Abstract
The INK4a/ARF tumor suppressor locus, a key executor of cellular senescence, is regulated by members of the Polycomb group (PcG) of transcriptional repressors. Here we show that signaling from oncogenic RAS overrides PcG-mediated repression of INK4a by activating the H3K27 demethylase JMJD3 and down-regulating the methyltransferase EZH2. In human fibroblasts, JMJD3 activates INK4a, but not ARF, and causes p16INK4a-dependent arrest. In mouse embryo fibroblasts, Jmjd3 activates both Ink4a and Arf and elicits a p53-dependent arrest, echoing the effects of RAS in this system. Our findings directly implicate JMJD3 in the regulation of INK4a/ARF during oncogene-induced senescence and suggest that JMJD3 has the capacity to act as a tumor suppressor.
Keywords: Polycomb, senescence, histone methylation, INK4a, JmjC proteins
The INK4b–ARF–INK4a tumor suppressor locus encodes two members of the INK4 family of cyclin-dependent kinase inhibitors, p16INK4a and p15INK4b, and an unrelated protein termed ARF that exploits the second exon of INK4a in an alternative translational reading frame (Sharpless 2005; Gil and Peters 2006). All three proteins are capable of causing cell cycle arrest. Whereas the INK4 proteins do so by blocking the phosphorylation and inactivation of the retinoblastoma protein (pRb), ARF blocks the ubiquitylation and degradation of p53. Given the properties of the encoded proteins, expression of the locus is tightly regulated and this is achieved in part by epigenetic silencing by Polycomb group (PcG) proteins (Gil and Peters 2006).
The PcG proteins participate in two types of multiprotein complexes, termed PRC2 and PRC1, that, respectively, establish and bind to H3K27me3 (histone H3 trimethylated on Lys 27). PcG-mediated repression is important for maintaining pluripotency, by blocking the expression of lineage specific transcription factors (Sparmann and van Lohuizen 2006). Loss of particular PcG genes results in premature senescence, due primarily to derepression of Ink4a/Arf (Jacobs et al. 1999). Senescence is a state of irreversible growth arrest that is engaged when primary cells are exposed to various forms of stress and guards against the effects of oncogenic mutations (Campisi and d'Adda di Fagagna 2007). Importantly, oncogene-induced senescence (OIS) can be observed in premalignant lesions in vivo and is recognized as an important facet of tumor suppression (Collado and Serrano 2006). Here we explore how signaling from an activated RAS oncogene can induce INK4a/ARF and senescence in the face of epigenetic silencing by PcG proteins.
Results and Discussion
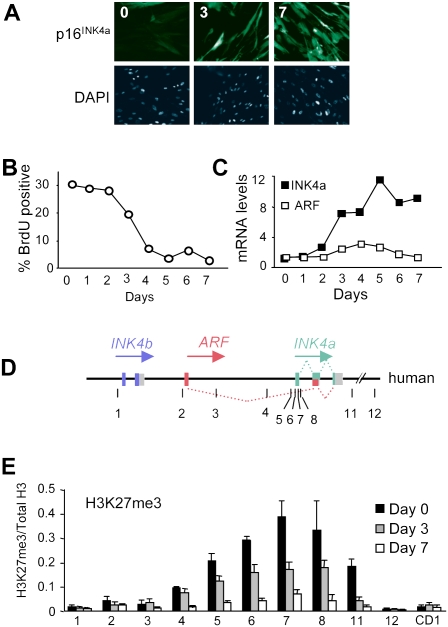
Based on the premise that activation of INK4a by oncogenic RAS is likely to require reversal of PcG-mediated repression, we investigated the extent of H3K27me3 modification at the locus in human diploid fibroblasts (HDFs) undergoing OIS. To facilitate these analyses, the IMR90 strain of fetal lung fibroblasts and a newly derived strain of breast fibroblasts (BF) were transduced with a tamoxifen-regulated form of activated H-RAS (Tarutani et al. 2003). Upon the addition of 4-hydroxytamoxifen (OHT), these cells develop characteristic features of senescence accompanied by up-regulation of p16INK4a and reduced BrdU incorporation (Fig. 1A–C; Supplemental Figs. S1A, S2A). In contrast to the situation in mouse embryo fibroblasts (MEFs), there is little change in the expression of ARF, as judged by quantitative RT–PCR (qRT–PCR) (Fig. 1C). Chromatin immunoprecipitation (ChIP) was used to compare the ratio of H3K27me3 to total H3 throughout the INK4a–ARF locus using previously described primer sets (Fig. 1D). Consistent with published findings (Bracken et al. 2007; Kia et al. 2008), the H3K27me3 mark was present in a relatively broad peak centered on the first exon of INK4a. Following the addition of OHT and activation of RAS, there was a time-dependent reduction in the levels of H3K27me3 at the locus (Fig. 1E; Supplemental Fig. S1B).
