Abstract
In Escherichia coli, ATP-DnaA, unlike ADP-DnaA, can initiate chromosomal replication at oriC. The level of cellular ATP-DnaA fluctuates, peaking at around the time of replication initiation. However, it remains unknown how the ATP-DnaA level increases coordinately with the replication cycle. In this study, we show that two chromosomal intergenic regions, herein termed DnaA-reactivating sequence 1 (DARS1) and DnaA-reactivating sequence 2 (DARS2), directly promote regeneration of ATP-DnaA from ADP-DnaA by nucleotide exchange, resulting in the promotion of replication initiation in vitro and in vivo. Coordination of initiation with the cell cycle requires DARS activity and its regulation. Oversupply of DARSs results in increase in the ATP-DnaA level and enhancement of replication initiation, which can inhibit cell growth in an oriC-dependent manner. Deletion of DARSs results in decrease in the ATP-DnaA level and inhibition of replication initiation, which can cause synthetic lethality with a temperature-sensitive mutant dnaA and suppression of overinitiation by the lack of seqA or datA, negative regulators for initiation. DARSs bear a cluster of DnaA-binding sites. DnaA molecules form specific homomultimers on DARS1, which causes specific interactions among the protomers, reducing their affinity for ADP. Our findings reveal a novel regulatory pathway that promotes the initiation of chromosomal replication via DnaA reactivation.
Keywords: DnaA reactivation, nucleotide exchange, initiation regulation, functional DNA sequence, protein complex, replication cycle
In prokaryotes and eukaryotes, chromosomal replication is initiated by specific proteins at a specific time during the cell cycle. In Escherichia coli, the occurrence of replicational initiation is strictly coupled with the growth rate of a cell (Cooper and Helmstetter 1968). During a cell cycle, the time taken from replicational initiation to cell division is principally constant at a certain temperature in various growth rates. Thus, the time of replicational initiation is the crucial determinant for the overall control of cell cycle. However, mechanisms to regulate occurrence of replicational initiation in a cell cycle-coordinated manner remain unclear. To address this issue, mechanisms to activate the initiator DnaA protein during the cell cycle have to be revealed.
The DnaA protein, the initiator of E. coli chromosomal replication, is a member of the AAA+ ATPase family and has an exceptionally high affinity (Kd of 10–100 nM) for ATP/ADP (Sekimizu et al. 1987; Neuwald et al. 1999; Messer 2002; Kaguni 2006; Kawakami et al. 2006). The minimal origin (oriC) region contains several 9-mer DnaA-binding sites (DnaA boxes). ATP-DnaA forms a specific multimeric complex on oriC in a cooperative manner (McGarry et al. 2004; Keyamura et al. 2007). In the resultant complexes, an ATP-dependent interaction between specific DnaA AAA+ domains occurs, resulting in local duplex unwinding within oriC (Kawakami et al. 2005; Erzberger et al. 2006; Ozaki et al. 2008). DnaB helicase is loaded onto the single strands generated, which promotes loading of DnaG primase and DNA polymerase (pol) III holoenzyme. ADP-DnaA also forms a homomultimer on oriC, but it cannot unwind the DNA duplex. Thus, generation of ATP-DnaA is prerequisite for replicational initiation and its regulation.
The level of cellular ATP-DnaA oscillates during the replication cycle, peaking at around the time of initiation (Kurokawa et al. 1999). At the post-initiation stage, DnaA-ATP is hydrolyzed in a DNA-loaded β sliding clamp-dependent and ADP-Hda protein-dependent manner, yielding inactive ADP-DnaA (Katayama et al. 1998; Kato and Katayama 2001; Su'etsugu et al. 2005, 2008). The β clamp is a subunit of the pol III holoenzyme, which is loaded onto DNA during DNA replication and remains attached to DNA after the creation of Okazaki fragments. This DnaA-inactivation system (RIDA, regulatory inactivation of DnaA) decreases the level of cellular ATP-DnaA in a replication-coordinated manner (Katayama et al. 1998; Kurokawa et al. 1999; Kato and Katayama 2001).
RIDA-defective cells have significantly elevated levels of ATP-DnaA and overinitiate chromosomal replication (Kato and Katayama 2001; Nishida et al. 2002; Fujimitsu et al. 2008). Thus, the control of DnaA-bound nucleotides is crucial for the regulation of the chromosomal replication cycle. RIDA functions independently of the temporal inactivation of oriC by SeqA and the reduction of oriC-accessible DnaA molecules by DnaA titration on the datA locus (Kurokawa et al. 1999; Katayama et al. 2001).
Little is known about the mechanism that elevates ATP-DnaA levels in a cell cycle-coordinated manner. We speculate that regeneration of ATP-DnaA from ADP-DnaA could efficiently provide ATP-DnaA to cells in the preinitiation stage. We reported previously that ADP-DnaA can indeed be converted to ATP-DnaA in vivo (Kurokawa et al. 1999). A conceivable mechanism of DnaA reactivation is nucleotide exchange. Given that cells contain ∼3 mM ATP, ∼10-fold that of ADP (Bochner and Ames 1982), release of ADP from ADP-DnaA could be directly followed by ATP binding, which would reactivate DnaA.
Acidic phospholipids, such as phosphatidylglycerol and cardiolipin, were reported previously to reactivate ADP-DnaA in vitro by nucleotide exchange in the presence of ATP and oriC (Sekimizu and Kornberg 1988; Crooke et al. 1992). Restricted expression of the pgsA gene, which encodes phosphatidylglycerophosphate synthetase, reduces the cellular levels of those acidic phospholipids and inhibits cell growth (Heacock and Dowhan 1989; Xia and Dowhan 1995). Although growth of these cells is rescued by a dnaA-independent chromosomal replication system, there is no direct evidence that acidic phospholipids promote regeneration of ATP-DnaA from ADP-DnaA, especially in vivo.
While searching for a novel DnaA-reactivating factor, we found previously that a specific DNA sequence can catalytically promote the release of ADP from ADP-DnaA, followed by ATP binding and reactivation of replication initiation in vitro (Fujimitsu and Katayama 2004). This specific sequence (termed DARS, for DnaA-reactivating sequence) lies in the ColE1 plasmid replication origin and contains three DnaA boxes. The arrangement of these DnaA boxes, two of which are oppositely oriented, is crucial for DnaA reactivation. However, it remains unknown whether a specific DNA sequence reactivates ADP-DnaA and controls replicational initiation in vivo, as the DARS of ColE1ori does not reactivate ADP-DnaA in vivo.
In this study, we demonstrate that the E. coli genome contains two intergenic regions, herein called DARS1 and DARS2, capable of reactivating ADP-DnaA in vitro and in vivo. These regions share a DnaA box cluster similar to the previously identified DARS (Fujimitsu and Katayama 2004). ADP-DnaA complexes are dissociated by these DARSs in vitro, and initiation-competent ATP-DnaA is generated in the presence of ATP. RIDA-inactivated DnaA is also reactivated by DARSs in vitro. The activity of DARS2 is stimulated by a heat-liable soluble factor extracted from E. coli cells. We further demonstrate that DARS1 and DARS2 can enhance the level of cellular ATP-DnaA and the initiation of chromosomal replication in vivo. Our analyses suggest that specific DnaA homomultimers form on DARS1, and that specific interactions occur between their AAA+ domains, thereby reducing their affinity for ADP. Our findings reveal a novel mechanism that is required to activate the initiator of replication in a manner coordinated with the cell cycle.
Results
Intergenic regions upstream of uvrB and mutH can dissociate ADP-DnaA complexes in vitro
To identify genomic sites capable of promoting ADP-DnaA dissociation, we searched for sequences carrying two DnaA boxes (with the typical 9-mer TTATNCACA sequence or with only one mismatch) that are closely located and oppositely orientated. We identified 51 sites that met these criteria and amplified these sequences by PCR (Supplemental Table 1) or by cloning in plasmid pACYC177, which has a moderate copy number (∼10 copies per cell). The ADP-releasing activities of the resultant DNA fragments were assessed by incubating them with [3H]ADP-DnaA for 15 min at 30°C and conducting a filter-binding assay.
