Abstract
Formation of the neural network requires concerted action of multiple axon guidance systems. How neurons orchestrate expression of multiple guidance genes is poorly understood. Here, we show that Drosophila T-box protein Midline controls expression of genes encoding components of two major guidance systems: Frazzled, ROBO, and Slit. In midline mutant, expression of all these molecules are reduced, resulting in severe axon guidance defects, whereas misexpression of Midline induces their expression. Midline is present on the promoter regions of these genes, indicating that Midline controls transcription directly. We propose that Midline controls axon pathfinding through coordinating the two guidance systems.
Keywords: Axon guidance, midline, frazzled, robo, slit
Axons navigate long distances to their targets by responding to a succession of molecular guidance cues expressed along the pathway (Tessier-Lavigne and Goodman 1996; Dickson 2002). Each neuron expresses multiple axon guidance receptors and also acts as a secretion source of guidance molecules. Two major evolutionarily conserved ligand-receptor systems are involved in controlling the axonal behavior: ligand Netrin and its receptor Frazzled/DCC, and ligand Slit and its receptor ROBO (for review, see Dickson 2002). Receptors of these guidance factors are localized not only on the growth cone, but also on the axon shaft, suggesting that they exert autonomous as well as nonautonomous functions (e.g., Hiramoto et al. 2000; Hivert et al. 2002). Because these two systems are thought to act in an antagonistic way, proper axonal behavior depends on the balance between these two systems; how such coordination is accomplished is largely unknown.
Coordination of the two guidance systems is likely achieved, at least in part, via regulating the expression levels of these ligands and receptors. Forced transcription of guidance genes often causes pathfinding errors (Harris et al. 1996; Mitchell et al. 1996; Kidd et al. 1998, 1999), indicating that transcriptional regulation plays an important role in determining the expression levels. Identifying a transcriptional regulator of axon guidance genes is thus an essential step in understanding how the coordination of axon guidance systems is established and maintained. Here, we show that Drosophila T-box transcription factor Midline is a key regulator controlling the expression of multiple components of the two major guidance systems: Frazzled, ROBO, and Slit.
Results and Discussion
Identification of midline gene
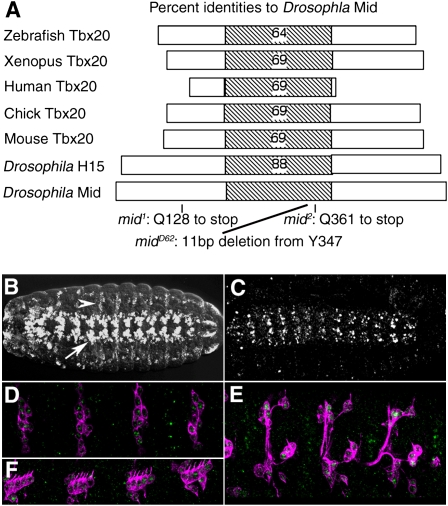
midline (mid) was initially found as one of the genes affecting the larval cuticle pattern, having deletions in pattern elements along the ventral midline (Nüsslein-Volhard et al. 1984). We identified mid as a gene encoding a novel T box protein of tbx20 subclass (Fig. 1A; Ahn et al. 2000; Yamagishi et al. 2004; Reim et al. 2005; Takeuchi et al. 2005; Song et al. 2006). T box proteins are transcription factors that play key roles during development (Stennard and Harvey 2005). Of eight T-box genes present in the Drosophila genome, CG6634 maps close to the location of mid at the cytological location 25E2. Three independently isolated mid alleles all contained point mutations in the ORF of CG6634, indicating that this gene corresponds to the mid locus. mid1 and mid2 harbor a nonsense mutation before and within the T box domain, respectively, and midD62 carries an 11-base-pair (bp) deletion in the T box region (Fig. 1A). Identification of mid as CG6634 was also made independently by Buescher et al. (2004), Reim et al. (2005), and Gaziova and Bhat (2009).