Figure 1.
Loss of H3K27me3 at the INK4a/ARF locus in cells undergoing OIS. (A) Induction of senescence and p16INK4a expression in IMR90-ER:RAS cells treated with OHT for 0, 3, and 7 d. (B,C) Percentage of BrdU-positive cells and relative levels of INK4a and ARF mRNAs as measured by qRT–PCR in IMR90-ER:RAS cells treated with OHT as indicated. The results are representative of three independent experiments. (D) Schematic representation of the human INK4b–ARF–INK4a locus (not to scale). Numbered lines show the approximate location of primer sets used for ChIP (Supplemental Table S1). (E) Degree of H3K27me3 modification at the INK4b–ARF–INK4a locus in IMR90-ER:RAS cells treated with OHT for 0, 3, and 7 d. A primer set amplifying the cyclin D1 gene (CD1) was used as a negative control. The results are presented as the ratio of H3K27me3 to total H3 as judged by ChIP analyses.
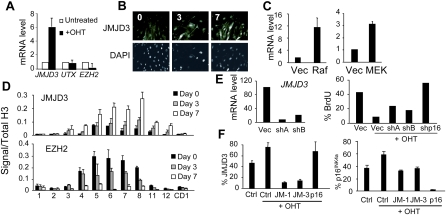
We reasoned that, besides nucleosome exchange, the most likely explanation for the changes in H3K27me3 levels would be either down-regulation of the methyltransferase EZH2 (Sparmann and van Lohuizen 2006) or an increase in one of the H3K27me3 demethylases, UTX or JMJD3 (Agger et al. 2007; De Santa et al. 2007; Hong et al. 2007; Jepsen et al. 2007; Lan et al. 2007; Lee et al. 2007; Xiang et al. 2007). Expression of the relevant mRNAs was assessed by qRT–PCR. Increased JMJD3 levels were observable within 3 d following RAS activation and the levels continued to rise up to 7 d (Fig. 2A,B; Supplemental Fig. S2B). Immunofluorescence revealed accumulation of the protein, predominantly in the cell nucleus, as early as days 1 and 2 after addition of OHT (Fig. 2B; Supplemental Fig. S2D). The effects were observed in different strains of HDFs and irrespective of the strategy used to activate the RAS–RAF–MEK pathway (Fig. 2C; Supplemental Fig. S1C). We saw little change in the expression of UTX but the expression of EZH2 declined substantially following RAS activation (Fig. 2A). It was shown previously that EZH2 is an E2F-dependent gene (Bracken et al. 2003) and that its levels decline during replicative senescence or upon various genotoxic stresses (Bracken et al. 2007). Interestingly, quantitative immunofluorescence and immunoblotting suggested that RAS initially caused a slight increase in EZH2 protein levels followed by a marked decrease, presumably reflecting the loss of mRNA expression (Supplemental Fig. S2C–F). In view of these complications, it is difficult to conclude whether there is any temporal order or hierarchy to the effects of EZH2 and JMJD3 on the INK4a locus.
Figure 2.
Up-regulation of JMJD3 by oncogenic RAS. (A) qRT–PCR assessment of the relative levels of JMJD3, UTX, and EZH2 mRNAs in IMR90-ER:RAS cells treated with OHT for 7 d normalized to untreated cells. (B) Increased levels of endogenous JMJD3 following induction of RAS. Nuclear staining using DAPI is shown in the bottom panel. (C) Increased expression of JMJD3 mRNA in IMR90 cells transduced with constitutively active forms of RAF or MEK1. (D) ChIP analyses showing enhanced binding of JMJD3 and reduced binding of EZH2 at the INK4a/ARF locus in IMR90-ER:RAS cells. (E, left) Knockdown of JMJD3 mRNA with two shRNAs. Controls and shRNAs were used to infect IMR90-ER:RAS cells. (Right) After selection, cells were treated with OHT for 48 h and pulsed with BrdU for 16 h. These results are representative of three independent experiments. (F) IMR90-ER:RAS cells were transfected with siRNAs or controls. Two days after, cells were treated with OHT for four more days, fixed, and subjected to immunofluorescence. Percentages of JMJD3-positive (left) or p16INK4a-positive (right) cells are shown.
