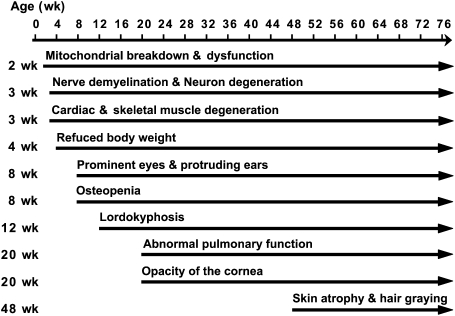
Figure 6.
Summary of the aging-related phenotypes as a function of age in the Cisd2−/− mice. The timing of the onset of each phenotype approximates the average age of onset for that phenotype; (wk) week. The onset age for each mouse for each phenotype shows variation around the average onset age to a limited degree.
Materials and methods
Generation of the Cisd2 knockout mouse
Mouse Cisd2 genomic DNA was obtained by screening a BAC library (Research Genetics, Inc.) derived from the C57BL/6 (B6) mouse strain. A SpeI–BamHI 6.4-kb DNA fragment, which contains part of intron 1, exon 2, and part of exon 3 of the Cisd2 gene, was used as the homologous recombination arms for construction of an insertion-type targeting vector (Supplemental Fig. 4). The Cisd2 targeting vector, containing the puromycin selection cassette, was linearized with ApaI and transfected into AB2.2 ES cells using electroporation. Targeted ES cell clones were screened by Southern blot analysis using a 3′-flanking probe, specifically a 1.7-kb BamHI–EcoRI fragment from exon 3. Targeted ES cells were injected into B6 blastcysts. Chimeric male mice were bred with B6 females. Germline transmission was obtained from their agouti progeny. The mice were bred in a specific pathogen-free facility and treated according to the National Research Council's Guide for the Care and Use of Laboratory Animals. Heterozygous males were backcrossed with B6 females for nine successive generations to introduce the Cisd2 targeted allele onto the B6 congenic background.
Histopathology
Various mouse tissues were collected, fixed with 10% formalin buffered with phosphate, and embedded in paraffin. Tissue sections (3–4 μm) were subjected to hematoxylin-eosin (H&E) and Masson's trichome staining by standard procedures (Young and Heath 2003).
Skeleton and bone density analyses
The bone specimens were fixed in 10% formalin buffered with phosphate and stored in 70% ethanol. The bone density per square centimeter was quantified from two-dimensional images of the bones using a DEXA apparatus (Northern Radiology). For micro-CT scanning, knockout and wild-type specimens were fixed in 10% formalin buffered with phosphate, stored in 70% ethanol, and then examined by eXplore Locus SP Preclinical Specimen Micro-CT (GE Healthcare). Whole-body and femur scans were performed in the axial plane with the specimens mounted in a cylindrical sample holder. Three-dimensional images of the skeletons were reconstructed from the micro-CT scanning slices and used for analyses of the skeletal structure and morphology. Quantitative data were calculated by eXplore MicroView version 2.0 Software Guide (GE Healthcare).
Cutaneous and skin analyses
Tissue sections of the dorsal skin were stained with H&E and Masson's trichome staining. The thicknesses of the dermal, adipose, and muscle layers were quantified by random measurements of the length of individual skin samples using SPOT Imaging Software Advance (Diagnostic Instruments, Inc.).
TEM
Various mouse tissues were fixed in a mixture of glutaraldehyde (1.5%) and paraformaldehyde (1.5%) in phosphate buffer (pH 7.3). They were post-fixed in 1% OsO4 and 1.5% potassium hexanoferrate, then rinsed in cacodylate and 0.2 M sodium maleate buffers (pH 6.0), and block-stained with 1% uranyl acetate. Following dehydration, the various tissues were embedded in Epon and sectioned for TEM as described previously (Kao et al. 1995).