Figure 1.
Molecular analysis of the mid gene. (A) The domain of Drosophila MID, aligned with other T-box proteins, showing the percentage of identical amino acid residues within the T-box domain. The domain consists of the amino acid residues 194–381 of MID. mid1 contains a nonsense mutation at amino acid 128. mid2 contains a nonsense mutation at amino acid 361 in the T-box domain of MID. midD62 contains an 11-bp deletion in the ORF, which results in deletion of Y347 and V348, and a frame shift. (B) In situ hybridization analysis of mid expression in a stage 16 whole-mount normal embryo. mid mRNA expression is confined to a subset of CNS neurons (arrow), epidermis, and sensory neurons in the periphery (arrowhead). (C) A normal stage 16 embryo stained with anti-MID. MID is detected in essentially the same pattern as in B. mid1, mid2, midD62, or Df(2L)cl-h1 mutants embryos exhibited no anti-MID immunoreactivity (data not shown). (D–F) Staining of MID (green) and 22C10 (magenta). MID is expressed in the 22C10-positive sensory neurons, including dp and dda (D), vp and vch (E), lch1 and lch5 (F).
MID is expressed in post-mitotic neurons and is required for the axon pathfinding
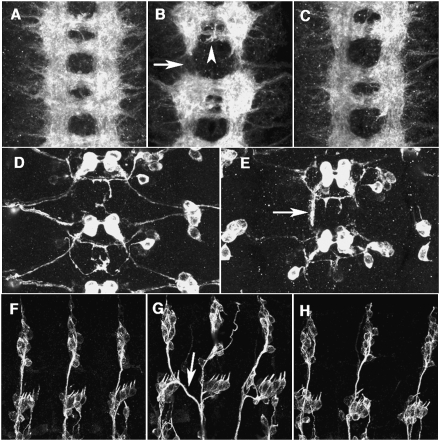
mid mRNA is initially expressed in a pattern of 14 stripes in stage 6 embryos (data not shown). Following gastrulation and germband extension, the developing nervous system exhibits a segmentally repeated striped pattern. In stage 16 embryos, high levels of mid mRNA and MID protein are seen in a subset of CNS neurons (Fig. 1B,C; Buescher et al. 2006). Expression of mid was also observed in sensory neurons of the peripheral nervous system (Fig. 1D–F). Previous studies on mid function focused on its role in segmentation and cell fate specification during neurogenesis (Nüsslein-Volhard et al. 1984; Buescher et al. 2004, 2006; Gaziova and Bhat 2009; Leal et al. 2009). Since MID is highly expressed in post-mitotic neurons during axonogenesis, we investigated the axon wiring phenotype of mid (Jacobs 1993) that might reflect the role at this stage. In normal embryos, axon tracts in the CNS are organized in a ladder-like scaffold, with longitudinal axon bundle running on both sides of the midline and two commissures, anterior and posterior, connecting the bilateral neuromeres. In mid mutant, longitudinal axon tracts were often interrupted (65%, n = 96), and the posterior commissure was much thinner than normal (71% of the commissures, n = 96), suggesting defects in pathway selection and/or axon elongation (Fig. 2B). These defects were largely rescued by pan-neuronal expression of mid (Fig. 2C). In the periphery, afferent axon of the sensory neurons often crossed the segment border (38%, n = 108) (Fig. 2G).
Figure 2.