In line with the loss of H3K27me3 following activation of RAS, there was a time-dependent increase in the binding of JMJD3 at the locus, coupled with a corresponding loss of EZH2 (Fig. 2D). Both proteins were concentrated around the first exon of INK4a, analogous to the distribution of PcG complexes and H3K27 methylation in untreated cells, whereas no significant binding was noted at the ARF and INK4b promoters. These changes were accompanied by decreased binding of the PcG proteins CBX7 and BMI1, which are recruited to sites of H3K27 methylation, and SUZ12, which along with EZH2 and EED is an integral component of the PRC2 complex (Supplemental Fig. S3). We also observed a reciprocal increase in the association of RNA polymerase II (Pol II) and of H3K4 trimethylation. The latter are consistent with reports that UTX and JMJD3 associate with H3K4 methyltransferase complexes (De Santa et al. 2007; Issaeva et al. 2007). However, we did not detect UTX at the INK4a locus using reagents and protocols that confirm the binding of UTX at other genes (Supplemental Fig. S4).
An implication of these findings is that JMJD3 might contribute to the implementation of OIS downstream from RAS/RAF/MEK. To address this, we asked whether shRNAs against JMJD3 alleviate OIS. In IMR90-ER:RAS cells, stable expression of JMJD3 shRNAs partially restored BrdU incorporation relative to control cells (Fig. 2E). Similar results were obtained with independent siRNAs and shRNAs and in cells that were freshly infected with a RAS retrovirus (data not shown). In addition, we confirmed that JMJD3 knockdown reduced p16INK4a expression but could not block completely the activation of p16INK4a by RAS (Fig. 2F; Supplemental Fig. S5).
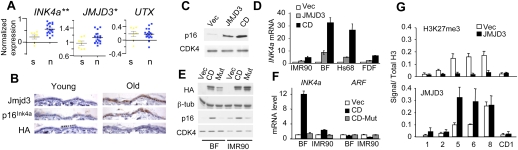
It is now appreciated that OIS is an important barrier to tumorigenesis in vivo and a number of studies have shown that preneoplastic lesions express markers and effectors of senescence (Collado and Serrano 2006). Examples include benign melanocytic nevi, which frequently contain an activated B-RAFV600E allele and show features of senescence, including elevated p16INK4a levels (Michaloglou et al. 2005; Gray-Schopfer et al. 2006). Interestingly, data mining of published profiles (Talantov et al. 2005) using Oncomine (http://www.oncomine.org) indicates that JMJD3 levels are significantly higher in melanocytic nevi compared with normal skin (P = 0.041) (Fig. 3A), whereas UTX expression is essentially the same in both (P = 0.9359). Attempts to validate this correlation at the protein level have been precluded by the lack of antibodies working reliably for immunohistochemistry (IHC) in human tissues. However, using antibodies that permit IHC in mouse tissues, we observed positive staining for Jmjd3 in mouse epidermis. Despite some variability, the proportion of Jmjd3-positive cells is markedly increased in skin from aged mice, mirroring the age-related increase in p16INK4a (Fig. 3B).
Figure 3.
Ectopic expression of JMJD3 activates p16INK4a in HDFs. (A) Expression of INK4a, JMJD3, and UTX in melanocytic nevi versus normal skin from Talantov et al. (2005). (**) P < 0.001; (*) P < 0.05. The P-values correspond to a nonparametric unpaired t-test. (B) Immunohistochemistry of Jmjd3 and p16Ink4a proteins in paraffin-embedded sections of mouse back skin. HA staining was used as a negative control. Similar results were obtained in mouse tail skin (not shown). (C) Immunoblot of p16INK4a in Hs68 fibroblasts transduced with JMJD3 or the CD construct. CDK4 was used as a loading control. (D) qRT–PCR showing that expression of full-length JMJD3 or the CD up-regulates INK4a in the indicated strains of the HDFs. (E) Immunoblot showing up-regulation of p16INK4a by the HA-tagged CD of JMJD3 (CD) but not by a catalytically inactive mutant (Mut). Equivalent results were obtained in BF and IMR90 cells. (F) Ectopic expression of JMJD3 CD in BF and IMR90 cells up-regulates INK4a but not ARF. (G) Expression of JMJD3 reduces the extent of H3K27me3 modification and increases the density of JMJD3 at the INK4a locus in IMR90 cells.