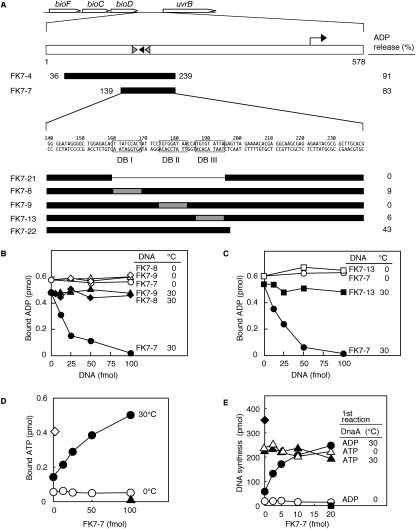
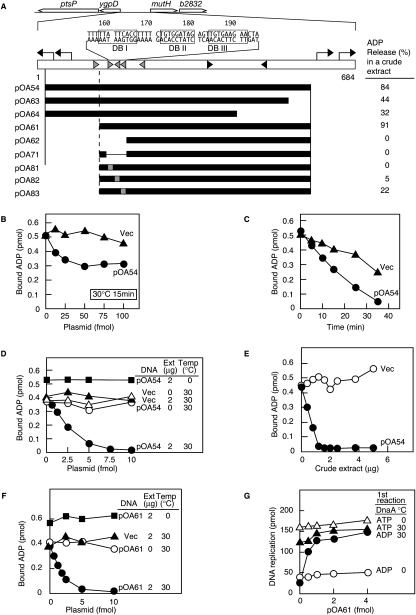
Two of the isolated DNA segments (FK7-4 and pOA54 in Figs. 1A and 2A–C, respectively) exhibited ADP-releasing activity. The FK7-4 and pOA54 sequences are located in intergenic regions between bioD and uvrB and between ygpD and mutH, respectively. Notably, each of these two regions contains three similarly arranged DnaA boxes.
Figure 1.
Structure and in vitro activity of DARS1. (A) Identification of DARS1. Open arrows indicate the coding regions. The right-angled arrow indicates the promoter. Black- and gray-filled arrowheads represent DnaA boxes that completely match the 9-mer consensus and that differ from the consensus by a single base, respectively. Numbers above the sequence of FK7-7 indicate the position from the stop codon of bioD. FK7-8, FK7-9, and FK7-13 contain nonsense sequence (AACTATATC; gray-filled square) (Schaper and Messer 1995) instead of DnaA box I (DB I), DnaA box II (DB II), and DnaA box III (DB III), respectively. [3H]ADP-DnaA (2 pmol) was incubated for 15 min at 30°C in buffer containing 2 mM ATP in the presence (50 fmol) or absence of the indicated DNA fragment (filled bars). DNA-dependent release of ADP is shown for each fragment (ADP release %). (B,C) ADP-releasing activity of DARS1 mutants. The indicated amounts of FK7-7 (○, ●) or its derivatives FK7-8 (◇, ♦), FK7-9 (△, ▲), and FK7-13 (□, ■) were incubated at 0°C (unfilled symbols) or 30°C (filled symbols) for 15 min with [3H]ADP-DnaA (2 pmol). (D) Nucleotide-exchanging activity of DARS1. ADP-DnaA (2 pmol) was incubated for 15 min at 0°C (○) or 30°C (●) with 1.5 μM [α-32P]ATP and the indicated amounts of FK7-7. [α-32P]ATP-DnaA (2 pmol) was similarly incubated at 0°C (◇). (▲) DnaA was not included. (E) Reactivation of DnaA by DARS1 for replication initiation. ADP-DnaA (○, ●) and ATP-DnaA (△, ▲) (0.4 pmol) were incubated (first reaction) for 15 min at 0°C (○, ▵) or 30°C (●, ▲) with 2 mM ATP and the indicated amounts of FK7-7. The samples were then further incubated for 20 min at 30°C in a minichromosome replication system. (■) minichromosome was not included; (♦) DNA synthesis by 1 pmol of ATP-DnaA.
Figure 2.
Structure and in vitro activity of DARS2. (A) Identification of DARS2. Symbols are the same as those in Figure 1A, with the exception that the numbers above the sequence indicate the positions from the start codon of ygpD. The ADP-releasing activities were assessed using the indicated regions on pACYC177 (5 fmol), as described in Figure 1A (ADP release %), except that a crude extract (2 μg of protein) was included. (B,C) ADP-releasing activity of DARS2 in the absence of a crude protein extract. (B) [3H]ADP-DnaA (2 pmol) was incubated for 15 min at 30°C with 2 mM ATP and the indicated amounts of pACYC177 (Vec; ▲) or pOA54 (●). (C) Similar reactions were performed at 30°C using 40 fmol of each plasmid for the indicated incubation time. (D,E) ADP-releasing activity of DARS2 in the presence of a crude protein extract. (D) [3H]ADP-DnaA (2 pmol) was incubated for 15 min at 0°C (■) or 30°C with the indicated amounts of pACYC177 (Vec; ▵, ▲) or pOA54 (○, ●, ■), and the indicated amounts of crude protein extract (Ext) (0 μg, ○ and ▵; 2 μg, ●, ▲, and ■). (E) Similar experiments were performed by incubating the indicated amounts of crude extract and each plasmid (5 fmol) for 15 min at 30°C. (F) ADP-releasing activity of minimal DARS2. [3H]ADP-DnaA (2 pmol) was incubated for 15 min at 0°C (■) or 30°C in the buffer containing the indicated amounts of pACYC177 (Vec; ■) or pOA61 (○, ●, ■) and the indicated amounts of crude extract (0 μg, ○; or 2 μg, ●, ▲, ■). (G) Reactivation of DnaA in replication initiation activity by DARS2. ADP-DnaA (○, ●) or ATP-DnaA (▵, ▲) (0.4 pmol) was incubated with the indicated amounts of pOA61 in the presence of a crude protein extract (0.4 μg of protein) at 0°C (○, △) or 30°C (●, ▲) (first reaction), followed by the minichromosomal replication assay as described in Figure 1E.
DARS1 reactivates ADP-DnaA by nucleotide exchange in vitro
Deletion analysis of FK7-4 identified the 101-base-pair (bp) minimal region bearing the DnaA box cluster required for ADP-releasing activity (FK7-7 in Fig. 1A). Based on this and further analyses (see below), we termed this region DARS1. Each DnaA box within DARS1 as well as the DnaA box cluster was required for ADP-releasing activity (cf. FK7-7 and FK7-21, FK7-8, FK7-9, FK7-13 in Fig. 1A–C). In addition to these DnaA boxes, a flanking region (42 bp spanning from base number 198 to 239) was required for full ADP-releasing activity (FK7-22 in Fig. 1A). These results suggest that the DnaA box cluster is the functionally crucial core element and that the flanking region is a stimulatory element.
FK7-7 promoted dissociation of ADP-DnaA at 30°C but not at 0°C (Fig. 1C). The defect at 0°C would not result from the lack of DnaA binding, as DnaA binds to FK7-7 at 0°C (see below). Basically, DnaA binds to DnaA boxes even at 0°C (Obita et al. 2002).
When ADP-DnaA was incubated at 30°C in the presence of FK7-7 and 1.5 μM ATP, ATP-DnaA was formed by nucleotide exchange (Fig. 1D). The resultant ATP-DnaA was fully active in minichromosomal replication in vitro (Fig. 1E).
DARS2 reactivation of ADP-DnaA and a role for a stimulating factor in vitro
pOA54 exhibited a moderate level of ADP-DnaA-releasing activity, even when present at excessive amounts (Fig. 2B). Under the same conditions, FK7-7 exhibited a full activity (data not shown). Longer incubation at 30°C increased pOA54-dependent ADP releasing (Fig. 2C). We further found that a crude protein fraction extracted from wild-type cells stimulated the DnaA-ADP-dissociating activity of pOA54 (Fig. 2D,E).
Next, we performed deletion analyses of pOA54 in the presence of the stimulatory fraction and determined the minimal region needed for ADP-releasing activity (pOA61 in Fig. 2A,F). We thus named the chromosome-derived region of pOA61 DARS2. Since pOA63 showed only moderate activity compared with pOA54, the deleted region at the right end of pOA63 is likely a stimulatory element (Fig. 2A). Activity of pOA64 was only slightly less than that of pOA63, suggesting that the DnaA box that completely matches the 9-mer consensus (rightmost box in Fig. 2A) is not crucial in this reaction. DnaA boxes I and II within DARS2 as well as the DnaA box cluster were crucial for the ADP-releasing activity (pOA62, pOA71, pOA81, and pOA82 in Fig. 2A). DnaA box III was required for full ADP-releasing activity (pOA83 in Fig. 2A). Full reactivation of DnaA for replication initiation was also seen in vitro in a DARS2-dependent manner (Fig. 2G). Like DARS1 (Fig. 1B–E), DARS2 required incubation at 30°C for the ADP releasing (Fig. 2C–F), although DnaA binds DARS2 at 0°C (data not shown).