MID is required for CNS and PNS pathfinding. (A–C) A stage 16 embryo stained with the monoclonal antibody (mAb) BP102 to reveal the axonal scaffold of the CNS. Anterior is up. (A) Normal CNS. (B) In mid1 mutant, the longitudinal tracts were interrupted (arrow) and posterior commissures were thinner (arrowhead). (C) Normal axonal scaffold is restored by pan-neural expression of MID (elav-Gal4/+; mid1/mid1; UAS-mid/+ embryo). (D,E) Visualization of lim3A+ motor neurons in a stage16 embryo using the lim3A-tau-myc transgene (anti-Myc epitope staining). (D) Normal. (E) Motor axons stall and follow a longitudinal path in mid1 mutant (arrow). Similar motoneuron projection defects were observed in mid2 and midD62 embryos (data not shown). (F–H) A stage16 embryo stained with mAb 22C10 to reveal the sensory axons in the PNS. Anterior is left and dorsal up. (F) Normal PNS. (G) In mid1 mutant, sensory axons branch excessively or cross the segment borders (arrow). (H) Restored dorsal sensory axons in an elav-Gal4/+; mid1/mid1; UAS-mid/+ embryo. Similar phenotypes of CNS and PNS were also observed in mid2 or midD62 mutant embryos (data not shown).
To identify the cellular defects of mid mutant embryos during axonogenesis we examined the axonal projections of several identified neurons, motoneurons RP1 and RP3, and a longitudinal pioneer neuron dMP2 (Hidalgo and Brand 1997), which do not express MID. Using specific cellular markers such as anti-Eve antibody, Lim3A-tau-myc transgene (Thor et al. 1999), or MAb 22C10, no alterations in gene expression pattern or cell position could be detected in mid mutants, suggesting that these cells do not have cell fate alterations caused by early MID function in segmentation or neurogenesis. In normal embryos RP1 and RP3 motor axons exit the CNS toward the periphery. In mid mutant embryos, however, axons of these neurons stalled or followed longitudinal axons (Fig. 2E). Axon of the dMP2 neuron also showed a severe stall phenotype, failing to connect to the next neuromere (Supplemental Fig. S1). The absence of endogenous MID expression in these neurons suggests that these cellular phenotypes in mid mutant are caused by defects in axon guidance environments that depend on mid function. One possibility is that MID controls expression of secreted guidance factors, or their receptors that possess not only cell-autonomous functions but also nonautonomous activities.
MID controls the expression of frazzled
The axon scaffold phenotype of mid mutant embryo, such as the thinning of the commissure and interrupted longitudinal axons, contains features of defects in the two major axon guidance systems: Netrin/Frazzled and Slit/ROBO. To identify potential transcriptional target genes of MID in axon guidance, we examined whether or not mid exhibits genetic interactions with components of these two axon guidance systems. While animals heterozygous for mid-null allele were normal, trans-heterozygotes of mid and frazzled and trans-hetrozygotes of mid and slit showed strong defects in the longitudinal tracts (Supplemental Fig. S2). This raised the possibility that MID is involved in the transcriptional regulation of components of the Netrin/Frazzled and Slit/ROBO systems.