As a more direct test of the ability of JMJD3 to regulate INK4a/ARF, we infected HDFs with retroviral vectors encoding HA-tagged versions of the full-length protein or a truncated form corresponding to the catalytic domain (CD). As noted previously, the CD was expressed at higher levels than the full-length protein (data not shown), but both caused a substantial increase in the expression of p16INK4a mRNA and protein (Fig. 3C,D). Similar effects were observed in other strains of HDFs (Fig. 3D) and were dependent on JMJD3 demethylase activity. Thus, point mutations that impair JMJD3 activity (Agger et al. 2007) also impair its ability to activate INK4a (Fig. 3E,F). Similar results were obtained with the catalytically inert full-length protein (data not shown). Note that ectopic expression of UTX did not activate INK4a (Supplemental Fig. S6). The low levels of p14ARF in HDFs precluded detection by immunoblotting, but we observed no change in the expression of ARF mRNA (Fig. 3F). Consistent with this, p53, which is stabilized by p14ARF, appeared unaffected by JMJD3 (Supplemental Fig. S7A). Moreover, whereas exogenous JMJD3 was recruited to the INK4a locus and caused a reduction in local H3K27 methylation (Fig. 3G), there was no discernible change in the already low levels of H3K27me3 at the ARF promoter (primer set 2). Similar conclusions apply in cells transduced with the CD version of JMJD3 (Supplemental Fig. S7B).
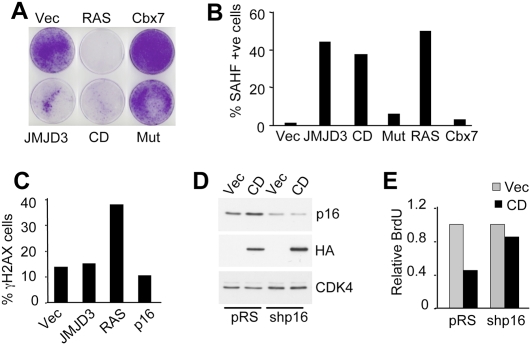
As anticipated from the accumulation of p16INK4a, ectopic expression of JMJD3 in HDFs induced features of senescence, including an enlarged cell and nuclear size, reduced BrdU incorporation, growth arrest, and the formation of senescence-associated heterochromatin foci (SAHFs) (Fig. 4A,B; Supplemental Fig. S7C–G). Whereas the full-length protein and CD induced all of these changes, the catalytically inactive version did not. RAS was included as a positive control in these experiments, whereas Cbx7 had a negative effect on p16INK4a expression and senescence. Interestingly, while RAS also elicited a DNA damage response, as judged by the appearance of γH2AX foci (Fig. 4C; Supplemental Fig. S7H), we did not detect an increase in the intensity of γH2AX staining or in the number of γH2AX-positive cells upon transduction with JMJD3. The effects of JMJD3 are more akin to those of exogenous p16INK4a. This suggested that JMJD3-mediated arrest might depend on INK4a, rather than being a consequence of global H3K27me3 demethylation. To address this possibility, we coexpressed JMJD3 with the E6 and E7 proteins of HPV16, which inactivate p53 and pRb, respectively. As shown in Supplemental Fig. S8, A and B, coexpression of E7 alone or E6 + E7 alleviated the effects of JMJD3 and enabled proliferation, whereas the cells transduced with E6 or empty vector were arrested by JMJD3. Along similar lines, cells in which the expression of p16INK4a was reduced using shRNA (Fig. 4D) were significantly less sensitive to arrest by JMJD3 than the controls (Fig. 4E). Moreover, the Leiden strain of p16INK4a-deficient fibroblasts, which are resistant to RAS-induced arrest (Drayton et al. 2003) were also resistant to arrest by JMJD3 (Supplemental Fig S8C,D), implying that in HDFs the arrest induced by JMJD3 is dependent on the INK4a/pRb rather than the ARF/p53 axis.