Western blotting and IHC staining
Tissue samples were homogenized in lysis buffer (20 mM Tris at pH 7.4, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100 with Complete protease inhibitor cocktail [(Roche]) and denatured by boiling for 5 min. The extracted proteins were separated on a 13% SDS–polyacrylamide gel (Bio-Rad) and electro-transferred to an Amersham Hybond N+ membrane (GE Healthcare). The membranes were blocked with 5% (w/v) nonfat dry milk, incubated with primary antibody, washed, and then detected using a Visualizer Kit (Upstate Biotechnologies, 64-201BP). The following antibodies were used for Western blotting: LC3B (1:1000; Cell Signaling, 2755); Gapdh (1:5000; Abcam, ab9482); Hsp60 (1:2000; Chemicon, AB3497); Hsp70 (1:2000; BD Transduction Laboratories, 610608); VDAC-1 (1:1000; Calbiochem, 529532); ATP5B (1:2000; Molecular Probe, A21351); PDH (1:1000; Santa Cruz Biotechnologies, sc65242). IHC staining of insulin protein was performed using paraffin-embedded pancreas sections (3 μm). Pancreas sections were soaked in antigen retrieval buffer containing 10 mM sodium citrate (pH 6.0) and heated in a microwave oven twice for 10 min (Sunpentown, SM-1220, 650W). The sections were then incubated with primary antibody against insulin (1:100; Abcam, ab7842 guinea pig polyclonal antibodies) for 18–24 h at 4°C, detected by biotinylated secondary antibodies (1:500; Abcam, ab6907), and visualized by the LSAB Kit (DakoCytomation, K0690).
Rabbit anti-mouse Cisd1 and Cisd2 polyclonal antibodies
Mouse cDNA fragments of Cisd1 (corresponding to amino acids 27–108) and Cisd2 (corresponding to amino acids 52–135) were amplified by PCR and cloned into the pQE-31 (Qiagen) vector, which contains a His tag sequence. The expression plasmids for His-Cisd1 and His-Cisd2 were transformed into M15 bacteria, induced by 2 mM IPTG (isopropylthio-β-D-galactoside), and purified using Nickel-resin (Novagen). These proteins were injected into rabbits to generate antisera containing polyclonal antibodies against the mouse Cisd1 and Cisd2, respectively.
Subcellular localization
The EGFP-tagged Cisd2 expression plasmids were transfected into NIH/3T3 cells using lipofectamin 2000 (Invitrogen, 11668-019). Cells transiently expressing EGFP-Cisd2 fusion proteins were plated on gelatin-coated glass coverslips, stained with various organelle probes including mitochondria (MitoTracker Red CMXRos; Invitrogen Life Technologies), ER (anti-calnexin; Sigma, C7617), and Golgi apparatus (anti-Golgi 97; Molecular Probe). The coverslips were then fixed and visualized by confocal microscopy (Olympus FluoView FV300). Nuclei were counterstained with DAPI (4′-6-diamidino-2-phenylindole; Sigma).
Isolation of mitochondria from skeletal muscle
Fresh skeletal muscles were washed twice with PBS and homogenized immediately in ice-cold SEH buffer (0.25 M sucrose, 1 mM EGTA, 3 mM HEPES, protease inhibitor cocktail at pH 7.2). The mitochondrial pellet was obtained by low-speed centrifugation (800g) of the homogenate, followed by high-speed centrifugation (10,000g) of the supernatant. Finally, the mitochondrial pellet was resuspended in an appropriate volume of SEH buffer. The isolated mitochondria were used immediately for evaluation of mitochondrial respiration and oxidative phosphorylation.
Mitochondrial subfractionation
Mitochondrial subfractionation was performed according to the method described by Pagliarini et al. (2005) with some modifications. Briefly, 0.5 mL of mitochondrial suspension (10 mg/mL) was incubated with 10 mg of purified digitonin on ice for 30 min, and the mixture was gently inverted every 10 min. After high-speed centrifugation (12,000g), the mitoplast (containing the IM and matrix) was pelleted, and the supernatant was subjected to ultracentrifugation (150,000g) to separate the OM and IMS of the mitochondria.
