Abstract
UVB-induced lesions in mammalian cellular DNA can, through the process of mutagenesis, lead to carcinogenesis. However, eukaryotic cells have evolved complex mechanisms of genomic surveillance and DNA damage repair to counteract the effects of UVB radiation. We show that following UVB DNA damage, there is an overall inhibition of protein synthesis and translational reprogramming. This reprogramming allows selective synthesis of DDR proteins, such as ERCC1, ERCC5, DDB1, XPA, XPD, and OGG1 and relies on upstream ORFs in the 5′ untranslated region of these mRNAs. Experiments with DNA-PKcs-deficient cell lines and a specific DNA-PKcs inhibitor demonstrate that both the general repression of mRNA translation and the preferential translation of specific mRNAs depend on DNA-PKcs activity, and therefore our data establish a link between a key DNA damage signaling component and protein synthesis.
Keywords: DNA damage, translation, upstream ORF
The DNA of an organism can be structurally altered by both intra- and extracellular agents, and damaged DNA, if it is not repaired, can lead to genomic instability, mutagenesis, and cell death. Therefore, eukaryotic cells have developed efficient genomic surveillance and DNA repair mechanisms to maintain their genetic integrity. These mechanisms act in conjunction with DNA damage checkpoints to ensure that cells are growth-arrested until DNA repair is completed (d'adda di Fagagna et al. 2004; Sancar et al. 2004; Bartek and Lukas 2007; Harper and Elledge 2007; Hakem 2008).
UV radiation introduces cyclopyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPDs) into DNA. These bulky DNA lesions are removed by the nucleotide excision repair (NER) pathway (Shuck et al. 2008). Initially, CPDs and 6-4PPDs are recognized through either specific DNA-binding complexes (DDB or XPC-RAD23B) or stalled RNA polymerase complexes. Subsequently, the concerted actions of the TFIIH–XPA–RPA complex unwind the damaged region of DNA, and the defective strand is excised by structure-specific endonucleases (ERCC1-XPF and XPG). The DNA duplex is then repaired through gap polymerization and DNA ligation (Fousteri and Mullenders 2008; Shuck et al. 2008). Both NER intermediates and stalled replication forks resulting from UV DNA damage activate the DNA damage checkpoint kinases ATR, ATM, and DNA-PKcs (Zou and Elledge 2003; Stiff et al. 2006; Yajima et al. 2006). These kinases, with the aid of mediator proteins such as 53BP1 and MDC1, initiate the assembly of damage checkpoint complexes at repair sites (Rappold et al. 2001; Goldberg et al. 2003; Harper and Elledge 2007). More than 700 ATM and ATR target proteins have been identified recently, including proteins involved in cell cycle arrest, DNA replication, chromatin remodeling, and NER (Matsuoka et al. 2007; Stokes et al. 2007). Thus it appears that checkpoint complexes containing ATM, ATR, and DNA-PKcs orchestrate the DNA damage response through multiple pathways, including coordinating processes such as growth arrest and replication arrest with DNA repair.
Signaling to gene expression plays a major role in the DNA damage response. Both UV and ionizing radiation increase the transcription of hundreds of genes, including genes directly involved in NER such as XPC, DDB2, PCNA, and Gadd45A (Rieger and Chu 2004; Boerma et al. 2005; da Costa et al. 2005). However, it has also been reported that cellular protein synthesis is markedly inhibited after UV irradiation (Deng et al. 2002; Wu et al. 2002). This apparent paradox could be explained by the differential regulation of mRNA translation, which has been observed under other conditions of cell stress (Spriggs et al. 2008). Large-scale profiling of mRNA translation efficiencies during cell stress has revealed that certain mRNAs evade the global inhibition of protein synthesis (Johannes et al. 1999; Blais et al. 2004; Bushell et al. 2006; Thomas and Johannes 2007; Spriggs et al. 2008). Furthermore, many of these mRNAs are translated using alternative mechanisms of translation initiation, such as internal ribosome entry (Johannes et al. 1999; Bushell et al. 2006). In general, these mRNAs encode proteins essential to the stress response. For example, selective mRNA translation results in increased synthesis of chromatin remodeling proteins during apoptosis, whereas during hypoxia, mediators of the unfolded protein response are preferentially translated (Blais et al. 2004; Bushell et al. 2006).
Here we show that UVB DNA damage reduces the global rate of protein synthesis and increases phosphorylation of the translation initiation factor eukaryotic initiation factor α (eIF2α). However, despite the general repression of translation, mRNAs encoding NER proteins are selectively recruited to the polysomes, and moreover, these mRNAs are efficiently translated. In addition, we established that upstream ORFs (uORFs) in the 5′ untranslated regions (UTRs) of these mRNAs play a vital role in the mechanism of selective mRNA translation. Both the inhibition of protein synthesis and the selective synthesis of NER proteins depend on UVB-induced DNA-PKcs activity. Therefore we demonstrated for the first time that signaling through the DNA damage checkpoint kinase, DNA-PKcs, coordinates the reprogramming of mRNA translation in response to UVB DNA damage.
Results
Exposure of HeLa cells to UVB light causes a reduction in protein synthesis
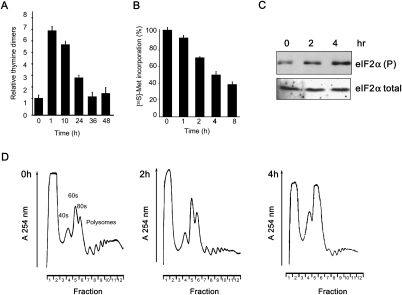
HeLa cells were exposed to a nonlethal dose of UVB light (275 J/m2) (see Supplemental Fig. S1A,B) or mock-treated, lysed, and the DNA examined to determine the extent of production of thymidine dimers as described previously (Mori et al. 1991). One hour after exposure, there was an approximately sevenfold increase in the thymidine dimers present in the DNA that were repaired by the cell after 36 h (Fig. 1A). To assess the effect that exposure to UVB light had on translation, global protein synthesis rates were measured, and the data show that these are reduced to ∼35% after 8 h (Fig. 1B). The levels and phosphorylation status of eIFs were determined, and in agreement with other studies of irradiation (Deng et al. 2002; Jiang and Wek 2005), the decrease in global translation rates appears to be mediated, in the most part, by a change in the phosphorylation state of the α subunit of eIF2 (Fig. 1C; Supplemental Fig. S1C). There was no change in the levels of eIF4G (or any increase in the cleavage products consistent with the nonapoptotic state of the cells) (Supplemental Fig. S1A,B) or in phosphorylation status of the eIF4E inhibitor 4EBP1 (Supplemental Fig. S1C). Moreover, 4 h following UVB exposure, no cell cycle arrest was detected (Supplemental Fig. S2). To examine the association of ribosomes with the total cellular population of mRNAs following UVB irradiation, cytoplasmic extracts prepared from both control and treated HeLa cells were subjected to sucrose density gradient analysis. Exposure of HeLa cells to UVB light resulted in a considerable decrease in the amount of polysomes and a corresponding increase in the abundance of the 40S and 60S complexes (Fig. 1D). There was no significant RNA degradation at these time points, and Northern analysis to compare the steady-state levels of ribosomal RNA, actin, and ribosomal protein S16 (rpS16) mRNAs in control cells and UVB-exposed cells demonstrated that there was no change after 8 h (Supplemental Fig. S1D–F). Taken together, these data are consistent with inhibition of protein synthesis at the initiation stage.