Figure 4.
JMJD3 causes p16INK4a-dependent growth arrest in HDFs. (A) IMR90 cells were infected with the indicated retroviruses. Crystal violet staining at 10–15 d post-infection. (B) JMJD3, CD, and activated RAS cause the appearance of SAHFs, whereas Cbx7 delays senescence in HDFs. Quantification was performed on at least 200 nuclei for each. (C) Immunofluorescence for γH2AX indicates that RAS causes a DNA damage response in IMR90 cells, whereas JMJD3 or p16INK4a do not. The results presented in A–C are representative of three independent experiments. (D,E) shRNA knockdown of p16INK4a alleviates the arrest caused by ectopic JMJD3. Immunoblot showing p16INK4a and HA-CD. BrdU incorporation was quantified by immunofluorescence and relative numbers were plotted. Two independent experiments showed similar results.
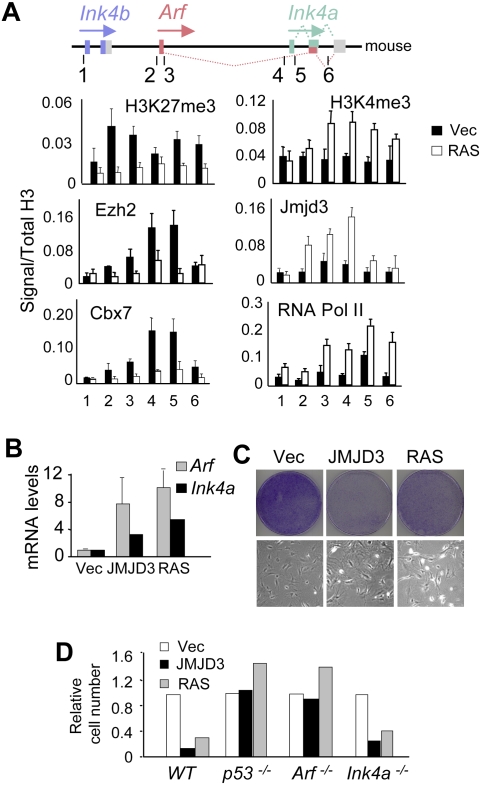
There are significant differences in the implementation of senescence in MEFs as opposed to HDFs, including a more prominent role for Arf (Sharpless 2005; Gil and Peters 2006). Both p19Arf and p16Ink4a accumulate as MEFs are passaged and both are induced by oncogenic RAS. However, Arf−/− cells escape from senescence, whereas Ink4a−/− cells do not (Kamijo et al. 1997; Krimpenfort et al. 2001; Sharpless et al. 2001, 2004). It was therefore important to explore the role of Jmjd3 in the regulation of the mouse Ink4a/Arf locus. In MEFs, H3K27me3 was present throughout the Ink4b–Arf–Ink4a locus, including the Arf promoter and first exon (Fig. 5A). Importantly, the signals were markedly decreased following expression of RAS, accompanied by increased binding of endogenous Jmjd3 and decreased binding of Ezh2 and Cbx7. We did not find evidence for Utx binding at the locus (Supplemental Fig. S4B). Marks of active transcription, H3K4me3 and RNA Pol II, also increased throughout the locus. The most pronounced changes occurred around the first exons of Ink4a and Arf (Fig. 5A). In line with these changes, ectopic expression of JMJD3 or RAS up-regulated the levels of Arf as well as Ink4a mRNA (Fig. 5B), causing a senescence-like growth arrest (Fig. 5C). Importantly, the effects were not apparent in MEFs nullizygous for either p53, Ink4a/Arf (data not shown), or Arf, whereas Ink4a−/− MEFs remained sensitive to growth inhibition by both JMJD3 and RAS (Fig. 5D). Comparison of BrdU incorporation in wild-type and Arf−/− MEFs confirmed this observation (Supplemental Fig. S9B). As reported previously (Bracken et al. 2007), we also observed a gradual decline in the level of H3K27me3 at the Ink4a/Arf locus when normal MEFs were passaged continuously, accompanied by a gradual increase in Jmjd3 and its association with the Ink4a/Arf locus (Supplemental Fig. S9C,D). These findings are consistent with the central role of the Arf/p53 pathway in senescence of murine cells.
Figure 5.