The ADP-releasing activity of DARS1 was not stimulated by the same crude extract as above (data not shown), suggesting the presence of a DARS2-specific regulatory factor. The activity of the crude extract was heat-liable (Supplemental Fig. 1) and RNaseA-resistant (data not shown).
DARS1 can reactivate ADP-DnaA resulting from RIDA in vitro
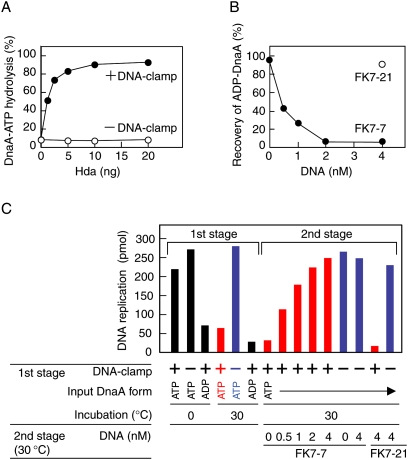
ADP-DnaA resulting from RIDA must be reactivated prior to the next round of replication initiation in vivo. We thus investigated if DARS1 could reactivate DnaA that was inactivated by RIDA in vitro.
In the first stage, [α-32P]ATP-DnaA was converted into [α-32P]ADP-DnaA using a staged reconstituted RIDA system. DnaA-ATP hydrolysis depended on Hda and the DNA-loaded clamp (Fig. 3A), consistent with a previous observation (Su'etsugu et al. 2005, 2008). In the second stage, the resultant [α-32P]ADP-DnaA was further incubated with DARS1 (FK7-7) or the DARS1-ΔDnaA box mutant (FK7-21). DnaA-bound nucleotides were efficiently released in a manner dependent on FK7-7 but not FK7-21 (Fig. 3B), consistent with the data presented in Figure 1A. These results indicate that DARS1 promotes the release of DnaA-bound ADP that was yielded by RIDA.
Figure 3.
DARS1 can reactivate ADP-DnaA resulting from RIDA in vitro. (A) DnaA-ATP hydrolysis by RIDA. [α-32P]ATP-DnaA (1 pmol) was incubated for 20 min at 30°C with the indicated amounts of Hda in the presence (+DNA-clamp) or absence (−DNA-clamp) of the DNA-loaded β sliding clamp (20 fmol of clamp). Nucleotides bound to DnaA were recovered and analyzed as described in the Materials and Methods. (B) Release of DnaA-bound ADP that was produced by RIDA. The RIDA reaction was first performed using [α-32P]ATP-DnaA (1 pmol), Hda (10 ng), and DNA-loaded β clamp (20 fmol of clamp) under the same conditions as in A. Products were further incubated for 15 min at 30°C with the indicated amounts of FK7-7 or FK7-21, followed by determination of DnaA-bound nucleotides. ADP-DnaA constituted 96% of ATP-DnaA/ADP-DnaA after the RIDA reaction. (C) DARS1-driven reactivation of RIDA-produced ADP-DnaA. In the first stage, ATP-DnaA or ADP-DnaA (0.4 pmol) was incubated for 20 min at 0°C or 30°C with Hda (4 ng) in the presence (+) or absence (−) of the DNA-loaded β clamp (8 fmol). In the second stage, the samples that had been incubated at 30°C with (red) or without (blue) the DNA-clamp complexes in the first stage were further incubated for 15 min at 30°C with the indicated amounts of FK7-7 or FK7-21. After the first or second stage, replication activity of DnaA was assessed in a minichromosomal replication system as described in Figure 1E. Incorporation of nucleotides was 6 pmol or 5 pmol in the absence of DnaA or minichromosome, respectively (data not shown).
We further assessed the replication initiation activity of DnaA after each stage in the above experiments. In the first stage, the level of ATP-DnaA activity was reduced to that of ADP-DnaA in a RIDA-dependent manner (Fig. 3C). Notably, in the second stage, the inactivated DnaA was fully reactivated by DARS1 (FK7-7) (Fig. 3C), indicating that DARS1 can reactivate ADP-DnaA yielded by RIDA. Similar results were obtained using DARS2 (Supplemental Fig. 2). Coordinated regulation of RIDA and DARS activities could cause the level of ATP-DnaA to oscillate during the cell cycle.
DARS1 or DARS2 multicopies increase the level of ATP-DnaA independently of de novo protein synthesis
To assess in vivo function, we tested whether an increase in the copy number of DARS1 or DARS2 would stimulate the regeneration of ATP-DnaA from ADP-DnaA in vivo. Previously, we observed the ADP-to-ATP regeneration of DnaA in vivo under conditions in which RIDA and de novo DnaA synthesis were inhibited (Kurokawa et al. 1999). To inhibit DnaA-ATP hydrolysis by RIDA, we used dnaN59, a temperature-sensitive mutant defective in β clamp that is unable to grow at 42°C. We inhibited de novo synthesis of DnaA using chloramphenicol, as newly translated DnaA molecules would predominantly bind to ATP. Cells were grown in a medium containing ortho-[32P], nucleotide-bound DnaA was isolated, and the bound nucleotides were analyzed using thin-layer chromatography.
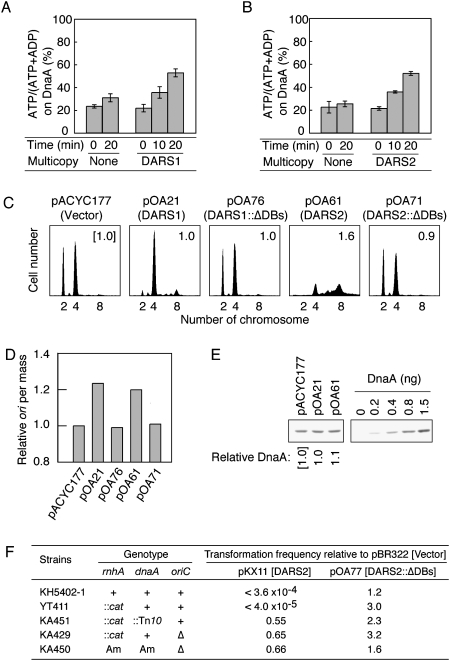
In dnaN59 cells bearing pOA21 (pACYC177-DARS1), the ATP-DnaA level increased at 42°C in the presence of chloramphenicol (Fig. 4A). Similar results were obtained for the cells bearing pOA61 (pACYC177-DARS2) (Fig. 4B). These results indicate that DARS1 and DARS2 can stimulate the regeneration of ATP-DnaA from ADP-DnaA in vivo.
Figure 4.
Regeneration of ATP-DnaA and reactivation of DnaA by DARS1 or DARS2 multicopies in vivo. (A) KA474 (dnaN59) cells bearing pACYC177 (None) or pOA21 (DARS1) were incubated in a medium containing [32P]orthophosphate at 28°C until the optical density (A660) reached 0.2, and shifted to 42°C in the presence of chloramphenicol. After incubation for the indicated amount of time, DnaA was immunoprecipitated using anti-DnaA serum, and DnaA-bound nucleotides were analyzed by thin-layer chromatography. Error bars represent the standard deviation from three independent experiments. (pOA21) pACYC177 derivative carrying FK7-7. (B) KA474 (dnaN59) cells bearing pACYC177 (None) or pOA61 (DARS2) were similarly analyzed. (C) Flow cytometry analysis. MG1655 cells bearing pACYC177 (vector), pOA21 (DARS1), pOA76 (DARS1∷ΔDnaA boxes [DARS1∷ΔDBs]), pOA61 (DARS2), or pOA71 (DARS2∷ΔDnaA boxes [DARS2∷ΔDBs]) were grown at 37°C in M9 medium containing ampicillin, followed by incubation for 4 h in the presence of rifampicin and cephalexin. Cell size and DNA content were analyzed by flow cytometry. The numbers inserted in each histogram are relative ratios of mean cell mass to that of cells bearing pACYC177. (pOA76) pACYC177 derivative bearing FK7-21. (D) Relative ratios of the number of origins (ori) per cell mass in cells bearing the indicated plasmids (obtained from C) to those in cells bearing pACYC177. (E) Immunoblot analysis using anti-DnaA antiserum. Cells were grown under the same conditions as those described in C. Portions of the cultures equivalent to the total cell volume in 200 μL at an optical density (A660) of 0.1 were subjected to immunoblot analysis. Relative ratios of DnaA content of cells bearing the indicated plasmids to that of cells carrying pACYC177 are shown. Results are representative of three independent experiments. (F) oriC- and dnaA-dependent colony formation inhibition by DARS2 oversupply. Cells were transformed with pBR322 (Vector), pKX11 (pBR322-DARS2), and pOA77 (pBR322-DARS2∷ΔDnaA boxes [DARS2∷ΔDBs]) and incubated at 30°C (KH5402-1, 22 h; YT411, 26 h; KA451, KA429, and KA450; 36 h) on LB agar plates containing ampicillin. Colonies with a diameter of >1.0 mm were counted. Relative ratios of transformation efficiency by pKX11 and pOA77 to that by pBR322 are shown. Transformation efficiency of the indicated strains by pBR322 was 104 to 107 per microgram of DNA. (Am) Amber mutation; (Δ) del-1017.