Figure 1.
Inhibition of protein synthesis following UVB-induced DNA damage. HeLa cells were mock- or UVB-irradiated (275 J/m2) and harvested at the times shown following exposure. (A) The relative amount of cyclo-pyrimidine dimers (CPDs) produced by this treatment were measured by ELISA using a monoclonal antibody specific to CPDs. Measurements are the mean of four independent experiments, and error bars represent 1 standard deviation from the mean. (B) Protein synthesis rates were determined by liquid scintillation counting of newly incorporated [35S]-methionine at the time points indicated. Measurements are the mean of three independent experiments normalized to that of the unirradiated cells. Error bars represent 1 standard deviation from the mean. (C) Cell lysates derived from control and UVB-irradiated cells were separated by SDS-PAGE, immunoblotted, and probed with antibodies against eIF2α. There was a significant change in the phosphorylation status of eIF2α but no other eIFs (see Supplemental Fig. S1C). (D) Cell lysates were separated on (10%–50%) sucrose density gradients and the absorbance across the gradient read at 254 nm. Positions of the 40S, 60S ribosomal subunits and polysomal fractions are indicated.
cDNA microarray analysis shows that a subset of mRNAs are subject to differential translational regulation following UVB irradiation
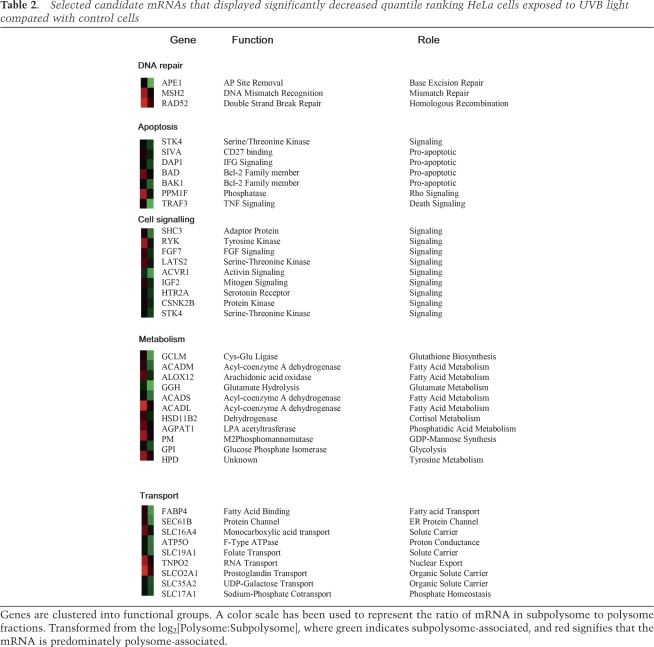
It was then important to identify those individual mRNAs that still remain polysomally associated following UVB exposure. Thus the polysome/subpolysome distribution of mRNAs was analyzed in untreated and UVB-irradiated cells using cDNA microarray (this type of analysis rules out changes in transcription that may occur; see Spriggs et al. 2008 for further information). Sucrose density gradients were fractionated into ∼12 fractions, and RNA derived from sucrose gradient fractions 7–12 (polysomes) was compared with RNA from fractions 1 to 6 (subpolysomes) to obtain a measure of the translational status of mRNAs in both control and UVB-exposed cells (Fig. 1D). The majority of mRNAs displayed reduced association with the polysomes following UVB exposure (Fig. 1D). However ∼8% of mRNAs were resistant to the UVB-induced translational inhibition and did not relocate from the polysomes into the subpolysomes. Together these data imply that certain mRNAs are subject to differential translational regulation following UVB exposure. Distinct categories of mRNAs that displayed either increased or decreased relative translational efficiency following UVB exposure were identified (Tables 1, 2). Within the group of genes whose relative translational efficiency was increased following UVB irradiation, ∼9% encode proteins with roles in DNA damage repair processes (Supplemental Table S3). These include several genes encoding members of the NER pathway including XPA, ERCC5, ERCC1, and DDB1, and interestingly, these were found to be unchanged when transcriptional profiling was performed following UVC exposure (Rieger and Chu 2004). Other mRNAs whose protein products have a survival role and anti-apoptotic role—e.g., TEGT, Bcl-2, and Mcl-1 (Table 1)—were also shown to be associated with the polysomes. The mRNAs whose translational efficiency was substantially reduced in UVB-irradiated cells include those with proapoptotic functions (e.g., BAD, BAK1, and SIVA [Table 2]) and death signaling (e.g., TRAF3 [Table 2]). These two sets of data are different from those obtained when cells were treated with TNF-related apoptosis-inducing ligand (TRAIL) to induce apoptosis (Supplemental Tables S1, S2; Bushell et al. 2006), showing that there is coordinated recruitment of a specific subset of mRNAs to the polysomes following UVB exposure.
Table 1.
Selected candidate mRNAs that displayed significantly increased quantile ranking HeLa cells exposed to UVB light compared with control cells
Genes are clustered into functional groups. A color scale has been used to represent the ratio of mRNA in subpolysome to polysome fractions. Transformed from the log2[Polysome:Subpolysome], where green indicates subpolysome-associated, and red signifies that the mRNA is predominately polysome-associated.
Table 2.
Selected candidate mRNAs that displayed significantly decreased quantile ranking HeLa cells exposed to UVB light compared with control cells
Genes are clustered into functional groups. A color scale has been used to represent the ratio of mRNA in subpolysome to polysome fractions. Transformed from the log2[Polysome:Subpolysome], where green indicates subpolysome-associated, and red signifies that the mRNA is predominately polysome-associated.