In MEFs, JMJD3 activates both Arf and Ink4a, causing Arf-dependent growth arrest. (A) MEFs were infected with a control vector or with activated RAS. After selection, cells were subjected to ChIP using the indicated antibodies and primer sets (Supplemental Table S1). The approximate location of the primer sets used is shown. (B) In normal MEFs, ectopic expression of JMJD3 or RAS causes up-regulation of both Arf and Ink4a transcripts. (C) Ectopic expression of JMJD3 or RAS in MEFs causes a senescence-like arrest. The top panel shows crystal violet staining at 10–15 d post-infection. The bottom panel shows typical senescence-like morphology. (D) JMJD3 and RAS cause growth arrest in normal and Ink4a−/− MEFs but not in p53−/− or Arf−/− MEFs. Relative cell numbers were assessed by crystal violet staining. The data are representative of between two and six independent experiments for each genotype.
In summary, our data confirm the dynamic nature of histone methylation and illustrate how signal transduction pathways can impact the epigenetic control of gene expression. Specifically, we identify JMJD3 as a major component in the activation of the INK4a/ARF locus by oncogenic RAS and the implementation of OIS. Signaling via the RAS–RAF–MEK pathway reduces the extent of H3K27me3 at the locus by up-regulating JMJD3 and down-regulating EZH2, leading to a loss of PcG-mediated repression. As with RAS itself, the effects of JMJD3 on the INK4a/ARF locus are context-dependent, causing up-regulation of mouse Arf but not of human ARF. This raises the possibility that the differences between mouse and human ARF are inherent in the respective genomic loci.
The other inference from our work is that JMJD3 has properties compatible with tumor suppression. Analogous to p16INK4a itself, one might expect to see elevated levels of JMJD3 in benign and premalignant lesions, reflecting its role in the implementation of OIS. Preliminary evidence suggests that this is indeed the case in melanocytic nevi, and it will obviously be important to broaden these observations to other benign lesions. Conversely, one might expect to see loss of JMJD3 expression in more aggressive tumors that have escaped from OIS. The Oncomine database (http://www.oncomine.org) contains evidence for reduced JMJD3 expression in a variety of tumor types, and inspection of the CONAN (copy number analysis) database (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/conan/search.cgi) indicates that loss of heterozygosity at JMJD3 has been recorded in 478 out of 770 tumors of multiple origin (data not shown). However, as noted by Cloos et al. (2008), JMJD3 resides on chromosome 17p13.1, within 0.15 Mb of TP53, making it difficult to distinguish whether these deletions confer an advantage to the tumor or are a by-product of TP53 deletions. The overexpression of EZH2 (Varambally et al. 2002; Bracken et al. 2003) and the negative impact that inhibition of H3K27 methylation has on tumor cells (Tan et al. 2007), coupled with the influence of JMJD3 on differentiation (De Santa et al. 2007; Jepsen et al. 2007; Burgold et al. 2008; Sen et al. 2008) and senescence (Agger et al. 2009; this study), all argue for a role for JMJD3 in tumorigenesis.
Materials and methods
Cell culture and retroviral infection
IMR90 and Hs68 were obtained from the American Type Culture Collection. The FDF strain was described (Brookes et al. 2004) and the BF strain was derived from a sample of reduction mammoplasty. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum. MEFs of Ink4a/ARF−/−; Ink4a−/−; Arf−/−, or p53−/− genotypes were obtained from S. Lowe, M. Serrano, C. Sherr, and A. Berns. Methods used for retrovirus production and infection, for crystal violet staining and BrdU incorporation, are detailed elsewhere (Brookes et al. 2004; Gil et al. 2004; Acosta et al. 2008).
Plasmids and vectors
Plasmids encoding human JMJD3 and shRNAs are described in detail in the Supplemental Material. A plasmid encoding ER:RAS was obtained from Paul Khavari, and was used to generate LNCX-ER:RAS. Knockdown of p16INK4a was achieved using a validated shRNA (Drayton et al. 2003).
qRT–PCR analysis
Total RNA was extracted using the RNeasy minikit (Qiagen) or the High Pure RNA kit (Roche). Reverse transcription was carried out using either SuperScript II (Gibco) or TaqMan (Applied Biosystems). PCR reactions were performed on ABI 7900HT (Applied Biosystems) using Power SYBR Green Master Mix or TaqMan Universal PCR Master Mix (Applied Biosystems). The primer sets and TaqMan 6-carboxy-fluorescein (FAM) Gene Expression Assays (Applied Biosystems) used are listed in Supplemental Table S1.