Measurement of oxygen consumption
The oxygen consumption rate was measured using a 782 Oxygen Meter (Strathkelvin Instruments). An aliquot of 300 μL of assay buffer (125 mM sucrose, 65 mM KCl, 2 mM MgCl2, 20 mM Na+,K+-phosphate buffer at pH 7.2) containing ~0.2~0.5 mg of mitochondria was delivered into the closed chamber of the oxygen meter at 37°C to measure the steady-state oxygen consumption rate of the mitochondria (Chen et al. 2008). In order to further estimate the respiratory function of mitochondria, we measured the glutamate-malate-supported respiration and RCR of mitochondria. First, we used a Hamilton syringe (Strathkelvin) to add 10 mM glutamate and 10 mM malate (Sigma-Aldrich) into the chamber as the electron donor and recorded the glutamate–malate-supported oxygen consumption rate. After 5 min, we injected 3 μL of 100 mM ADP to attain a final ADP concentration of 1 mM in the assay medium. The rate of activated respiration was recorded to measure the RCR of the mitochondria.
Respiratory enzyme complex activity
The following activity assays were performed according to the method described by Wei et al. (1998). The activities of NCCR (which represents complex I–III activity) and SCCR (which represents complex II–III activity) were measured by following the reduction of exogenous oxidized cytochrome c. An aliquot of 20~50 μg of submitochondrial particles (SMP) was preincubated with the assay buffer (1.5 mM KCN, 50 mM K2HPO4 at pH 7.4) containing β-NADH or succinate for 15 min at 37°C. After addition of cytochrome c to the mixture, the change in the absorbance at 550 nm was recorded on a UV/visible spectrophotometer. CCO (which represents complex IV) activity was determined by following the oxidation of exogenous reduced cytochrome c. An aliquot of 20~50 μg SMP was preincubated in the assay buffer (5 mM K2HPO4 at pH 7.4) for 10 min at 30°C. After addition of ferrocytochrome c to the assay mixture, the change in absorbance at 550 nm was recorded on a UV/visible spectrophotometer.
Oral glucose tolerance test and insulin tolerance test
Mice after a 10-h fast (10 p.m. to 8 a.m.) were orally administrated with glucose solution (1.5 g/kg body weight) using a feeding needle (Juan et al. 2004). Blood samples were collected from tail tips before (0 min) and after glucose load at the indicated time points. The blood glucose levels were measured using glucose test strips (LifeScan; Johnson & Johnson) and SureStep Brand Meter. Serum insulin levels were determined by an ELISA kit (Mercodia). The insulin tolerance test was performed after a 2-h fast (9 a.m. to 11 a.m.) and involved an intraperitoneal injection of insulin (0.75 U/kg body weight; Novolin human regular insulin; Novo Nordisk) (Tran et al. 2008). There were three mice for each group and three independent measurements for each mouse.
Statistics
Results are presented as means ± SD. Differences among multiple groups were analyzed by a one-way ANOVA (SPSS 14.0 statistical software). Comparisons between two groups were done using a Student's t-test. Mouse survival rates were calculated by the Kaplan-Meier method, and differences in the survival of different groups of mice were determined by the log-rank (Mental-Cox) test. When analyzing statistical differences between the knockout and wild-type mice, P < 0.05 was considered significant.
Acknowledgments
We thank Dr. Lian-Fu Deng (Affiliated Ruijin Hospital of Shanghai Second Medical University, Shanghai Institute of Traumatology and Orthopeadics); Dr. An-Guor Wang (Taipei Veterans General Hospital); Dr. Ming-Ling Kuo (Chang Gung University); Dr. Chih-Cheng Chen (Academia Sinica); and Dr. Alan M. Lin, Dr. Hen-Li Chen, Dr. Chun-Ming Chen, Dr. Chi-Chang Juan, Yi-Shin Lai, Ching-Wen Cheng, and Hui-Wen Zhuang (National Yang-Ming University) for their insight and technical assistance. We thank the Microarray and Gene Expression Analysis Core Facility of the National Yang-Ming University Genome Research Center. The Core Facility is supported by the National Research Program for Genomic Medicine (NRPGM), National Science Council. We acknowledge support from the National Science Council (NRPGM 95HC007, NSC96-2752-B-010-004-PAE, and NSC97-2320-B-010-015-MY3) and a grant from the Ministry of Education, Aim for the Top University Plan.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1779509.
Supplemental material is available at http://www.genesdev.org.