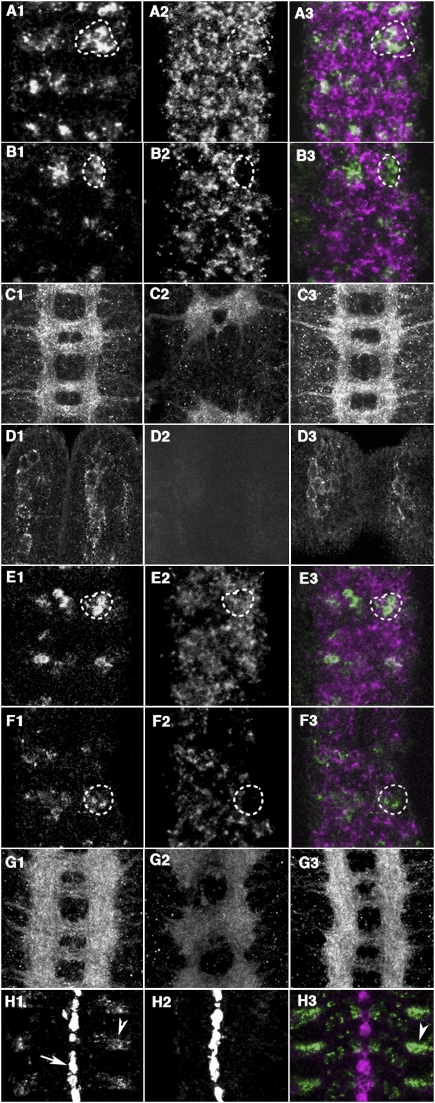
We first examined the Netrin system, Netrin encoded by NetrinA and NetrinB genes, and their receptor Frazzled, which has a dual function as a cell-autonomous guidance receptor and a nonautonomous ligand presentation receptor (Kolodziej et al. 1996; Hiramoto et al. 2000). In normal CNS, Frazzled mRNA is expressed in many neurons. In mid mutant embryo, cluster of mid-expressing neurons exhibited specific reduction of Frazzled mRNA (Fig. 3A,B; e.g., 47% reduction in the marked area). Reduction of Frazzled protein could also be detected in commissural and longitudinal axons in the CNS as well as in the mechanosensory neurons (ch and es) in the periphery (Fig. 3C,D). This reduction of Frazzled in either CNS or periphery was restored by pan-neuronal expression of mid (Fig. 3C,D). Moreover, misexpression of MID in salivary glands induced the expression of frazzled (Fig. 4A). These results indicate that MID can regulate the expression of frazzled. On the other hand, expression levels of NetrinA and NetrinB were not significantly altered, either in mid mutant or upon MID misexpression (Fig. 4A; data not shown). Thus, MID controls Netrin/Frazzled signaling through regulating frazzled expression.
Figure 3.
mid is required for the expression of multiple axon guidance molecules. (A1–B3) Stage 16 mid1 mutant embryos and their heterozygous siblings were examined for frazzled mRNA expression. (A1–A3) A mid/+ heterozygous embryo shows clear expression of frazzled mRNA (A2) in mid-positive cells (A1). (B1–B3) mid1 mutants show reduced or no expression of frazzled (B2) in mid mRNA-labeled cells (B1). (C1–D1) Stage 16 embryos stained with anti-Frazzled antibody. (C1) Normal CNS. (C2) mid1 CNS is weakly stained than normal embryos. (C3) Restored Frazzled expression in an elav-Gal4/+; mid1/mid1; UAS-mid/+ embryo. (D1) Wild-type mechanosensory neurons. (D2) No signal was detected in the mid1 mechnosensory neurons. (D3) Restored Frazzled expression in the mechnosensory neurons of an elav-Gal4/+; mid1/mid1; UAS-mid/+ embryo. (E1–F3) Stage 16 mid1 mutant embryos and their heterozygous siblings were examined for robo mRNA expression. (E1–E3) A mid/+ heterozygous embryo shows clear expression of robo mRNA (E2) in mid-positive cells (E1). (F1–F3) mid1 mutants shows reduced or no expression of robo mRNA (F2) in mid mRNA-labeled cells (F1). (G1–G3) Stage 16 embryos stained with anti-ROBO antibody. (G1) Normal CNS. (G2) mid1 CNS is weakly stained than normal. (G3) Restored ROBO expression in an elav-Gal4/+; mid1/mid1. (H1–H3) In situ hybridization analysis of slit expression in a stage 16 embryo. (H1) In normal embryo, the highest level of slit mRNA occurs in the midline glia (arrow), whereas low levels of slit mRNA are present in cells located lateral to the midline (arrowhead). (H2) In mid1 mutant, silt mRNA expression in lateral regions is weaker than normal. (H3) Colocalization of mid (green) and slit (magenta) mRNA in lateral cells (arrowhead).
Figure 4.