Changes in gene expression and the rate of synthesis of specific proteins following UVB exposure correlate with cDNA microarray analysis
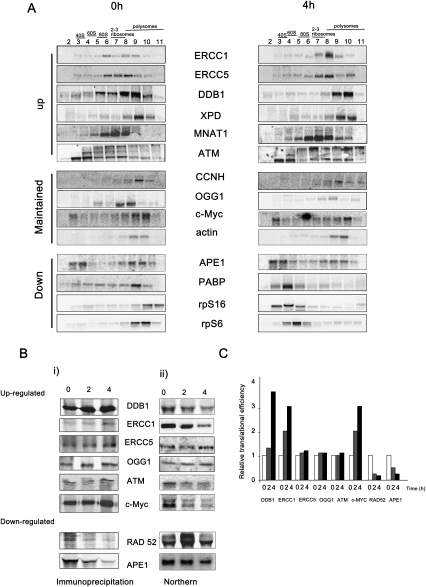
To verify the data from the microarray analysis, Northern analysis was also performed (Fig. 2A). According to the microarray analysis, APE1 (Table 2), PABP, rpS16, and rpS6 (data not shown) mRNAs showed a marked decrease in their relative translational efficiency following UVB exposure. In agreement with these data, Northern analysis of sucrose density gradient fractions showed a substantial reduction in the amount of these mRNAs that sedimented with the polysomes and a concomitant increase in the mRNA associated with the subpolysomal fractions (Fig. 2A). Conversely, according to the microarray analysis, ERCC5, DDB1, ERCC1 (Table 1), and XPD (data not shown) mRNAs all showed a marked increase in their relative translational efficiency following UVB exposure. In agreement, these mRNAs are all associated with fractions that contained more polysomes following UVB exposure (Fig. 2A; Supplemental Fig. S3 for quantification of these data). Interestingly, the mRNAs that relocated to heavier polysomes following UV-exposure were found to be associated with two or more ribosomes under control conditions (Fig. 2A). Thus, the mRNAs that encode ERCC1, ERCC5, DDB1, XPD, and MNAT1 are all found to be associated with ribosomes, with very little message associated with the 40S or 60S ribosomal fraction, in comparison with APE1 and PABP (Fig. 2; Supplemental Fig. S3). It has been shown previously that messages that contain uORFs in their 5′ UTRs are generally associated with two or more ribosomes (Zhou et al. 2008), suggesting that many of the mRNAs identified by the screen (Table 1) contain uORFs. The mRNAs encoding c-Myc and OGG1 remain associated with approximately the same number of ribosomes and showed no net change in their positions on the gradients (Fig. 2A; Supplemental Fig. S3). Collectively, these data show that mRNAs that encode proteins required for the DDR are subject to differential polysomal association following UVB exposure (Fig. 2; Tables 1, 2).
Figure 2.
Post-transcriptional changes in gene expression and changes in the rate of protein synthesis of a number of polypeptides confirm the polysome profiling data. (A) Untreated HeLa cells or HeLa cells exposed to UVB light were harvested after 4 h and applied to 10%–50% ([w/v] sucrose density gradients). mRNA was generated from the individual fractions from the gradients, and Northern analysis was performed to determine the polysome/subpolysome distribution of the mRNAs shown. Representative blots are shown of paired experiments carried out at the same time, but analysis was performed on three independent occasions. These data are in agreement with that obtained from polysome profiling. For example, the data would predict that DDB1 becomes more polysomally associated following UVB exposure while APE1 becomes less polysomally associated following UVB exposure. (B, panel i) To measure the rates of protein synthesis of DDB1, ERCC1, ERCC5, OGG1, ATM, c-Myc, RAD52, and APE1, untreated HeLa cells and HeLa cells 2 and 4 h following UVB exposure were incubated with [35S]-methionine for 10 min prior to harvesting. Total cell lysates were prepared from these samples, and the proteins shown were immunoprecipitated using specific antibodies. Immunoprecipitated proteins were resolved by SDS-PAGE and detected using PhosphorImager analysis. These experiments were performed on three independent occasions. (Panel ii) To measure total RNA levels, parallel samples were taken, and mRNA was extracted. These samples were subjected to Northern analysis, and DDB1, ERCC1, ERCC5, OGG1, ATM, c-Myc, RAD52, and APE1 mRNAs were detected using specific probes as indicated. These experiments were performed on three independent occasions. (C) The data obtained from the experiments shown in A and B (carried out in parallel) were analyzed using ImageQuant software to measure the fold-change for each of the mRNAs and proteins following UVB exposure. This allowed the calculation of the relative translational efficiency. The data show that there is an increase in translational efficiency for proteins that were found to be more polysomally associated (e.g., DDB1) but a marked decrease in translational efficiency in the proteins that were less polysomally associated (e.g., RAD52).
To determine whether the changes in polysome association reflected rates of synthesis of specific polypeptides, cells were exposed to UVB light and pulse-labeled for 5 min with [35S]-methionine before harvesting at the times indicated post-irradiation, cell lysates were prepared and proteins immunoprecipitated. The rate of synthesis of DDB1, ERCC1, ERCC5, OGG1, ATM, and c-Myc all increased following exposure of cells to UVB (Fig. 2Bi). For example, 4 h following exposure of cells to UVB light, there is a reduction in global protein synthesis rates to 50% (Fig. 1B); yet under these conditions, the rates of synthesis of c-Myc and DDB1 have increased by approximately twofold (Fig. 2B, panel i). Moreover these changes are not reflected by alterations in the total amount of mRNA present, and in the parallel samples that were taken for Northern analysis, there was a decrease in the corresponding mRNAs following UVB exposure (Fig. 2B, panel ii). In contrast, the rates of synthesis of APE-1 and RAD52 decreased to a greater extent than the global protein synthesis rates to 19% and 3%, respectively (Fig. 2B, panel i), and again, while there was a decrease in the levels of these mRNAs, these are insufficient to account for the changes in protein synthesis observed (Fig. 2B, panel ii). The images obtained from these paired experiments were scanned and quantified using ImageQuant, and these data were used to calculate the relative translational efficiency. The data show that in the case of DDB1, ERCC1, and c-Myc, there is an overall increase in translational efficiency, albeit to different extents (Fig. 2C). There was a small, but reproducible, increase in the synthesis of ERCC5 (∼1.8-fold), and an increase in the amount of ERCC5 mRNA that was polysomally associated mRNA following UVB exposure (Fig. 2A). In contrast, there was a large decrease in the translational efficiency of RAD52 and APE1 (Fig. 2C). These changes in the rate of protein synthesis correlate well with the microarray data and the polysome analysis with the exception of c-Myc. The increased translational efficiency of c-Myc following UV exposure is perhaps rather surprising given that there was no net change in position on the sucrose density gradient (Fig. 2A). However, we showed previously that c-myc mRNA contains an IRES within the 5′ UTR that is active following DNA damage (Subkhakulova et al. 2001), a target binding site for miR34C within the 3′ UTR, and that in control cells, c-myc mRNA is translationally repressed yet present on the polysomes (Kong et al. 2008). One explanation for these data is that following UVB exposure, a relief of miR repression allows the increased synthesis of c-Myc by cap-independent mechanisms (Fig. 2B,C).