DARS1 or DARS2 multicopies promote replication initiation
To assess whether multicopies of DARS1 and DARS2 promote replication initiation in vivo, we analyzed cells bearing pOA21 (DARS1) or pOA61 (DARS2) using flow cytometry. In rapidly growing E. coli cells, replication of the chromosome is initiated before completion of the previous round of replication, resulting in a single cell containing a set of two or four origins, or of four or eight origins (Skarstad et al. 1995). The number of origins per unit cell mass can be used as a parameter of the ability to initiate replication (Skarstad et al. 1989). To determine the number of origins in a cell, exponentially growing cells were incubated in the presence of rifampicin and cephalexin, inhibitors of replication initiation and cell division, respectively, to allow completion of ongoing replication. The number of chromosomes in cells, as determined by flow cytometry, corresponds to the number of origins at the time of addition of the drugs.
Cells bearing pACYC177 vector predominantly contained two or four origins (Fig. 4C). Introduction of pOA21 or pOA61 considerably promoted initiation: pOA21 resulted in most cells bearing four origins and pOA61 resulted in most cells bearing eight origins. Asynchronous initiations and increased mean cell mass were also induced by pOA61, resulting in a considerable number of cells bearing five, six, or seven origins. Both plasmids increased the number of origins per cell mass compared with pACYC177 (Fig. 4D), indicating that initiation was promoted in a DARS-dependent manner.
pACYC177 bearing DnaA box-deleted DARS derivatives did not stimulate initiation (pOA76 [DARS1∷ΔDnaA boxes] and pOA71 [DARS2∷ΔDnaA boxes] in Fig. 4C,D), consistent with the in vitro data (Figs. 1A, 2A). Also, the number of origins per unit cell mass was similar between pOA21 and pOA61 (Fig. 4D). The difference between pOA21 and pOA61 in the number of origins in a cell was thus attributed to the increased cell mass induced by pOA61 (Fig. 4C).
Cellular DnaA content was not significantly affected by the plasmids (Fig. 4E), which supports the hypothesis that DARS1 and DARS2 can promote replication initiation by stimulating DnaA reactivation in vivo. Transcription of the dnaA gene is autoregulated, and ATP-DnaA represses about twofold more tightly dnaA transcription than ADP-DnaA in vitro (Speck et al. 1999). The cellular DnaA molecules are present at a level of 1000–2000 molecules per cell and they are very stable (Torheim et al. 2000; Shimuta et al. 2004). Thus, fluctuations in dnaA transcription should not necessarily largely affect the total cellular DnaA levels. Indeed, the cellular DnaA level is not significantly increased even in hda mutant cells (Fujimitsu et al. 2008).
Oversupply of DARS2 using pKX11, a derivative of pBR322 (twofold to threefold higher in the copy number than pACYC177) inhibited colony formation in an oriC- and dnaA-dependent manner (Fig. 4F). oriC- or dnaA-disrupted cells replicate the chromosomes depending on rnhA disruption-dependent origins (Kogoma 1997) and were resistant to DARS2 oversupply. Activation of the alternative replication origins in the presence of the intact oriC and dnaA did not suppress the inhibition of colony formation caused by DARS2 oversupply (YT411 in Fig. 4F), which excludes the possibility that the colony formation inhibition could result from inhibition of the oriC function by titration of replication initiation factors such as DnaA. Instead, these results are consistent with the idea that DARS2 oversupply can cause overinitiation at oriC, resulting in growth inhibition of cells.
Inhibition of the colony formation of cells bearing the intact oriC and dnaA depended on the DnaA box cluster on DARS2 (pOA77 in Fig. 4F), which is consistent with the idea that DnaA binding to DARS2 is prerequisite for the inhibition. However, we cannot exclude the possibility that an unknown regulatory factor is titrated out in a DARS2 DnaA box cluster-dependent manner, thereby causing overinitiation.
DARS1-bearing pBR322 or pUC19 did not impair colony formation (data not shown), suggesting different regulations to DARSs (see the Discussion).
Deletion of DARS1 or DARS2 rescues growth of hda-defective cells
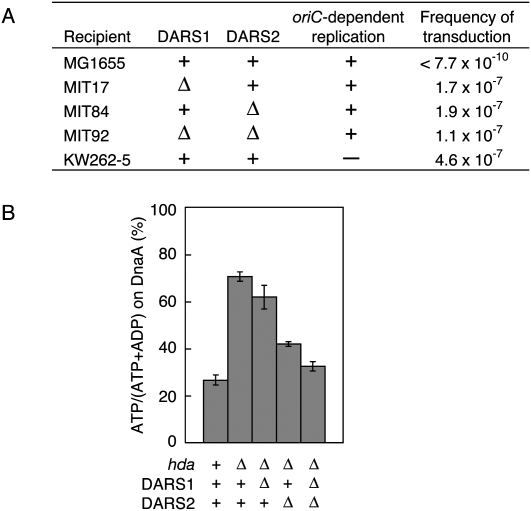
To investigate if DARSs are essential for cell growth, we first disrupted the chromosomal DARSs. The region corresponding to FK7-4 (Fig. 1A) and the DnaA box cluster (35-bp region containing DB I–DB III in Fig. 2A) were replaced with a drug-resistant gene in recD-disrupted cells or in cells carrying λ Red recombinase (Supplemental Material). Disrupted DARS1 and DARS2 were introduced into MG1655 (wild type) by P1 transduction. The resultant mutants (MIT17 [ΔDARS1∷kan], MIT78 [ΔDARS2∷cat], and MIT84 [ΔDARS2∷spec]) formed as many colonies as MG1655 on LB plates at 30°C, 37°C, and 42°C (data not shown). Similar results were obtained from MG1655 bearing disruptions in both DARS1 and DARS2 (MIT80 [ΔDARS1∷kan ΔDARS2∷cat] and MIT92 [ΔDARS1∷kan ΔDARS2∷spec]) (data not shown). These results indicate that DARS1 and DARS2 are dispensable for cell growth.
To investigate a role for DARSs in the in vivo regulation of DnaA nucleotide forms, we tested whether the DARS deletions could suppress the lethality caused by the hda deletion. Lack of Hda increases the cellular ATP-DnaA level, causing overinitiation of replication and inhibition of colony formation (Kato and Katayama 2001; Gon et al. 2006; Fujimitsu et al. 2008). When Δhda∷cat were introduced into MG1655 (wild type) via P1 phage, colonies failed to form (Fig. 5A; Supplemental Fig. 3; Kato and Katayama 2001). In contrast, transduction of Δhda∷cat into MIT17 (ΔDARS1∷kan), MIT84 (ΔDARS2∷spec), and MIT92 (ΔDARS1∷kan ΔDARS2∷spec) resulted in colonies forming at efficiencies similar to those of a control strain, KW262-5 (ΔoriC∷Tn10 rnhA∷Tn3) (Fig. 5A; Kogoma 1997; Kato and Katayama 2001). When we performed the plasmid complementation tests using the resultant Δhda ΔDARS double mutants and pACYC177, pOA21 (pACYC177-DARS1), or pOA61 (pACYC177-DARS2), colony formation of the transformants was severely inhibited in a manner dependent on plasmid-carried DARS (data not shown). This result means that no critical suppressor mutations occur in the mutant cells. Thus, DARS mutations suppressed the growth inhibition caused by hda deletion, consistent with the hypothesis that DARS stimulates oriC- and DnaA-dependent initiation. In further support of this hypothesis, we recently isolated a suppressor of the hda-185 (Cs) mutant that carries a transposon inserted within DARS2 (K Mazda, K Fujimitsu, and T Katayama, unpubl.).