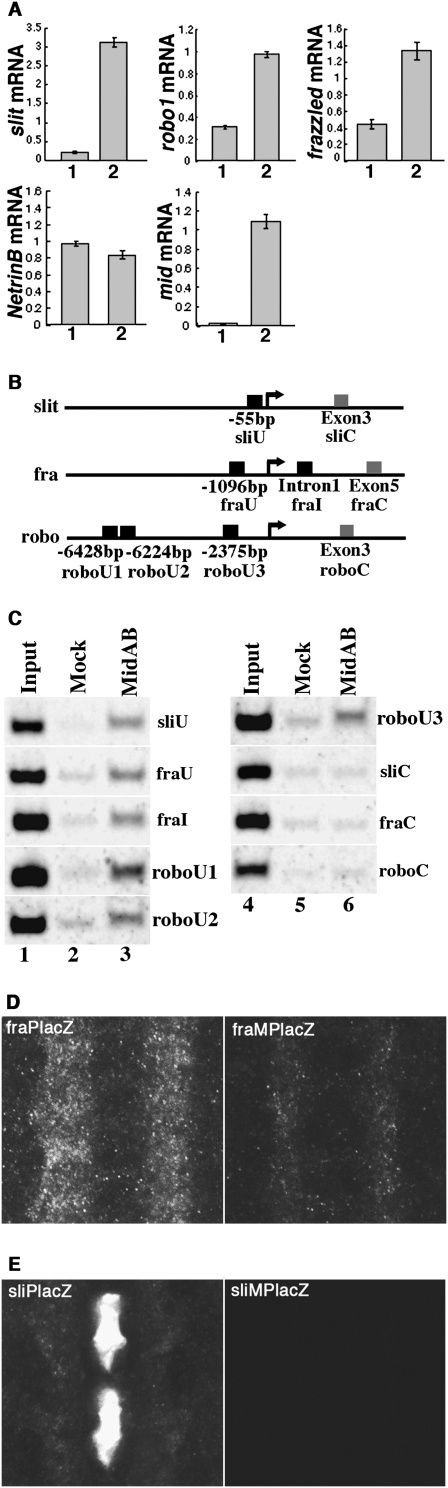
MID coordinates expression of key axon guidance genes. (A) Levels of mRNA in the salivary gland of normal third instar larvae (lane 1) or upon MID misexpression (sgP[Gal4]/+; UAS-mid/+) (lane 2). Shown are levels of transcripts normalized to the corresponding levels of tubulin mRNA in each sample, as determined by quantitative real time PCR. (B) Schematic representation of the MID-binding sites on the genomic sequence of slit, frazzled, and robo. The black boxes indicate the MID-binding site and the gray boxes indicate regions without the MID-binding site adopted as negative controls. Only the sites with perfect match with the consensus sequence were considered. The arrows represent transcription start sites and the numbers in base pairs are distance from the start sites. (C) ChIP with the antibody against MID revealed occupancy of MID at sliU, fraU, fraI, roboU1, roboU2, and roboU3, but not at the control regions sliC, fraC, and roboC (shown in lanes 3,6). The input was 0.1% (lanes 1,4). No primary antibody was used for mock (lanes 2,5). (D,E) Staining of stage 16 embryos with an anti-β-galactosidase antibody. (D) The reporter fraPlacZ was expressed in CNS and the mutant reporter fraMPlacZ exhibited significantly reduced expression in the CNS. (E) The reporter sliPlacZ but not the reporter sliMPlacZ was expressed in the midline glia and lateral cells.
MID controls the expression of robo and slit
Loss of MID function also affected the expression of components of the Slit/ROBO system. In situ hybridization and immunostaining revealed that expression of robo mRNA and protein was significantly reduced in mid mutant (Fig. 3E–G; e.g., 59% reduction in the marked area). This reduction of ROBO was restored by pan-neuronal expression of mid (Fig. 3G). Loss of mid activity also affected an aspect of slit expression. In normal embryos, slit mRNA is highly expressed in midline glia and weakly in neurons located lateral to the midline (Fig. 3H). Slit expression in these lateral cells also contributes to the guidance of the longitudinal axons (Hiramoto and Hiromi 2006). In mid mutant, slit mRNA expression in lateral cells was severely reduced (Fig. 3H; e.g., 90% reduction in the area marked by an arrowhead). As with frazzled, expression of slit and robo mRNA and Slit protein was induced upon MID overexpression in the salivary gland (Fig. 4A; Supplemental Fig. S3). Thus, mid induces the expression of Frazzled, ROBO, and Slit, components of the two major axon guidance systems.