uORFs allow translational reprogramming of DNA repair enzymes following UVB exposure
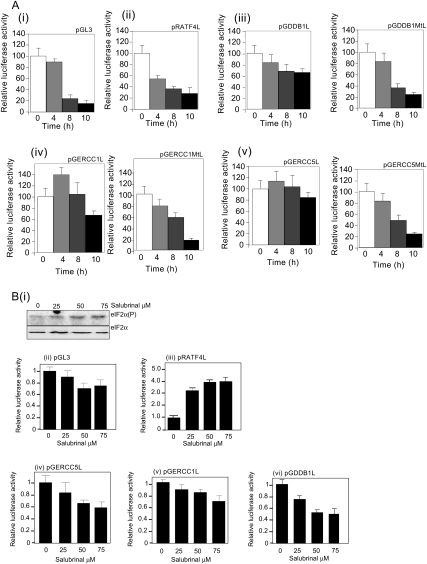
There are several mechanisms that could be responsible for the selective recruitment of mRNAs to the polysomes, and these could involve distinct sequence motifs in both the 3′ and 5′ UTRs of these mRNAs (Gebauer and Hentze 2004). Examination of the 5′ UTRs of the members of the repair pathway that remain polysomally associated following UVB show that seven out of eight of these mRNAs contain uORFs compared with ∼30% of the total genome (Matsui et al. 2007), which is approximately a threefold enrichment. The 5′ UTRs of ERCC1, ERCC5, and DDB1 (Supplemental Fig. S4A) were chosen for further study. To investigate whether these regions of RNA could be involved in the DDR, the cDNAs that corresponded to these 5′ UTRs were subcloned into pGL3 upstream and in-frame with the gene encoding firefly luciferase (for vector diagrams, see Supplemental Fig. S4B), and the effect of these 5′ UTRs on translation of the downstream reporter firefly luciferase tested (Supplemental Fig. S4C, panel i). To test whether these 5′ UTRs had a role in maintaining expression of the corresponding mRNAs following UVB exposure, these constructs (Supplemental Fig. S4B) were then transfected into HeLa cells, and after 48 h, cells were exposed to UVB light. Samples were harvested at the times shown following irradiation, and luciferase activity was determined (Fig. 3A, panel i–v). The data show that in the presence of UVB light, there is a reduction in expression of luciferase from the mRNAs encoded by pGL3 (Fig. 3A, panel i) and the mRNAs that contain the mutated versions of the 5′ UTRs encoded by the plasmids pGDDB1mtL, pERCC1mtL, and pERCC5mtL (Fig. 3A, panels ii,iii,iv; where the AUG codons have been mutated to AUA) by ∼85% after 10 h. The presence of the wild-type 5′ UTRs derived from ERCC1, ERCC5, and DDB1 allows luciferase expression to be maintained to approximately a four times higher level when compared with the mutated constructs, suggesting that these upstream regions do indeed have a role in maintaining synthesis of the related mRNAs following UVB exposure (Fig. 3A, panels iii,iv,v) at 10 h. Moreover, the data show that at 4 h, there is an increase in the synthesis of luciferase produced from the vectors that harbor the ERCC1 and ERCC5 5′ UTRs in agreement with the increased rate of synthesis of the endogenous proteins observed at this time point (Fig. 2C). However, other elements must also be required for DDB1 synthesis since endogenous synthesis rates of this protein increase 4 h after UVB exposure (Fig. 2C). Interestingly, there was no increase in the production of luciferase from the vector that contained the 5′ UTR of ATF4 (Fig. 3A, panel ii). This 5′ UTR contains a uORF that has been shown previously to be selectively translated during conditions of low ternary complex due to an increase in the phosphorylation of eIF2α (Harding et al. 2000; Lu et al. 2004; Hinnebusch 2005). Therefore, to determine whether the increase in phosphorylation of eIF2α alone was sufficient for the DNA damage uORFs identified to be functional, two alternative methods were used to induce an increase in the phosphorylation eIF2α, namely, exposure of cells to salubrinal (Fig. 3B) and thapsigargin (Supplemental Fig. S4C, panel i). The data show that, as expected, the uORF contained within the 5′ UTR of ATF4 was sufficient to increase the expression of luciferase fourfold following exposure of cells to salubrinal (Fig. 3B, panel ii); however, no such increase was observed from the cells transfected with vectors that contained the 5′ UTRs found in ERCC5, ERCC1, or DDB1 (Fig. 3B, panels iv–vi). These data would strongly suggest that phosphorylation of the α subunit of eIF2 alone is insufficient to elicit a full response to the translational reprogramming that occurs following the exposure of cells to UVB light, suggesting that additional signaling pathways and proteins must be involved.
Figure 3.
uORFs allow continued synthesis of DNA repair enzymes. (A) The plasmid constructs that contained the uORFs derived from DDB1, ERCC1, and ERCC5 (and the mutated versions) (S3A) were transfected into HeLa cells, which were then exposed to UVB light. After the times shown post-irradiation, the cells were lysed, and luciferase activity was determined. Since translation of the transfection control was also reduced following UVB exposure, mRNA was isolated from cell lysates, and the amount of firefly luciferase mRNA present in the cells was measured by quantitative PCR. The luciferase expression was then calculated relative to the level of mRNA present. The error bars represent one standard deviation from the mean. The data show that the presence of the 5′ UTRs derived from DDB1 (panel ii), ERCC1 (panel iii), or ERCC5 (panel iv) confer resistance to translational repression, and luciferase is still produced following UVB exposure. However, mutating the AUG codons is sufficient to inhibit the production of firefly luciferase following UVB exposure. More of the 5′ UTR derived from ATF4 was insufficient to increase the expression of luciferase. (B, panel iii) Cells were transfected with the constructs shown and then treated with salburinal for 12 h. (Panel i) This compound is a selective inhibitor of complexes that dephosphorylate eIF2α (Boyce et al. 2005), and in the presence of this compound, there was an increase in the phosphorylation of the α subunit of eIF2 as expected. However, the data show that in contrast to ATF4 (panel ii) the 5′ UTRs derived from ERCC5 (panel iv), ERCC1 (panel v), or DDB1 (panel vi) were not resistant to the effects of this compound, and the decrease in translation following treatment was similar to that found with the pGL3 reporter vector. Taken together, these data suggest that a reduction in ternary complex levels alone is insufficient to elicit a response.
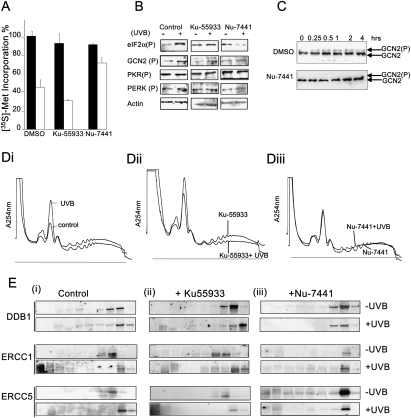
DNA-PKcs is required for translational repression following cellular UVB exposure
To identify the cellular sensors that are involved in the UVB-mediated translational response, the role of kinases involved in the DDR were investigated. Thus, specific inhibitors of two of the kinases were used: Ku-55933, which has been shown to inhibit ATM (Hickson et al. 2004), and Nu-7441, which inhibits DNA-PKcs (Zhao et al. 2006). Moreover, when Nu-7741 was tested in vitro at a concentration of 10 μM against a panel of 60 diverse kinases, no inhibition of kinase activity in this panel was observed (Hardcastle et al. 2005). HeLa cells were exposed to UVB light in the presence/absence of these inhibitors and then pulse-labeled with [35S]-methionine 4 h after exposure (Fig. 4A). The presence of Ku-55933 alone causes a small reduction in total protein synthesis rates, and these are further inhibited following UVB exposure. In contrast, while treatment of cells with Nu-7441 also had a small effect on protein synthesis in control cells, in the presence of this compound, the translational repression observed following UVB exposure was inhibited (Fig. 4A). Western analysis was performed to assess the effects of these compounds on the phosphorylation of the α subunit of eIF2 (Fig. 4B). Treatment of cells with Ku-55933 causes an increase in phosphorylation of the α subunit of eIF2 in nonirradiated cells; however, following UVB exposure, this is increased further. In the presence of UVB light and the DNA-PKcs-specific inhibitor NU-7441, there was no additional increase in the phosphorylation of the α subunit of eIF2, suggesting that this compound inhibits the translational repression that occurs following UVB exposure (Fig. 4B). It has been shown previously that signaling through GCN2 is required to inactivate translation following UV exposure (Deng et al. 2002; Jiang and Wek 2005). Therefore, the effect that these inhibitors had on the phosphorylation status of GCN2 and other eIF2 kinases, PRK and PERK, following UVB-exposure was investigated (Fig. 4B). The data show that while there is no increase in the phosphorylation of PKR and PERK following exposure of cells to UVB light, there is an increase in the phosphorylation of GCN2. Moreover, incubation of HeLa cells with the DNA-PKcs (Nu-7441) but not the ATM (Ku-55933) inhibitor blocks the activation of GCN2 (Fig. 4B). To examine this effect on GCN2 in the presence of inhibitors, further cells were transfected with cDNA encoding GCN2 protein that additionally encoded a Flag tag. Cells were exposed to UVB light in the presence/absence of the inhibitors, lysed, and the GCN2 protein was immunoprecipitated from cell extracts (Fig. 4C). It can be seen that following UVB exposure, there is an increase in the degree of phosphorylation of GCN2. However, in the presence of Nu-77441, there is no decrease in GCN2 phosphorylation, again suggesting that the inhibition of protein synthesis occurs via signaling from DNA-PKcs to GCN2 (Fig. 4C).