Figure 5.
DARS1 and DARS2 are required for the regulation of cellular ATP-DnaA levels. (A) hda∷cat was transduced into MG1655 (wild type), MIT17 (ΔDARS1∷kan), MIT84 (ΔDARS2∷spec), MIT92 (ΔDARS1∷kan ΔDARS2∷spec), and KW262-5 (rnhA∷Tn3 ΔoriC∷Tn10: oriC-independent replication) by P1 phage, followed by incubation at 37°C for 20 h or 36 h (only for KW262-5) on LB agar plates containing chloramphenicol (20 μg/mL). Colonies with diameters of >1 mm were counted. Frequency of transduction represents the number of colonies per P1 phage particle. (B) KW262-5 (rnhA∷Tn3 ΔoriC∷Tn10), MK86 (rnhA∷Tn3 ΔoriC∷Tn10 Δhda∷cat), MIT47 (rnhA∷Tn3 ΔoriC∷Tn10 Δhda∷cat ΔDARS1∷kan), MIT86 (rnhA∷Tn3 ΔoriC∷Tn10 Δhda∷cat ΔDARS2∷spec), and MIT88 (rnhA∷Tn3 ΔoriC∷Tn10 Δhda∷cat ΔDARS1∷kan ΔDARS2∷spec) were incubated in medium containing [32P]orthophosphate at 37°C until the optical density (A660) reached 0.2. DnaA-bound nucleotides were recovered and determined as in Figure 4A. Error bars represent the standard deviation from three independent experiments.
Cells bearing both ΔDARS1 and Δhda formed colonies smaller than cells bearing both ΔDARS2 and Δhda (Supplemental Fig. 3). Doubling times of these cells were 32 min (for cells bearing ΔDARS1 Δhda) and 25 min (for cells bearing ΔDARS2 Δhda) in LB medium at 37°C. These results suggest that DARS2 may have a higher activity than DARS1 in cells.
DARS1 and DARS2 increase the level of cellular ATP-DnaA
To determine directly if DARSs affect the ATP-DnaA level in vivo, we used Δhda ΔoriC∷Tn10 rnhA∷Tn3 cells bearing DARS deletions in a DnaA immunoprecipitation assay. In cultures of wild-type cells, the ATP-DnaA level constituted 25% of the ATP-DnaA/ADP-DnaA, whereas in cultures of Δhda cells, the level was elevated to 71% (Fig. 5B; Supplemental Fig. 4), consistent with our previous study (Kato and Katayama 2001). Introducing ΔDARS1 caused a 14% decrease in ATP-DnaA level in Δhda cells, and introducing ΔDARS2 caused a 30% decrease (Fig. 5B; Supplemental Fig. 4). This difference in ATP-DnaA levels is consistent with that observed in the growth rates of the mutant cells (Supplemental Fig. 3). In cells lacking both DARS1 and DARS2, the level of ATP-DnaA decreased by 41%, resulting in a level comparable with that in the wild-type hda control strain (Fig. 5B; Supplemental Fig. 4). These results indicate that DARS1 and DARS2 function independently and concordantly to elevate the ATP-DnaA level in vivo.
DARS1 and DARS2 stimulate replication initiation in vivo
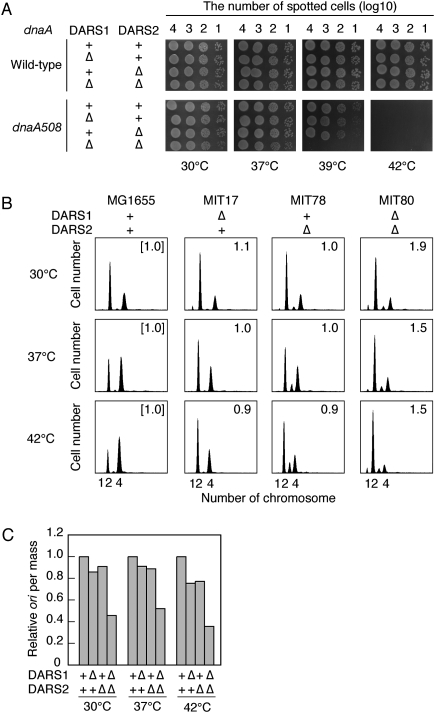
We first investigated whether deletion of DARSs on chromosomes enhanced the thermosensitivity of temperature-sensitive dnaA508 mutants. The dnaA508 allele bears mutations within domain I, which is important for inter-DnaA interaction (Hansen et al. 1992; Weigel et al. 1999; Felczak et al. 2005). Domains III and IV, which bind ATP/ADP and DnaA box (Roth and Messer 1995; Messer 2002; Fujikawa et al. 2003), are intact in this allele, which is consistent with the possibility that initiation activity of DnaA508 might be supported by ATP binding. Simultaneous deletion of both DARSs severely reduced the colony formation ability of dnaA508 mutant cells at 39°C (Fig. 6A). Flow cytometry analysis demonstrated that deletion of DARS1, DARS2, or both repressed extrainitiations that occur in datA or seqA mutant cells (data not shown). These results are consistent with the idea that DARSs support replicational initiation activity in vivo.
Figure 6.
DARS1 and DARS2 are required for the timely initiation of replication. (A) Enhancement of dnaA508 thermosensitivity by deletion of DARS. Cells were grown overnight at 30°C, serially diluted, spotted on LB plate, and incubated for 16 h at 30°C or for 11 h at 37°C, 39°C, or 42°C. (B) Flow cytometry analyses. Cells of MG1655 (wild type), MIT17 (ΔDARS1∷kan), MIT78 (ΔDARS2∷cat), and MIT80 (ΔDARS1∷kan ΔDARS2∷cat) were grown in M9 medium at 30°C, 37°C, or 42°C, followed by run-out replication, as described in Figure 4C. The numbers inserted in each histogram are ratios of mean cell mass to that of wild-type cells at each temperature. (C) Relative ratios of the number of origins (ori) per cell mass in the indicated cells (from B) to that of wild-type cells at the indicated temperatures.
To assess the role of chromosomal DARSs in replication initiation, we analyzed cells lacking DARS1 (MIT17), DARS2 (MIT78), or both (MIT80) by flow cytometry. Wild-type (MG1655) cells mainly contained two or four origins at 30°C, 37°C, or 42°C (Fig. 6B). The cells bearing four origins increased depending on growing temperatures. MIT17 (ΔDARS1) and MIT78 (ΔDARS2) had relatively fewer cells bearing four origins at all temperatures used than MG1655, which is especially evident at 37°C and 42°C. The number of origins per mass in the DARS mutants was less than in the wild-type cells, indicating an inhibition of initiation (Fig. 6C). Furthermore, MIT78 cells bearing only three origins per cell were detected at 37°C and 42°C (Fig. 6B), indicating that synchronous initiations were inhibited.
In the ΔDARS1 ΔDARS2 double mutant (MIT80), a reduction in the number of cells bearing four origins as well as an increase in mean cell mass were observed at each temperature compared with those of the wild type (Fig. 6B). These resulted in a severe reduction in the number of ori per cell mass (Fig. 6C). This indicates that initiation was further inhibited in the double mutant and suggests that the defects in DARS1 and DARS2 act synergistically, especially at 30°C and 37°C (Fig. 6C). At 42°C the synergistic effect was only slight, which might be related to the enhanced defects by a single ΔDARS (Fig. 6C). A moderate increase in mean cell mass in MIT80 allows an increase in the origin number in each cell, resulting in an apparently moderate inhibition in initiation in Figure 6B compared with the data in Figure 6C. In MIT80, inhibition of replication initiation may adversely affect cell division.
The DnaA content of these mutant cells was comparable with that of wild-type cells (Supplemental Fig. 5). Concordantly, these results are consistent with the hypothesis that DARS1 and DARS2 act cooperatively to stimulate the initiation of replication, and that this role of DARSs is required for regulating the initiation of chromosomal replication in coordination with the cell cycle.