References
- Amr S, Heisey C, Zhang M, Xia XJ, Shows KH, Ajlouni K, Pandya A, Satin LS, El-Shanti H, Shiang R. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81:673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TG, Bundey SE. Wolfram (DIDMOAD) syndrome. J. Med Genet. 1997;34:838–841. doi: 10.1136/jmg.34.10.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TG, Bundey SE, Fielder AR, Good PA. Optic atrophy in Wolfram (DIDMOAD) syndrome. Eye. 1997;11:882–888. doi: 10.1038/eye.1997.226. [DOI] [PubMed] [Google Scholar]
- Barrett TG, Scott-Brown M, Seller A, Bednarz A, Poulton K, Poulton J. The mitochondrial genome in Wolfram syndrome. J Med Genet. 2000;37:463–466. doi: 10.1136/jmg.37.6.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Casademont J, Saiz A, Cardellach F, Volpini V, Solans A, Tolosa E, Urbano-Marquez A, Estivill X, Nunes V. Autosomal recessive Wolfram syndrome associated with an 8.5-kb mtDNA single deletion. Am J Hum Genet. 1996;58:963–970. [PMC free article] [PubMed] [Google Scholar]
- Boucquey M, De Plaen E, Locker M, Poliard A, Mouillet-Richard S, Boon T, Kellermann O. Noxp20 and Noxp70, two new markers of early neuronal differentiation, detected in teratocarcinoma-derived neuroectodermic precursor cells. J Neurochem. 2006;99:657–669. doi: 10.1111/j.1471-4159.2006.04093.x. [DOI] [PubMed] [Google Scholar]
- Bu X, Rotter JI. Wolfram syndrome: A mitochondrial-mediated disorder? Lancet. 1993;342:598–600. doi: 10.1016/0140-6736(93)91416-j. [DOI] [PubMed] [Google Scholar]
- Bundey S, Poulton K, Whitwell H, Curtis E, Brown IA, Fielder AR. Mitochondrial abnormalities in the DIDMOAD syndrome. J Inherit Metab Dis. 1992;15:315–319. doi: 10.1007/BF02435965. [DOI] [PubMed] [Google Scholar]
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein (‘mitoNEET’) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- Coleman DL. The influence of genetic background on the expression of mutations at the diabetes (db) locus in the mouse. VI: Hepatic malic enzyme activity is associated with diabetes severity. Metabolism. 1992;41:1134–1136. doi: 10.1016/0026-0495(92)90299-p. [DOI] [PubMed] [Google Scholar]
- Domenech E, Gomez-Zaera M, Nunes V. Wolfram/DIDMOAD syndrome, a heterogenic and molecularly complex neurodegenerative disease. Pediatr Endocrinol Rev. 2006;3:249–257. [PubMed] [Google Scholar]
- Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, Urano F. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J Biol Chem. 2005;280:39609–39615. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: Lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Hofmann S, Bezold R, Jaksch M, Obermaier-Kusser B, Mertens S, Kaufhold P, Rabl W, Hecker W, Gerbitz KD. Wolfram (DIDMOAD) syndrome and Leber hereditary optic neuropathy (LHON) are associated with distinct mitochondrial DNA haplotypes. Genomics. 1997;39:8–18. doi: 10.1006/geno.1996.4474. [DOI] [PubMed] [Google Scholar]
- Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, Mueckler M, Marshall H, Donis-Keller H, Crock P, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome) Nat Genet. 1998;20:143–148. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Takeda S, Tamura A, Takahashi R, Yamaguchi S, Takei D, Yamada T, Inoue H, Soga H, Katagirl H, et al. Disruption of the WFS1 gene in mice causes progressive β-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13:1159–1170. doi: 10.1093/hmg/ddh125. [DOI] [PubMed] [Google Scholar]
- Juan CC, Shen YW, Chien Y, Lin YJ, Chang SF, Ho LT. Insulin infusion induces endothelin-1-dependent hypertension in rats. Am J Physiol Endocrinol Metab. 2004;287:E948–E954. doi: 10.1152/ajpendo.00536.2003. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CH, Chen JK, Kuo JS, Yang VC. Visualization of the transport pathways of low density lipoproteins across the endothelial cells in the branched regions of rat arteries. Atherosclerosis. 1995;116:27–41. doi: 10.1016/0021-9150(95)05519-3. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Davis T, Ostler EL, Faragher RG. What can progeroid syndromes tell us about human aging? Science. 