Figure 4.
DNA-PKcs is required for both translational inhibition and reprogramming of protein synthesis. (A) HeLa cells were pretreated with inhibitors of DNA-PKcs (Nu-7441), ATM (Ku-55933), or DMSO control and then exposed to UVB light. Protein synthesis rates determined by liquid scintillation counting of newly incorporated [35S]-methionine 4 h following exposure. Measurements are the mean of three independent experiments, and the values were normalized to that of the nonirradiated cells. Error bars represent one standard deviation from the mean. (B) To assess the effect that the ATM (Ku-55933) or DNA-PKcs (Nu-7441) inhibitors had on the phosphorylation status of eIF2α and the eIF2α kinases shown (GCN2, PRK, and PERK), cells were preincubated with these compounds and exposed to UVB light, and samples were harvested after 4 h. Cell lysates generated from these samples were separated by SDS-PAGE and immunoblotted with the antibodies shown. In the presence of the ATM inhibitor (Ku-55933), there is an increase in the degree of phosphorylation of eIF2α in nonirradiated cells; however, following UVB exposure, there is further phosphorylation of this protein. In contrast, in the presence of the DNA-PKcs inhibitor (Nu-7441), there is no increase in the phosphorylation of eIF2α following UVB exposure. The kinases that are known to phosphorylate eIF2α, GCN2, PKR, and PERK were also tested, and the DNA-PKcs inhibitor blocks the phosphorylation of GCN2 but not the other eIF2α kinases. (C) To assess the effect that UVB light and the DNA-PKcs inhibitor had on GCN2, cells were transfected with a plasmid that encodes GCN2 containing a Flag tag. Following exposure with UVB in the presence/absence of the DNA-PKcs inhibitor GCN2, protein was immunoprecipitated and separated by SDS-PAGE and immunoblotted for total GCN2. The data show that in the presence of the DNA-PKcs inhibitor GCN2 phosphorylation is inhibited. (D) HeLa cells were pretreated with the ATM inhibitor (Ku-55933) (panel ii) or the DNA-PKcs (Nu-7441) (panel iii) or untreated and were then either exposed to UVB light. Cell lysates were prepared 4 h following exposure and were applied to 10%–50% (w/v) sucrose density gradients. These were fractionated and the absorbance across the gradient measured at 254 nm. (E) Cell lysates from HeLa cells ±Ku-55933 or ±Nu-7441 and treated with ±UVB light were fractionated using sucrose density gradient analysis on 10%–50% (w/v) sucrose gradients. Northern analysis was performed on the mRNA isolated from the individual fractions to determine the polysome/subpolysome distribution of DDB1, ERCC1, and ERCC5.
To examine the association of ribosomes with the total cellular population of mRNAs in the presence of these inhibitors following UVB irradiation, cytoplasmic extracts prepared from both control and treated cells were subjected to sucrose density gradient analysis (Fig. 4D). The data show that in cells incubated with the ATM inhibitor (Ku-55933) alone there is a partial inhibition of total protein synthesis, since there is in an increase in the subpolysomes and a decrease in the amount of polysomal material on the gradient (in agreement with the increase in the phosphorylation of the α subunit of eIF2 observed) (Fig. 4D, panel ii). However, following UVB exposure, there is a further reduction in the amount of polysomal material, in agreement with the inhibition of protein synthesis observed (Fig. 4A,B). In the presence of the DNA-PKcs inhibitor (Nu-7441) alone, there is a slight increase in the amount of subpolysomally associated material (Fig. 4D, panel iii). However, following UVB exposure, there is no further decrease in the amount of polysomally associated mRNAs, and the gradient traces are virtually identical in agreement with the [35S]-methionine incorporation data (Fig. 4A). It was then necessary to determine whether signaling from DNA-PKcs was required for the relocalization of these mRNAs. Thus the effect that a reduction in the activity or levels of this protein had on polysomal association of mRNAs encoding DDB1, ERCC1, and ERCC5 was determined (Fig. 4E). In the presence of the ATM inhibitor (Ku-55933), all three mRNAs associated with an increased number of ribosomes following UVB exposure (Fig. 4E, panel ii), whereas following incubation with DNA-PKcs inhibitor (Nu-7441), this was prevented (Fig. 4E, panel iii).
These data would suggest that signaling from DNA-PKcs is required for the translational reprogramming that occurs following UVB exposure.