A direct control of frazzled, robo, and slit by MID
To address how MID activates expression of the three axon guidance genes, we first determined the binding sequence of MID using an in vitro binding site selection method (Liu et al. 2003). MID-binding sequence was selected from a pool of random oligonucleotides using MID protein affinity-purified from an embryonic extract. The consensus sequence deduced from the selected oligonucleotides was (G/A/T)NA(A/T)N(T/G)(A/G)GGTCAAG (Supplemental Fig. S4). This sequence was found in the upstream regions or an intron of slit, frazzled, and robo (Fig. 4B), and all of these sites were conserved among several Drosophila species (Supplemental Fig. S5). To determine whether MID binds to these regions in vivo, we performed chromatin immunoprecipitation (ChIP) using anti-MID antibody. In all three genes, MID was present around the MID-binding sites, but not on regions without the binding site (Fig. 4C; Supplemental Fig. S6). In contrast, a potential MID-binding site 32-kb upstream of the commisureless gene, whose expression is not affected in mid mutants, was not occupied by MID (data not shown).
We next assessed the importance of the MID-binding sites in frazzled and slit by transgenic reporter assays. To test the role of the MID site in frazzled, reporter genes were constructed that contain the transcription start site of frazzled and an upstream region including a wild-type MID-binding site (fraPlacZ) or a mutated site (fraMPlacZ). Compared with the wild-type reporter gene, the reporter with a mutated binding site showed reduced expression levels (33% reduction) (Fig. 4D). Thus MID-binding site is indeed required for the proper expression of frazzled. Mutating the MID-binding site in slit also caused a severe effect on slit expression. The lacZ expression in sliPlacZ is driven by the slit regulatory element and the endogenous promoter. While sliPlacZ with the wild-type binding site recapitulated the slit expression in the midline glia and lateral cells, base substitutions in the MID-binding site in sliMPlacZ abolished the lacZ expression (Fig. 4E). It is possible that the MID-binding site resides in an essential promoter element of slit, and hence, the base substitutions abolished slit transcription in all cells. However, the same results were obtained using sli4.5HHlacZ and sliM4.5HhlacZ in which the slit regulatory element is fused to a heterologous hsp70 promoter (Supplemental Fig. S7). As mid was expressed in the lateral cells but not in midline glia, these results suggest that MID-binding sites in slit control slit transcription via binding to multiple factors: MID in lateral cells and unknown factor(s) in midline glia. Taken together, these results demonstrate a direct role for MID in the regulation of frazzled and slit, and suggest that MID governs the expression of multiple axon guidance genes through directly binding of the MID sites in their regulatory regions.
MID directs the expression of components of two major axon guidance systems
We showed that MID directly controls transcription of key components of the two major axon guidance systems: the Netrin/Frazzled system and the Slit/ROBO system. Because these two systems are considered to have opposing outputs, it is interesting that the expression of both systems are induced by the same transcription factor, MID. Dynamic expression of Frazzled and ROBO is required for growth cones to simultaneously respond to both attractants and repellents, integrate these signals, and then respond to the relative balance of forces (Bashaw and Goodman 1999; Garbe and Bashaw 2007). These molecules also provide nonautonomous functions required for cell motility, such as mediating cell adhesion and promoting axon elongation (Hiramoto et al. 2000; Rhee et al. 2002). The coordination of axon guidance systems by MID may thus ensure cooperative actions of multiple guidance molecules in growth cone dynamics, axonal adhesion, and elongation. The role of MID in the transcriptional regulation of axon guidance might be a conserved function, because its orthologs of human, mouse, and zebrafish Tbx20 are also expressed in motor neurons (Ahn et al. 2000; Takeuchi et al. 2005).