We further assessed the total DNA content per cell mass in exponentially growing cells. MIT80 (ΔDARS1 ΔDARS2) cells contained substantially reduced DNA content per cell mass (Supplemental Table 2). In contrast, in cells that replicate the chromosomes independently of oriC, the DNA content was not affected by the simultaneous deletions of DARS1/2 (MIT168 in Supplemental Table 2). These results are consistent with the idea that DARSs promote chromosomal replication in an oriC-dependent manner.
DnaA domains III and IV are required for DARS function
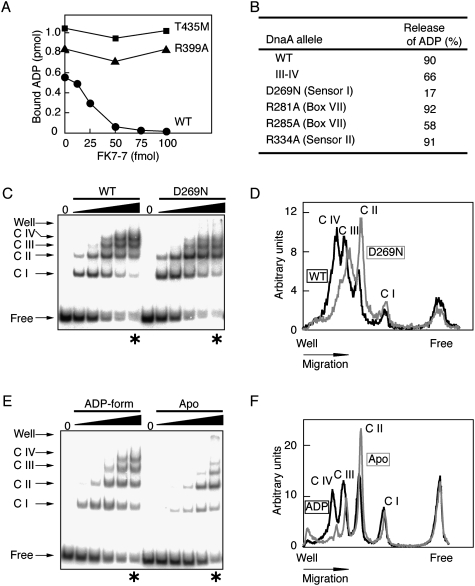
To determine how DARS promotes ADP-DnaA complex dissociation, we performed a biochemical analysis using DnaA mutants. Based on the observation that neither a single DnaA box nor the oriC region promotes the dissociation of ADP-DnaA (data not shown), we inferred that the DnaA multimers formed on DARS1 contain specific inter-DnaA interactions. First, DnaA R399A and DnaA T435M, DnaA domain IV mutants lacking activity of overall DNA binding and DnaA box binding, respectively (Sutton and Kaguni 1997; Blaesing et al. 2000), were defective in DARS1-mediated ADP release (Fig. 7A). These results are consistent with the results of DnaA box substitutions in DARS1 and DARS2 (Figs. 1A–C, 2A). Second, domain I–II-truncated DnaA (denoted DnaA III–IV) fully sustained the DARS1-dependent ADP-releasing activity (Fig. 7B). Domain II is a flexible linker (Abe et al. 2007). These results suggest the specific importance of domain III, which bears the AAA+ motifs, in addition to domain IV.
Figure 7.
Formation of highly ordered complexes on DARS1 is associated with the AAA+ domain and bound nucleotide. (A,B) Analysis of DARS1-dependent ADP-release activity of DnaA mutants. [3H]ADP-bound wild-type or mutant DnaA proteins (2 pmol) were incubated for 15 min at 30°C with 2 mM ATP and the indicated amounts (A) or 50 fmol (B) of FK7-7. In A, wild-type (●) and mutant DnaA (R399A, ▲; T435M, ■) proteins were assessed. In B, the amount of ADP released in a FK7-7-dependent manner is shown as a percentage of the total starting amount of DnaA-bound ADP. (III–IV) Truncated DnaA carrying only domain III (AAA+) and IV. The other DnaA proteins are full length, with amino acid substitutions in the AAA+ motifs indicated. (C) Complexes consisting of DARS1 and wild-type DnaA or DnaA D269N. Various amounts (0, 60, 150, 300, 600, and 1000 fmol) of wild-type DnaA (WT) or DnaA D269N (D269N) were incubated with 32P-labeled FK7-7 (30 fmol) and 2 mM ADP, followed by EMSA. (C I–IV) Complexes I–IV; (Free) protein-free FK7-7. Asterisks indicate the lanes scanned for D. (D) Intensities in the lanes indicated by asterisks in C were determined by densitometric scanning and are presented in arbitrary units. (Black line) Wild-type DnaA; (gray line) DnaA D269N. Migration positions for free DNA (Free) and complexes I–IV (C I–IV) are indicated. (E) Complexes consisting of DARS1 and ADP-DnaA or apo-DnaA. Various amounts (0, 60, 150, 300, 600, and 1000 fmol) of ADP-DnaA (ADP-form) or apo-DnaA (Apo) were incubated for 15 min on ice with 32P-labeled FK7-7 (30 fmol) in the presence or absence of 2 mM ADP, followed by EMSA. Asterisks indicate the lanes scanned for F. (F) Intensities in the lanes indicated by asterisks in E were determined and are presented similarly to D. (Black line) ADP-form; (gray line) Apo. Migration positions for free DNA (Free) and complexes I–IV (C I–IV) are indicated.
Asp-269, the AAA+ sensor 1 motif, is required for DARS function and formation of high-order complexes
First, we analyzed a DnaA AAA+ sensor 1 motif mutant, DnaA D269N. Asp-269 is located near the nucleotide-binding pocket and DnaA D269N has a reduced affinity for ATP/ADP, whereas DnaA box-binding activity is sustained (Kawakami et al. 2006). When ADP-bound DnaA D269N was incubated in the presence of DARS1 and 2 mM ATP, ADP was not substantially released (Fig. 7B), indicating that Asp-269 is required for DARS1 function.
We next analyzed complexes formed by ADP-DnaA and DARS1 using an electrophoretic mobility shift assay (EMSA). EMSA was performed in the presence of 2 mM ADP to detect the complex containing ADP-DnaA and DARS1. With increasing amounts of ADP-DnaA, the migration of DARS1 was sequentially retarded, resulting in at least four discrete bands in the gel (C I–IV in Fig. 7C). These results indicate that ADP-DnaA can form multimeric complexes containing four or more ADP-DnaA molecules on DARS1. Under the same buffer conditions as those used for EMSA, DARS1-dependent ADP release was seen (data not shown).
EMSA further demonstrated that DnaA D269N is impaired to form complexes containing three or more DnaA molecules (Fig. 7C,D), which is consistent with its inability to dissociate DnaA-ADP (Fig. 7B). Initial DARS1-binding rates of DnaA D269N were comparable with those of wild-type DnaA (see free DNA in Fig. 7C), which is consistent with a previous report demonstrating that the affinities of wild-type DnaA and DnaA D269N for a single DnaA box are similar (Kawakami et al. 2006). These results suggest that the conformation of the AAA+ domain is important for the inter-DnaA interaction in DARS complexes and for modulating the interaction with ADP. The sensor 1 motif might directly affect the conformation of the nucleotide-binding pocket in DARS complexes (see below; Discussion).
ADP-DnaA molecules assemble on DARS1 in a unique manner
DnaA R281A and DnaA R285A have substitutions in the arginine residues of the Box VII motif arginine fingers. Although the affinity of these mutant proteins for ATP/ADP is intact, they are defective in specific inter-DnaA interactions within the initiation complexes, and are thus unable to activate replication initiation (Felczak and Kaguni 2004; Kawakami et al. 2005). DnaA R281A and DnaA R285A were able to undergo DARS1-mediated ADP release (Fig. 7B), but DnaA R285A showed a moderate decrease in ADP release. Like wild-type DnaA, DnaA R285A formed multimeric complexes on DARS1 in EMSA (data not shown), which suggests that Arg-285 assists in the conformational change of the protomers. DnaA R334A, which has a substitution in the arginine residue of sensor 2, is defective in DnaA-ATP hydrolysis in RIDA-derived complexes, although its affinity for ATP/ADP is intact (Nishida et al. 2002). This protein was fully active in DARS1-dependent reactivation (Fig. 7B).
Apo-DnaA multimers on DARS1 are unstable
Several AAA+ family proteins form multimers, and nucleotides bound to these multimers mediate interactions between protomers, leading to an overall conformational change of the multimers (Gai et al. 2004; Iyer et al. 2004; Enemark and Joshua-Tor 2006). We thus speculated that the nucleotide status of DnaA might affect DARS-specific reactions. Indeed, formation of DARS complexes containing three or more DnaA molecules was inhibited for apo-DnaA, even when high levels of DnaA were input, and the remaining free DNA levels were similar between ADP-DnaA and apo-DnaA (Fig. 7E,F). Initial DARS1-binding rates of apo-DnaA were inhibited slightly compared with those of ADP-DnaA (see Free DNA in Fig. 7E). These observations suggest that apo-DnaA-DARS1 complexes are unstable. This is consistent with the hypothesis that apo-DnaA, a product of ADP release, efficiently dissociates from DARS complexes, allowing further association of ADP-DnaA and enhancing cyclic reactivation of DnaA (Supplemental Fig. 6).