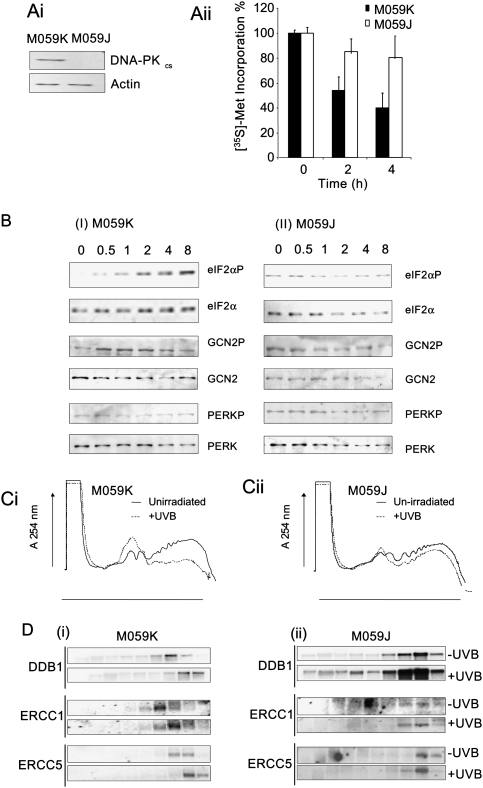
To confirm a role for DNA-PKcs in translational inhibition, RNAi was used to reduce the level of DNA-PKcs (Supplemental Fig. S5). However, although the data obtained agree with that obtained using the inhibitors (Fig. 4A–C), it was difficult to reduce the levels of DNA-PKcs to more than ∼50% via this technique. Therefore, a DNA-PKcs-deficient cell line, M059J, was used in conjunction with its related paired cell line, M059K (Allalunis-Turner et al. 1993) to further examine the role of this protein in translational inhibition (Fig. 5A, panel i). These cells were exposed to UVB light, pulse-labeled with [35S]-methionine, and harvested at the times shown following UVB exposure (Fig. 5A, panel ii). Four hours after UVB exposure, the rate of protein synthesis in M059J cells was reduced to 85% while in M059K cells, it was reduced to 40% in agreement with the data obtained with the DNA-PKcs inhibitor (Fig. 5A, panel ii). Cells were then exposed to UVB light, and the degree of phosphorylation of eIF2α, GCN2, and PERK was determined. The data show that in cells that contain DNA-PKcs there is an increase in the degree of phosphorylation of eIF2α and GCN2; however, in the cells that lack DNA-PKcs, there was no increase (Fig. 5B), in agreement with the data obtained with the selective DNA-PK inhibitors (Fig. 4). To determine the effect on recruitment of selective mRNAs to the ribosomes, cells were then exposed to UVB light and the total ribosomes analyzed (Fig. 5C, panels i,ii). These data show that in cells that contain DNA-PKcs, there is a reduction in the polysomally associated mRNAs following UVB exposure (Fig. 5C,panel i) but not in the cells that have no expression of DNA-PKcs (Fig. 5C, panel ii). The RNA obtained from these gradients was probed to assess changes in polysomal location for DDB1, ERRC1, and ERCC5 (Fig. 5D). The data show that, in M059K cells upon UVB exposure, all three mRNAs become associated with an increased number of ribosomes (Fig. 5D, panel i), yet in the DNA-PKcs-deficient M059J cells, this was again inhibited (Fig. 5D, panel ii). Taken together, these data provide further evidence to show that following UVB exposure, signaling via DNA-PKcs is required to mediate translational repression.
Figure 5.
(A, panel i) Cell lysates from M059K or M059J cells were prepared separated by SDS-PAGE and immunoblotted for DNA-PKcs. As expected, no DNA-PKcs was detected in M059J cells. (panel ii) M059K or M059J cells were exposed to UVB light, and protein synthesis rates were determined by liquid scintillation counting of newly incorporated [35S]-methionine. Measurements are the mean of three independent experiments normalized to that of the nonirradiated cells. Error bars represent one standard deviation from the mean. (B) Cell lysates from M059K (panel i) orM059J (panel ii) cells were exposed to UVB light and immunoblotted for eIF2α, GCN2, and PERK. The data show that there in no change in the degree of phosphorylation of eIF2α or GCN2 in cells that lack DNA-PKcs. (C) M059K (panel i) orM059J (panel ii) cells were exposed to UVB light and, after 4 h, cell lysates were prepared and fractionated using sucrose density gradient analysis on 10%–50% (w/v) sucrose gradients. (D) Cell lysates were generated from M059K (panel i) orM059J (panel ii) cells ± exposure to UVB light and applied to 10%–50% (w/v) sucrose gradients. mRNA was isolated from individual fractions, and Northern analysis was performed to determine the polysome/subpolysome distribution of DDB1, ERCC1, and ERCC5.
Discussion
Our data suggest that there are two separate effects on translation as a result of UVB exposure, a reduction in total protein synthesis rates, and a reprogramming of the translation of a subset of mRNAs, particularly those required for the DDR, and this allows the increased synthesis of proteins required to repair the damage (Fig. 2; Tables 1, 2). Importantly, the data show that the mRNAs that are recruited to the polysomes are specific to UVB exposure (Fig. 2; Tables 1, 2). Thus in parallel experiments to examine which mRNAs remain polysomally associated during apoptosis (Supplemental Tables 1, 2; Bushell et al. 2006), and in studies by others that have examined polysomal association of mRNAs following hypoxia and ER stress when translation is also reduced by increases in phosphorylation of the α subunit of eIF2 (Blais et al. 2004; Thomas and Johannes 2007), members of the excision repair pathway were not identified. To identify whether there was a common RNA motif among the mRNAs that were found to be up-regulated following UVB exposure whose protein products were associated with DNA repair processes, the 5′ and 3′ UTRs of these mRNAs were examined. The data show that there is a prevalence of uORFs in the 5′ UTRs of these mRNAs (Supplemental Fig. S4), and these elements help to maintain protein synthesis of the mRNAs tested following DNA damage (Fig. 3). In the case of ERCC1 and ERCC5 5′ UTRs 4 h following UVB exposure, there is an induction in luciferase expression that is similar to that observed with the endogenous protein (Figs. 2C, 3A [panels iv, v]). However, the presence of these elements is not sufficient to maintain protein synthesis under other conditions in which levels of ternary complex are reduced (Fig. 3B; Supplemental Fig. S4), and we hypothesize that certain trans-acting factors are also required, and as part of the UV damage response, the interaction of specific proteins with the uORFs identified could control their function.
We investigated the signaling pathways that could be involved in the translational response, and our data suggest a role for DNA-PKcs in both the translational shutdown and the reprogramming. These data were surprising for two reasons. Firstly, previous studies have suggested that DNA-PKcs plays a major role in nonhomologous end-joining, ATM responds primarily to double-strand breaks, and ATR reacts to UVB or stalled replication forks (Yang et al. 2003). However, it has been shown that following UV damage, ATR phosphorylates DNA-PKcs, and the data suggest that DNA-PKcs could be a direct downstream target of ATR (Yajima et al. 2006; Stokes et al. 2007). These data agree with earlier studies that suggest that DNA-PKcs has a role in NER (Muller et al. 1998) and cell cycle arrest following UV exposure (Park et al. 1999). The data shown herein support a role for this protein in the UV damage response and strongly suggest that signaling via DNA-PKcs is required to inhibit gene expression at the level of translation following UVB exposure. Secondly, a direct role for DNA-PKcs in the control of translation is difficult to reconcile with the nuclear localization of this protein. However, a subpopulation of DNA-PKcs has been shown to be associated with the plasma membrane (Feng et al. 2004), and it has been shown recently that cAMP regulates the nuclear/cytoplasmic trafficking of DNA-PKcs (Huston et al. 2008). While further studies are required to fully elucidate the mechanism of DNA-PKcs signaling to the translation machinery following UVB exposure, our data suggest that this protein either directly or indirectly controls the phosphorylation of GCN2 (Figs. 4, 5). These data agree with previous studies that show that in UVC-irradiated cells, GCN2 was responsible for phosphorylation of the α subunit of eIF2. These data showed that cells that lacked GCN2 were unable to repress translation following UV irradiation, whereas inhibition of translation was still observed in cells that were deficient for PKR or PERK (Deng et al. 2002; Jiang and Wek 2005).