Materials and methods
Sequencing of mid mutant alleles
Homozygous mid mutants were identified in embryo collections by the use of the CyOact-GFP balancer chromosome. For each mutant allele, we sequenced both genomic exons and the cDNA after PCR amplification and subcloning. For each PCR, three clones were sequenced on both strands. Mutations appeared in all three clones; no other areas of the ORF showed reproducible sequence alterations.
Generation of transformants
fraPlacZ was made by inserting a 4.4-kb genomic fragment containing the upstream region and transcription start site of frazzled with a MID-binding site (CTTGACCCACATTT) into CaSpeR-AUG-β-gal vecter. fraMPlacZ was made from fraPlacZ and contained a mutated MID-binding site (ACCTGTTAGACGGG). slit promoter reporter constructs were designed as described (Wharton and Crews 1993). sliPlacZ was constructed by placing 1.2NM to the upstream of 4.8 BV containing a MID-binding site (TCAGTTTGGTCAAG). sliMPlacZ was made from sliPlacZ and contained a mutated MID-binding site (CTGTCCCAACTCCA). sliM4.5HHlacZ containing the mutated MID-binding site was made from sli4.5HHlacZ. UAS-mid was made by inserting the mid cDNA into the pUAST plasmid (Brand and Perrimon 1993). Several independent transgenic lines were used, with identical results. The pan-neuronal expression of mid was achieved by driving UAS-mid transgene using elav-Gal4 (Lin and Goodman 1994). Misexpression in the salivary gland was done using sgP[Gal4]/+ driver strain (Liu et al. 2003).
Immunofluorescence staining
Embryos were fixed and processed as described (Liu et al. 2003). Rabbit anti-MID antibody was made using bacterially expressed C-terminal 83 amino acids of MID as an antigen, and was used at 1:1000 dilution. We obtained the following monoclonal antibodies from Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD: 22C10 (1:20; developed by S. Benzer), P102 (1:20; developed by C. Goodman); anti-Myc 9E10 (1:10; developed by J. M. Bishop); anti-Slit mAb C555.4 (1:50; developed by S. Artavanis-Tsakonas; Kidd et al. 1999); anti-ROBO mAb M13C9 (1:20; developed by C. Goodman; Kidd et al. 1998). Rabbit anti-Frazzled (1:500) was a gift from P. Kolodziej (Kolodziej et al. 1996); Rabbit anti-β-galactosidase (1:2000) was purchased from Cappel. Mutant embryos were identified using balancer chromosome carrying the β-galactosidase gene, or staining with anti-MID antibody.
DNA-binding selection and ChIP
In vitro DNA-binding selection was performed as described (Liu et al. 2003). ChIP was performed as described (Smith et al. 2001), using 10- to 20-h embryos. PCR primers were designed to amplify ∼100-bp fragments (Supplemental Material).
Acknowledgments
We thank John B. Thomas for lim3A-lacZ and lim3A-tau-myc flies, Stephen T. Crews for sli4.5HHlacZ line, and Bloomington Stock Center for providing stocks. This work was supported by Grants in Aid for Scientific Research (S.H. and Y.H.) and Research Grant for the Genome Network Project (T.G.) from the MEXT of Japan. Q.-X.L. was supported by a Center of Excellence Program of Japan.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1774209.
Supplemental material is available at http://www.genesdev.org.