ChIP
ChIP was performed as described (Ananthanarayanan et al. 2004) using the EZ-Magna ChIPTM G kit (catalog no. 17-409, Millipore). Immunoprecipitation of cross-linked chromatin was conducted with antibodies described in Supplemental Table S2. After immunoprecipitation, DNA was extracted using the QIAquick PCR purification kit (Qiagen) and an aliquot was amplified by real time qPCR using oligonucleotide primers described in Supplemental Table S1. To confirm target enrichment, each PCR product was evaluated by end-point PCR.
Immunofluorescence and immunoblotting
Antibodies used were human p16INK4a (JC-8, CRUK), mouse p16Ink4a (M-156, sc-1207, Santa Cruz Biotechnologies), β-tubulin (H-235, sc-9104, Santa Cruz Biotechnologies), p53 (DO-1, sc-126, Santa Cruz Biotechnologies), HA (16B12, MMS-101P, Covance; or Y-11, sc-805, Santa Cruz Biotechnologies), CDK4 (H-22, sc-601, Santa Cruz Biotechnologies), EZH2 (AC22, 3147, Cell Signaling), γH2AX (05-636, Upstate Biotechnologies), and JMJD3 (ARP40102, Aviva; and ab38113, Abcam). Immunofluorescence was performed using an automated high-throughput microscope (InCell Analyzer 1000, GE). Image processing and quantification was performed using the InCell Investigator software (GE).
Immunohistochemistry
Skin samples from young (6–12 wk) and old (14–18 mo) mice were fixed overnight in 4% PFA and embedded in paraffin wax. Antigen retrieval was performed by microwaving in 10 mM citrate buffer (pH 6.0) for 10 min. Nonspecific binding was blocked by incubation in Power Block Universal Blocking Reagent for 10 min (10× PowerBlock; Biogenex). Primary antibodies were diluted in PBS 1% BSA. The antibodies were detected using the EnVision+ System-HRP (DAB) kit for rabbit primary antibodies (Dako).
Database analyses
We used Oncomine, (http://www.oncomine.org) to analyze the expression of different genes in available microarray data comparing normal skin with nevi samples (Talantov et al. 2005) by using an unpaired t-test (two tailed). Graphics and statistic values were generated using Prism 5.
Acknowledgments
We are grateful to H. Peretz, R. Shafir, P. Kavhari, S. Lowe, M. Narita, M. Serrano, A. Berns, C. Sherr, R. Festenstein, and T. Rodríguez for supplying reagents. We thank D. Beach, M. da Costa, S. Raguz, and A. Muñoz for helpful comments; P. Moemeni for help; J. Tsao for technical support; E. Hernando, G. Stamp, and M. Nohadani for help with immunohistochemistry; and K. Helin for sharing results previous to publication. M.B. was funded by training awards from the Department of Defense Prostate Cancer Directive, and from a KO8 award from NIDDK/NIH. A.B. is funded by FCT. M.J.W. was supported by PHS awards from the NIH. G.P. is funded by CRUK. J.G. is funded by the MRC, grants from CRUK and the AICR, and by the EMBO Young Investigator Programme.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.511109.
Supplemental material is available at http://www.genesdev.org.
References
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PAC, Rudkjær L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A–ARF locus in response to oncogene- and stress-induced senescence. Genes & Dev. 2009 doi: 10.1101/gad.510809. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem. 2004;279:54348–54357. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res. 2004;298:549–559. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes & Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Drayton S, Rowe J, Jones R, Vatcheva R, Cuthbert-Heavens D, Marshall J, Fried M, Peters G. Tumor suppressor p16INK4a determines sensitivity of human cells to transformation by cooperating cellular oncogenes. Cancer Cell. 2003;4:301–310. doi: 10.1016/s1535-6108(03)00242-3. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, Marais R, Wynford-Thomas D, Bennett DC. Cellular senescence in naevi and immortalisation in melanoma: A role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b–ARF–INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Croce LD, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes & Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani M, Cai T, Dajee M, Khavari PA. Inducible activation of Ras and Raf in adult epidermis. Cancer Res. 2003;63:319–323. [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]