Finally, our data provide further evidence for control of gene expression by “RNA operons” (Keene 2007), whereby specific RNA motifs and their cognate RNA binding proteins organize RNA transcripts to ensure that the correct proteins are synthesized when required by the cell, particularly under conditions of patho-physiological cell stress (Keene 2007). Such RNA operons are coregulated and are therefore capable of forming higher-order “RNA regulons” (Keene 2007), and we hypothesize that it is likely that “DNA damage regulons” may exist to post-transcriptionally regulate gene expression following alternative types of DNA damage that is meditated, for example, by exposure of cells to alkylating agents or ionizing radiation.
Materials and methods
Plasmid constructs
The plasmids used are described in Coldwell et al. (2001). The 5′ UTRs of mRNA as indicated were obtained by PCR using primer pairs (Supplemental Fig. S5). These fragments were then cloned into the pGL3 vector at the 5′ end with either EcoRI or SpeI as indicated by sequence.
Antibodies
Antibodies were purchased from Cell Signaling XPG, DDB1, APE1, DNA-PKcs, RAD52, PARP, eIF2 α, eIF2α (P), β-actin, 4EBP1, and eIF4E and used at 1:2000 (with the exception of actin, 1:10,000) neomarkers, c-Myc, (1:2000), Gentex, ATM (1:5000), or were a gift from Dr. S. Morley (University of Sussex); eIF4G, eIF4E (P).
Cell culture and transient transfections
HeLa cells were cultured using conditions described on the American Type Culture Collection Web page (see http://www.atcc.org). Cell lines M059J and M059K were a kind gift (CRUK, Clare Hall Laboratories) and were grown as above but in a medium containing a 1:1 mixture of Dulbecco's Modified Eagle's Medium and Ham's F12 medium supplemented with 15 mM HEPES, 0.5 mM sodium pyruvate, 1.2 g/L sodium bicarbonate, 0.05 mM nonessential amino acids (Sigma), 2.5 mM L-glutamine (Sigma), and 10% FCS (Helena Biosciences).
Ultraviolet induction of DNA damage
Cells were typically grown to 80%–90% confluency, and their media was removed before being exposed to 275 J/m2 UVB-irradiation from a UVBM-57 lamp (peak wavelength 302 nm) (UVBP Products), and their media were then replaced. Consistent with previous studies, this degree of UVB exposure in HeLa cells did not cause a reduction in cell viability (Zhang et al. 1997).
Measurement of DNA damage by ELISA
Levels of CPD formation in native DNA were measured using an ELISA-based technique as described by Mori et al. (1991). Briefly, 96-well PVC microtiter plates were coated with 0.003% protamine sulfate. Genomic DNA was extracted using a Qiamp Blood Kit (Qiagen) according to the manufacturer's instructions at the time points indicated in the text. DNA was then diluted to 0.2 μg/mL concentration and heated for 10 min to 100°C and immediately chilled for 15 min on ice to prevent renaturation. Fifty microliters of each sample were distributed per well using five wells for each sample and then dried completely overnight at 37°C. Following washing with PBS-T (0.05% Tween 20 in PBS), plates were incubated with TDM-2 antibody (1:1000 dilution), which recognizes CPDs in ssDNA; secondary antibody was added at 1:2000 Biotin-F(ab′)2 fragment of anti-mouse IgG (H + L) (Zymed) diluted with PBS and developed using Peroxidase-Streptavidin (Zymed and buffer containing 8 mg of o-phenylene diamine, 4 μL of H2O2 [35%], 20 mL of citrate-phosphate buffer at pH 5.0). The absorbance at 492 nM was measured using a microtiter plate reader (Bio-Tek Instruments).
TRAIL-induced apoptosis
The TRAIL induces apoptosis via the death receptor–mediated pathway of apoptosis. HeLa cells were grown to 90% confluency and treated with 1 μg/mL His-tagged TRAIL (a kind gift from Marion MacFarlane, University of Leicester). Cells were harvested at 4 h post-treatment.
Treatment of cells with small molecule inhibitors
Cells were grown to 80% confluence as described and, 20 min before initial exposure to ultraviolet radiation, were incubated with the small chemical inhibitors Ku-55933 (10 μM) and Nu-7741 (1 μM), before harvesting after 2, 4, or 8 h.
Transient transfections
HeLa cells were transfected using FuGene 6 (Roche) following the manufacturer's instructions. The activities of firefly luciferase in lysates prepared from transfected cells were measured using a Luciferase reporter assay system (Promega), and light emission was measured over 10 sec using an OPTOCOMP I luminometer. Activity was calculated as the average of firefly luciferase expression relative to the level of mRNA (measured by quantitative PCR) and expressed relative to the value obtained in untreated cells. All experiments were performed in triplicate on at least three independent occasions. Errors were calculated as the standard deviation of the three activities and expressed as a percentage of the average activity.
Determination of protein synthesis rates
For determination of protein synthesis rates following UVB exposure, the method used has been described in detail previously (Clemens et al. 1998).
SDS-PAGE and Western blotting
For the detection of eIF4E, eIF4G, 4EBP1, and PARP S10, cell extracts were prepared as described previously (Clemens et al. 1998; Morley et al. 1998) and subjected to electrophoresis on SDS–polyacrylamide gels, and the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Proteins were detected with the relevant antisera using chemiluminescent reagents (West et al. 1998).
Immunoprecipitation
HeLa cells were radiolabeled with [35S]-methionine, and immunoprecipitations were performed as described previously (Stoneley et al. 2000). Samples were subjected to SDS-PAGE, and the amount of radiolabel incorporated was visualized and quantified on a PhosphorImager (Molecular Dynamics). Experiments were performed on three independent occasions, and a representative gel is shown.
Sucrose gradient density centrifugation and RNA detection
Sucrose gradient density centrifugation was used to separate ribosomes into polysomal and subpolysomal forms. Gradients were then fractionated with continuous monitoring at 254 nm, and RNA was isolated from each fraction as described previously (Johannes et al. 1999).
RNA analysis
Northern analysis of RNA isolated from sucrose density gradients was performed as described previously (Johannes and Sarnow 1998). Radiolabeled DNA hybridization probes were generated using the RadPrime kit according to the manufacturer's instructions (Invitrogen). Northern analyses on total RNA samples obtained following UVB exposure were performed as described on at least three independent occasions.
Preparation of fluorescently labeled cDNA for microarray hybridization and data analysis
The human cDNA microarrays contained a set of ∼10,000 human cDNA clones, manufactured in MRC Unit Leicester. Fluorescently labeled DNA probes were generated from equal proportions of RNAs (∼7 μg) of pooled polysomal fractions (fractions 1–6 F, Cy5) and pooled nonpolysomal fraction (fractions 7–11, Cy3). Microarray slides were scanned using a GenePix 4200A microarray scanner and GenePix Pro 5.1 software (Axon Instruments).