References
- Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Goodman CS. Chimeric axon guidance receptors: The cytoplasmic domains of slit and netrin receptors specify attraction versus repulsion. Cell. 1999;97:917–926. doi: 10.1016/s0092-8674(00)80803-x. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buescher M, Svendsen PC, Tio M, Miskolczi-McCallum C, Tear G, Brook WJ, Chia W. Drosophila T box proteins break the symmetry of Hedgehog-dependent activation of wingless. Curr Biol. 2004;14:1694–1702. doi: 10.1016/j.cub.2004.09.048. [DOI] [PubMed] [Google Scholar]
- Buescher M, Tio M, Tear G, Overton PM, Brook WJ, Chia W. Functions of the segment polarity genes midline and H15 in Drosophila melanogaster neurogenesis. Dev Biol. 2006;292:418–429. doi: 10.1016/j.ydbio.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Bashaw GJ. Independent functions of Slit-Robo repulsion and Netrin-Frazzled attraction regulate axon crossing at the midline in Drosophila. J Neurosci. 2007;27:3584–3592. doi: 10.1523/JNEUROSCI.0301-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziova I, Bhat KM. Ancestry-independent fate specification and plasticity in the developmental timing of a typical Drosophila neuronal lineage. Development. 2009;136:263–274. doi: 10.1242/dev.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R, Sabatelli LM, Seeger MA. Guidance cues at the Drosophila CNS midline: Identification and characterization of two Drosophila Netrin/UNC-6 homologs. Neuron. 1996;17:217–228. doi: 10.1016/s0896-6273(00)80154-3. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Brand AH. Targeted neuronal ablation: The role of pioneer neurons in guidance and fasciculation in the CNS of Drosophila. Development. 1997;124:3253–3262. doi: 10.1242/dev.124.17.3253. [DOI] [PubMed] [Google Scholar]
- Hiramoto M, Hiromi Y. ROBO directs axon crossing of segmental boundaries by suppressing responsiveness to relocalized Netrin. Nat Neurosci. 2006;9:58–66. doi: 10.1038/nn1612. [DOI] [PubMed] [Google Scholar]
- Hiramoto M, Hiromi Y, Giniger E, Hotta Y. The Drosophila Netrin receptor Frazzled guides axons by controlling Netrin distribution. Nature. 2000;406:886–889. doi: 10.1038/35022571. [DOI] [PubMed] [Google Scholar]
- Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V. Robo1 and Robo2 are homophilic binding molecules that promote axonal growth. Mol Cell Neurosci. 2002;21:534–545. doi: 10.1006/mcne.2002.1193. [DOI] [PubMed] [Google Scholar]
- Jacobs JR. Perturbed glial scaffold formation precedes axon tract malformations in Drosophila mutants. J Neurobiol. 1993;24:611–626. doi: 10.1002/neu.480240507. [DOI] [PubMed] [Google Scholar]
- Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the Robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN. frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87:197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- Leal SM, Qian L, Lacin H, Bodmer R, Skeath JB. Neuromancer1 and Neuromancer2 regulate cell fate specification in the developing embryonic CNS of Drosophila melanogaster. Dev Biol. 2009;325:138–150. doi: 10.1016/j.ydbio.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Liu Q-X, Jindra M, Ueda H, Hiromi Y, Hirose S. Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development. 2003;130:719–728. doi: 10.1242/dev.00297. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Doyle JL, Serafini T, Kennedy TE, Tessier-Lavigne M, Goodman CS, Dickson BJ. Genetic analysis of Netrin genes in Drosophila: Netrins guide CNS commissural axons and peripheral motor axons. Neuron. 1996;17:203–215. doi: 10.1016/s0896-6273(00)80153-1. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wiechaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Rouxs Arch Dev Biol. 1984;193:267–283. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- Reim I, Mohler JP, Frasch M. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech Dev. 2005;122:1056–1069. doi: 10.1016/j.mod.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Rhee J, Mahfooz NS, Arregui C, Lilien J, Balsamo J, VanBerkum MF. Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat Cell Biol. 2002;4:798–805. doi: 10.1038/ncb858. [DOI] [PubMed] [Google Scholar]
- Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- Song MR, Shirasaki R, Cai CL, Ruiz EC, Evans SM, Lee SK, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Crews ST. CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- Yamagishi T, Nakajima Y, Nishimatsu S, Nohno T, Ando K, Nakamura H. Expression of tbx20 RNA during chick heart development. Dev Dyn. 2004;230:576–580. doi: 10.1002/dvdy.20076. [DOI] [PubMed] [Google Scholar]