Discussion
In this study, we demonstrated that specific genomic sequences, DARS1 and DARS2, reactivate DnaA for replication initiation by exchanging bound ADP with ATP in vitro. Further analyses confirmed that similar events also occur in vivo. These findings provide the direct evidence that ATP-DnaA regeneration in vivo can indeed promote initiation of replication, and that specific sequences, DARS1 and DARS2, are required for the novel pathway ensuring the initiation to occur in a cell cycle-coordinated manner. DARS can catalytically regenerate DnaA. The direct regulation of DnaA by the specific DNA regulator would allow a rapid catalytic reaction to occur without the energy used for the synthesis of a protein regulator. The timely initiation of chromosomal replication is crucial for the growth rate-coordinated regulation of the cell cycle in E. coli. The present findings thus serve as an indispensable basis toward uncovering dynamic regulations for the bacterial cell cycle as well as the initiation of chromosomal replication.
Analyses of cells either oversupplied with or lacking DARS1, DARS2, or both revealed that DARSs are important for ensuring the timely initiation of chromosomal replication during the cell cycle. We infer that cell cycle-coordinated regulation of initiation largely depends on DARS driving reactivation of ADP-DnaA. The importance of a single DARS increases at 42°C especially (Fig. 6B,C), suggesting a correlation between the rates of cell growth and DnaA reactivation. Simultaneous deletion of both DARSs synergistically inhibited replication initiation. Thus, DARSs likely play a redundant role in cells and are able to compensate for each other.
Simultaneous deletion of both DARSs decreased the ATP-DnaA level from ∼70% to ∼30% of ATP-DnaA/ADP-DnaA in Δhda cells (Fig. 5B). This supports the hypothesis that DARS is the predominant factor that provides ATP-DnaA. The remaining ∼30% of ATP-DnaA may be provided by de novo synthesis of DnaA or by other pathways (see below). De novo synthesized DnaA would predominantly bind ATP.
Whereas deletions of DARSs synergistically inhibited initiation of replication (Fig. 6C), they exhibited additive effects on the decrease in the level of ATP-DnaA (Fig. 5B). When the level of ATP-DnaA increases prior to initiation in cells lacking either DARS1 or DARS2, the remaining DARS may activate DnaA, which would slightly increase the amount of time needed for ATP-DnaA levels to reach the threshold needed for initiating replication. The simultaneous deletion of both DARSs would further severely inhibit initiation.
Cells lacking both DARS1 and DARS2 can form colonies that are, however, impaired in replication initiation. This suggests that de novo DnaA synthesis and possible regeneration by alternative pathways can provide the minimum amount of ATP-DnaA required for initiation of replication. Acidic phospholipids are candidates for alternative regeneration factors. In addition, there might be unidentified DARSs in the genome. The presence of multiple pathways to provide ATP-DnaA might be advantageous in cell cycle controls responding to various growth conditions.
When DARS was not oversupplied in dnaN(Ts) cells, the ATP-DnaA level was not elevated (Fig. 4), suggesting the presence of a tight regulatory system for DARS activity. Moreover, DARS1 and DARS2 are likely regulated by distinct mechanisms. DARS2 has a specific activator (Fig. 2D,E). Unlike DARS1, DARS2 activity during the cell cycle may be regulated by the dynamics of the DARS2 activator. Oversupply and deletion of DARS2 inhibit cell division and synchronous replication initiations more severely than those of DARS1 (Figs. 4C, 6B). Regulation of DARS2 might be more important in the cell cycle control than that of DARS1. In addition, the chromosomal positions of DARSs may be related to their regulation. These issues remain to be addressed.
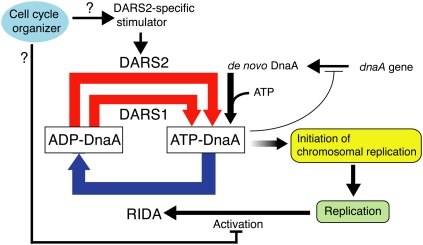
Based on the findings and these analyses of chromosomal DARSs, we propose a model for regulation of initiation linked with the DnaA cycle (Fig. 8). ATP-DnaA is regenerated from ADP-DnaA largely by DARSs. Newly synthesized DnaA would provide ATP-DnaA, which assists the increase of the ATP-DnaA level. Acidic phospholipids might play a similar assisting role. Cells should have a DARS regulatory system including a DARS2-specific stimulator, which could be regulated to function in a cell cycle-coordinated manner. As a result of concordant actions of these factors, the ATP-DnaA level reaches a threshold required for replicational initiation at a timely manner. After replicational initiation, RIDA is activated by DNA loading of the pol III clamps, thereby promoting DnaA-ATP hydrolysis and repression of reinitiations. In fast growing cells, RIDA could be down-regulated in a cell cycle-coordinated manner by an unidentified pathway to assist the increase of the ATP-DnaA level (Kurokawa et al. 1999; Su'etsugu et al. 2008). Moreover, DnaA functions as a transcription repressor, and this activity is also stimulated by ATP binding (Messer and Weigel 1997; Gon et al. 2006); e.g., ATP-DnaA represses dnaA transcription more tightly than ADP-DnaA (Speck et al. 1999). DARS system would affect such transcriptional controls.
Figure 8.
Model for the DnaA cycle in regulation of the replicational initiation. A considerable amount of ATP-DnaA is regenerated from ADP-DnaA by DARSs in a cell cycle-coordinated or constitutive manner, leading to replicational initiation. After this, ATP-DnaA is converted to ADP-DnaA by RIDA. See the text for details.
DARS1 and DARS2 share the DnaA box cluster essential for DARS activity. DARSs probably facilitate a specific inter-DnaA interaction that leads to the release of bound ADP. Indeed, ADP-DnaA molecules form high-order complexes on DARS1 via specific AAA+ domain-dependent interactions (Fig. 7C). Furthermore, DnaA-bound ADP promotes the formation of DARS1 complexes (Fig. 7E), suggesting that a nucleotide-dependent conformational change of the AAA+ domain affects interactions among protomers. Similar examples are reported for AAA+ helicases, such as SV40 Large T antigen and papillomavirus E1 protein (Gai et al. 2004; Enemark and Joshua-Tor 2006).
Based on these and other analyses, we propose a mechanical model for DARS1-mediated reactivation of DnaA (Supplemental Fig. 6). ADP-DnaA molecules assemble on DARS1 in a manner dependent on both DnaA box binding and nucleotide-mediated interaction of AAA+ domains. In these complexes, a heat-dependent conformational change of the AAA+ domain in DnaA occurs, resulting in reduction in ADP affinity, production of apo-DnaA, release of apo-DnaA from the complexes, and further binding of new ADP-DnaA, which thereby allows a catalytic reaction to occur. Released apo-DnaA binds ATP. The flanking region of the DnaA box cluster promotes DARS activity, possibly by facilitating DnaA multimer formation or apo-DnaA dissociation by nonspecific binding.
DnaA sensor 1 Asp-269 is required for DARS1-dependent ADP release and formation of highly ordered complexes on DARS1. Sensor 1 motif is proposed to be an analog of the switch II motif, which is conserved in G-protein families such as the Ras family (Davey et al. 2002; Rossman et al. 2005). Switch II directly binds GEF (guanidine nucleotide exchange factor), thereby promoting the release of GDP bound to G-protein. Similarly, the Asp-269 residue might directly contact an adjacent protomer during complex formation, causing stabilization of the complexes and a conformational change of the DnaA AAA+ domain that decreases its affinity for nucleotides. Alternatively, a mutation in Asp-269 could indirectly affect the interaction between AAA+ domains by introducing an overall conformational change in the nucleotide-binding pocket (Kawakami et al. 2006).
DARS-like DnaA box clusters are seen in genomic sequences of many eubacterial species. In Shigella flexneri, Salmonella typhimurium, Yersinia pestis, Photorhabdus luminescens, Erwinia carotovora atroseptica, and Vibrio cholerae, the DnaA box clusters upstream of uvrB and mutH are highly conserved (Supplemental Fig. 7), suggesting that DARS1 and DARS2 are conserved among E. coli-related species. In other proteobacterial species, a DnaA box cluster occurs in various intergenic regions. In species other than proteobacterial species, DnaA box clusters occur near the dnaA gene or other genes with arrangements of DnaA boxes different from those of E. coli DARS, which might interact with the cognate DnaA orthologs and function as DARS.