2004;305:1426–1431. doi: 10.1126/science.1102587. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao HZ, Roussos ET, Péterfy M. Genetic analysis of the diabetes-prone C57BLKS/J mouse strain reveals genetic contribution from multiple strains. Biochim Biophys Acta. 2006;1762:440–446. doi: 10.1016/j.bbadis.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov S, Hernandez L, Sullivan T, Stewart CL. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- Mtanda AT, Cruysberg JR, Pinckers AJ. Optic atrophy in Wolfram syndrome. Ophthalmic Paediatr Genet. 1986;7:159–165. doi: 10.3109/13816818609004133. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic β cells. Mol Cell. 2005;19:197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, Contursi C, Minucci S, Mantovani F, Wieckowski MR, et al. Protein kinase C β and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- Puca AA, Daly MJ, Brewster SJ, Matise TC, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, Murray J, Schmidt RE, Herrera PL, Permutt MA. Mice conditionally lacking the Wolfram gene in pancreatic islet β cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48:2313–2321. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- Rötig A, Cormier V, Chatelain P, Francois R, Saudubray JM, Rustin P, Munnich A. Deletion of mitochondrial DNA in a case of early-onset diabetes mellitus, optic atrophy and deafness (DIDMOAD, Wolfram syndrome) J Inherit Metab Dis. 1993;16:527–530. doi: 10.1007/BF00711672. [DOI] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of Jurkat cells. EMBO J. 1999;18:2040–2048. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Strom TM, Hörtnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, Gerbitz KD, Meitinger T. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998;7:2021–2028. doi: 10.1093/hmg/7.13.2021. [DOI] [PubMed] [Google Scholar]
- Takeda K, Inoue H, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y, Shinoda K, Oka Y. WFS1 (Wolfram syndrome 1) gene product: Predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet. 2001;10:477–484. doi: 10.1093/hmg/10.5.477. [DOI] [PubMed] [Google Scholar]
- Terauchi Y, Matsui J, Suzuki R, Kubota N, Komeda K, Aizawa S, Eto K, Kimura S, Nagai R, Tobe K, et al. Impact of genetic background and ablation of insulin receptor substrate (IRS)-3 on IRS-2 knock-out mice. J Biol Chem. 2003;278:14284–14290. doi: 10.1074/jbc.M211045200. [DOI] [PubMed] [Google Scholar]
- Torraco A, Diaz F, Vempati UD, Moraes CT. Mouse models of oxidative phosphorylation defects: Powerful tools to study the pathobiology of mitochondrial diseases. Biochim Biophys Acta. 2009;1793:171–180. doi: 10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Yamamoto Y, Gesta S, Kahn CR. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Vora AJ, Lilleyman JS. Wolfram syndrome: Mitochondrial disorder. Lancet. 1993;342:1059. [PubMed] [Google Scholar]
- Wallace DC. Mouse models for mitochondrial disease. Am J Med Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- Wei YH, Lu CY, Lee HC, Pang CY, Ma YS. Oxidative damage and mutation to mitochondrial DNA and age-dependent decline of mitochondrial respiratory function. Ann N Y Acad Sci. 1998;854:155–170. doi: 10.1111/j.1749-6632.1998.tb09899.x. [DOI] [PubMed] [Google Scholar]
- Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci. 2007;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, Tokita A, Satake C, Tashiro F, Katagiri H, et al. WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic β-cells. Hum Mol Genet. 2006;15:1600–1609. doi: 10.1093/hmg/ddl081. [DOI] [PubMed] [Google Scholar]
- Young B, Heath JW. Wheater's functional histology, a text and colour atlas. 4th ed. Churchill Livingstone, London: 2003. [Google Scholar]
- Zatyka M, Ricketts C, da Silva Xavier G, Minton J, Fenton S, Hofmann-Thiel S, Rutter GA, Barrett TG. Sodium–potassium ATPase 1 subunit is a molecular partner of Wolframin, an endoplasmic reticulum protein involved in ER stress. Hum Mol Genet. 2008;17:190–200. doi: 10.1093/hmg/ddm296. [DOI] [PMC free article] [PubMed] [Google Scholar]