Analysis of microarray data
GenePix Pro 5.1 was used to quantify fluorescence intensities for individual spots on the microarray. The data obtained were then normalized using Normalise version NorTT (MRC Toxicology Unit; http://www.systems-toxicology.com). Data were ranked from most polysome-associated to least polysome-associated, and the rank difference was calculated. Genes selected for further study were at least 2 standard deviations from the mean rank difference. Only genes for which the fluorescence signal in each channel exceeded certain cutoffs (including regression correlation, signal–noise ratio, signal intensity, and the tolerance of missing data among multiple measurements) were retrieved from SMD for further analysis. The measure Ii(x) of each gene x defining the polysomal association status was calculated as follows (Qin and Sarnow 2004): 2816 genes passed the SMD filter and were used for the ranking analysis.
Log2 control ratio of gene:
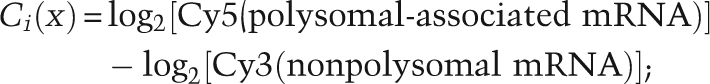 |
(i replicates of control experiment, i = 1:5)
Log2 apoptosis ratio of gene x:
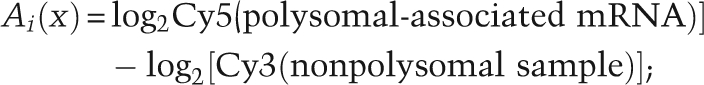 |
(i replicates of UV-exposed experiment, i = 1:5)
The relative standing of values mean[Ai(x)] or mean[Ci(x)] were calculated, respectively, in the ordinal and percentage rank (Fig. 2B). The rank change in percentile of gene x between UVB and control is defined as
The mRNAs remaining associated with polysomes are expected to have a high value of D(x), while the mRNAs shifted dramatically out of polysomes have a relatively low D(x). We used Northern blot analysis to determine the D(x) cutoff value of positive candidate genes for further experimental analyses; approximately the top 3% of the list test positive in this analysis. Microarray experiments were conducted three times, and the results shown are an average of three experiments. Data were further visualized and clustered by using TIGR MeV 4.0 (http://www.tm4.org/mev.html).
Acknowledgments
Thank you to Professor S. Jackson, CRUK Gurdon Institute, Cambridge, for helpful discussions. This work was supported by Cancer Research UK (A.K.), BBSRC (Biotechnology and Biological Sciences Research Council; I.R.P., H.C.D, and L.Y.), and the Wellcome Trust (M.S. and K.A.S.). M.B. is a David Phillips Fellow (BBSRC). A.E.W. is a BBSRC Professorial fellow.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.516509.
Supplemental material is available at http://www.genesdev.org.
References
- Allalunis-Turner MJ, Barron GM, Day RS, III, Dobler KD, Mirzayans R. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat Res. 1993;134:349–354. [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerma M, van der Wees CGC, Vrieling H, Svensson JP, Wondergem J, van der Laarse A, Mullenders LHF, van Zeeland AA. Microarray analysis of gene expression profiles of cardiac myocytes and fibroblasts after mechanical stress, ionising or ultraviolet radiation. BMC Genomics. 2005;6:6. doi: 10.1186/1471-2164-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousee C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. A selective inhibitor of eIF2a dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong Y-W, Hamilton T, Spriggs KA, Dobbyn HC, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Clemens MJ, Bushell M, Morley SJ. Degradation of eukaryotic polypeptide chain initiation factor eIF 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, deSchoolmeester ML, Fraser CA, Pickering BM, Packham G, Willis AE. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. doi: 10.1038/sj.onc.1204547. [DOI] [PubMed] [Google Scholar]
- da Costa RMA, Riou L, Paquola A, Menck CFM, Sarasin A. Transcriptional profiles of unirradiated or UVB-irradiated human cells expressing either the cancer-prone XPB/CS allele or the noncancer-prone XPB/TTD allele. Oncogene. 2005;24:1359–1374. doi: 10.1038/sj.onc.1208288. [DOI] [PubMed] [Google Scholar]
- d'adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes & Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- Deng J, Harding H, Raught B, Gingras A, Berlanga J, Scheuner D, Kaufman R, Ron D, Sonenberg N. Activation of GCN2 in UVB-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- Feng J, Park J, Cron P, Hess D, Hemmings B. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze M. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle IR, Cockcroft X, Curtin NJ, El-Murr MD, Leahy JJ, Stockley M, Golding BT, Rigoreau L, Richardson C, Smith GC, et al. Discovery of potent chromen-4-one inhibitors of the DNA-dependent protein kinase (DNA-PK) using a small-molecule library approach. J Med Chem. 2005;48:7829–7846. doi: 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hickson I, Yan Z, Richardson CJ, Green SJ, Martin NMB, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GCM. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Huston E, Lynch MJ, Mohamed A, Collins DM, Hill EV, MacLeod R, Krause E, Baillie GS, Houslay MD. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci. 2008;105:12791–12796. doi: 10.1073/pnas.0805167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wek R. GCN2 phosphorylation of eIF2α activates NF-κB in response to UVB irradiation. Biochem J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap-binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yachie N, Okada Y, Saito R, Tomita M. Bioinformatic analysis of post-transcriptional regulation by uORF in human and mouse. FEBS Lett. 2007;581:4184–4188. doi: 10.1016/j.febslet.2007.07.057. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or 6-4 photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Morley SJ, McKendrick L, Bushell M. Cleavage of translation initiation factor 4G eIF4G during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein MAP kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- Muller C, Calsou P, Frit P, Cayrol C, Carter T, Salles B. UV sensitivity and impaired nucleotide excision repair in DNA-dependent protein kinase mutant cells. Nucleic Acids Res. 1998;26:1382–1389. doi: 10.1093/nar/26.6.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Park SJ, Peng X, Wang M, Yu MA, Lee SH. Involvement of DNA-dependent protein kinase in UV-induced replication arres. J Biol Chem. 1999;274:32520–32527. doi: 10.1074/jbc.274.45.32520. [DOI] [PubMed] [Google Scholar]
- Qin XL, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger K, Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 2004;32:4786–4803. doi: 10.1093/nar/gkh783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: From understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subkhankulova T, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated initiation of c-Myc protein synthesis following genotoxic stress. Biochem J. 2001;359:183–192. doi: 10.1042/0264-6021:3590183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–1131. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- Wu S, Hu Y, Wang JL, Chatterjee M, Shi Y, Kaufman RJ. Ultraviolet light inhibits translation through activation of the unfolded protein response kinase PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2002;277:18077–18083. doi: 10.1074/jbc.M110164200. [DOI] [PubMed] [Google Scholar]
- Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol. 2006;26:7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yu Y, Hamrick H, Duerksen-Hughes P. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24:1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rosentein BS, Wang Y, Lebwohl M, Mitchell D, Wei H. Induction of 8-oxo-7, 8 dihydro-2′-deoxyguanosine by ultra violet radiation in calf thymus DNA and HeLa cells. Photochem Photobiol. 1997;65:119–124. doi: 10.1111/j.1751-1097.1997.tb01886.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Thomas H, Batey M, Cowell I, Richardson C, Griffin R, Calvert A, Newell D, Smith G, Curtin N. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]