Materials and methods
Protein, primers, strains, and plasmids
Protein, primers, strains, and plasmids are provided in the Supplemental Material.
ADP-releasing activity, exchange of DnaA-bound nucleotides, and initiation activity of DnaA in vitro
These in vitro experiments were performed as described previously (Fujimitsu and Katayama 2004), with the following minor modifications. Reaction solutions for ADP release and exchange of the DnaA-bound nucleotide included 100 mM potassium glutamate. Reactions assessing the activity of pACYC177-DARS2 included poly dI–dC (150 ng).
Preparation of the crude extract capable of stimulating DARS2 activity
Exponentially growing cells were collected by centrifugation, suspended to an optical density (A595) of 250 in chilled buffer (50 mM HEPES-KOH at pH 7.6, 300 mM KCl, 2 mM dithiothreitol), incubated for 30 min on ice in the presence of lysozyme (300 μg/mL), and frozen in liquid nitrogen. After thawing at 0°C–4°C, cleared lysates were obtained by centrifugation at 48,000 rpm for 30 min in a Beckman Ti 50-2 rotor. Ammonium sulfate (0.33 g/mL) was added slowly, with stirring, to the supernatant, followed by further stirring for 30 min. Precipitates were collected by centrifugation as above, and dissolved in buffer (50 mM HEPES-KOH at pH 7.6, 10% [v/v] glycerol, 1 mM EDTA, 2 mM dithiothreitol).
Determination of DnaA nucleotide form and cellular DnaA content, and flow cytometry
The nucleotide forms of cellular DnaA were determined as described previously (Kurokawa et al. 1999; Fujimitsu et al. 2008). DnaA content in the cells was determined by immunoblot analysis, as described previously (Fujimitsu et al. 2008). Flow cytometry analysis was performed as described previously (Kawakami et al. 2006; Keyamura et al. 2007; Fujimitsu et al. 2008).
EMSA using DARS1
FK7-7 was PCR-amplified using pOA21 and primers (32P-5′-end-labeled SIS-6 and SIS-7) (Supplemental Table 3), followed by purification using a Wizard SV spin column (Promega) and a CENTRI-SEP column (Princeton Separations). ADP-DnaA was prepared by incubation of apo-DnaA with 2 μM ADP for 15 min at 0°C, with the exception of DnaA D269N, which was incubated with 2 mM ADP. ADP-DnaA was incubated for 5 min at 0°C or 30°C in 12.5 μL of buffer (20 mM HEPES-KOH at pH 7.6, 10 mM magnesium acetate, 1 mM EDTA, 8 mM dithiothreitol, 0.1 mg/mL bovine serum albumin, 5% glycerol, 50 mM potassium glutamate, 2 mM ADP, 21 ng poly dI–dC, 2.1 ng [30 fmol] of 32P-FK7-7). The mixtures were separated by 6% PAGE using 90 mM Tris-borate buffer at room temperature and were analyzed using BAS2500 (FujiFilm). When apo-DnaA was included in experiments, 125 mM potassium chloride was included in the DARS1-binding reaction, ADP was excluded in the DARS1-binding reaction and electrophoresis was performed at 4°C.
The reconstituted RIDA system
The reconstituted RIDA system was used as described previously (Su'etsugu et al. 2005, 2008). Briefly, [α-32P]ATP-DnaA (1 pmol) was incubated for 20 min at 30°C in buffer (12.5 μL) containing ADP-Hda-cHis, the DNA-loaded clamp, and 2 mM ATP. Nucleotides bound to DnaA were recovered on nitrocellulose filters and analyzed using thin-layer chromatography and BAS2500.
Acknowledgments
This work was supported mainly by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Technology, and Science of Japan.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1775809.
Supplemental material is available at http://www.genesdev.org.
References
- Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282:17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- Blaesing F, Weigel C, Welzeck M, Messer W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol Microbiol. 2000;36:557–569. doi: 10.1046/j.1365-2958.2000.01881.x. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Crooke E, Castuma CE, Kornberg A. The chromosome origin of Escherichia coli stabilizes DnaA protein during rejuvenation by phospholipids. J Biol Chem. 1992;267:16779–16782. [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O'Donnell M. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J Biol Chem. 2004;279:51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Katayama T. Reactivation of DnaA by DNA sequence-specific nucleotide exchange in vitro. Biochem Biophys Res Commun. 2004;322:411–419. doi: 10.1016/j.bbrc.2004.07.141. [DOI] [PubMed] [Google Scholar]
- Fujimitsu K, Su'etsugu M, Yamaguchi Y, Mazda K, Fu N, Kawakami H, Katayama T. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J Bacteriol. 2008;190:5368–5381. doi: 10.1128/JB.00044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Gon S, Camara JE, Klungsoyr HK, Crooke E, Skarstad K, Beckwith J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen FG, Koefoed S, Atlung T. Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol Gen Genet. 1992;234:14–21. doi: 10.1007/BF00272340. [DOI] [PubMed] [Google Scholar]
- Heacock PN, Dowhan W. Alteration of the phospholipid composition of Escherichia coli through genetic manipulation. J Biol Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kaguni JM. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- Katayama T, Fujimitsu K, Ogawa T. Multiple pathways regulating DnaA function in Escherichia coli: Distinct roles for DnaA titration by the datA locus and the regulatory inactivation of DnaA. Biochimie. 2001;83:13–17. doi: 10.1016/s0300-9084(00)01206-2. [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- Kawakami H, Ozaki S, Suzuki S, Nakamura K, Senriuchi T, Su'etsugu M, Fujimitsu K, Katayama T. The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol Microbiol. 2006;62:1310–1324. doi: 10.1111/j.1365-2958.2006.05450.x. [DOI] [PubMed] [Google Scholar]
- Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su'etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes & Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Messer W, Weigel C. DnaA initiator—Also a transcription factor. Mol Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Nishida S, Fujimitsu K, Sekimizu K, Ohmura T, Ueda T, Katayama T. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: Evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- Obita T, Iwura T, Su'etsugu M, Yoshida Y, Tanaka Y, Katayama T, Ueda T, Imoto T. Determination of secondary structure in solution of Escherichia coli DnaA DNA-binding domain. Biochem Biophys Res Commun. 2002;299:42–48. doi: 10.1016/s0006-291x(02)02590-1. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Roth A, Messer W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995;14:2106–2111. doi: 10.1002/j.1460-2075.1995.tb07202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J Biol Chem. 1995;270:17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Kornberg A. Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1988;263:7131–7135. [PubMed] [Google Scholar]
- Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Shimuta T, Nakano K, Yamaguchi Y, Ozaki S, Fujimitsu K, Matsunaga C, Noguchi K, Emoto A, Katayama T. Novel heat shock protein HspQ stimulates the degradation of mutant DnaA protein in Escherichia coli. Genes Cells. 2004;9:1151–1166. doi: 10.1111/j.1365-2443.2004.00800.x. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Lobner-Olesen A, Atlung T, von Meyenburg K, Boye E. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol Gen Genet. 1989;218:50–56. doi: 10.1007/BF00330564. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Bernander R, Boye E. Analysis of DNA replication in vivo by flow cytometry. Methods Enzymol. 1995;262:604–613. doi: 10.1016/0076-6879(95)62047-8. [DOI] [PubMed] [Google Scholar]
- Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su'etsugu M, Shimuta T, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- Su'etsugu M, Nakamura K, Keyamura K, Kudo Y, Katayama T. Hda monomerization by ADP binding promotes replicase clamp-mediated DnaA-ATP hydrolysis. J Biol Chem. 2008;283:36118–36131. doi: 10.1074/jbc.M803158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD, Kaguni JM. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J Biol Chem. 1997;272:23017–23024. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- Torheim KN, Boye E, Lobner-Olesen A, Stokke T, Skarstad K. The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol Microbiol. 2000;37:629–638. doi: 10.1046/j.1365-2958.2000.02031.x. [DOI] [PubMed] [Google Scholar]
- Weigel C, Schmidt A, Seitz H, Tungler D, Welzeck M, Messer W. The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol Microbiol. 1999;34:53–66. doi: 10.1046/j.1365-2958.1999.01568.x. [DOI] [PubMed] [Google Scholar]
- Xia W, Dowhan W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]