Abstract
Protein translocation begins with the efficient targeting of secreted and membrane proteins to complexes embedded within the membrane. In Eukarya and Bacteria, this is achieved through the interaction of the signal recognition particle (SRP) with the nascent polypeptide chain. In Archaea, homologs of eukaryal and bacterial SRP-mediated translocation pathway components have been identified. Biochemical analysis has revealed that although the archaeal system incorporates various facets of the eukaryal and bacterial targeting systems, numerous aspects of the archaeal system are unique to this domain of life. Moreover, it is becoming increasingly clear that elucidation of the archaeal SRP pathway will provide answers to basic questions about protein targeting that cannot be obtained from examination of eukaryal or bacterial models. In this review, recent data regarding the molecular composition, functional behavior and evolutionary significance of the archaeal signal recognition particle pathway are discussed.
Keywords: protein targeting, protein translocation, ribonucleoprotein complex, RNA, signal sequence
Introduction
A first step in the biogenesis of nascent polypeptide chains destined for translocation into or across the plasma membrane involves the correct recognition and delivery of these extracytoplasmic proteins to membranous translocation sites. Analysis of available archaeal genomes as well as the examination of individual genes has revealed that Archaea contain components of the signal recognition particle (SRP), a ribonucleoprotein complex involved in protein targeting in Eukarya and Bacteria (Herskovits et al. 2000, Eichler and Moll 2001, Keenan et al. 2001). In many aspects, the archaeal SRP represents a mosaic of components and properties associated with the bacterial and eukaryal SRP complexes. Certain aspects of the archaeal SRP-mediated translocation pathway are, however, apparently unique to members of this domain of life, possibly reflecting the distinctive biological strategies developed by Archaea. In this review, recent findings related to the structure, function and assembly of the archaeal SRP are discussed.
The signal recognition particle: linking protein translation to protein translocation
Most of our understanding of the SRP pathway has come from examination of the eukaryal system. In terms of molecular composition and functional behavior, the SRP of higher Eukarya remains the best-understood example of the particle to date. Consisting of six proteins (SRP72, SRP68, SRP54, SRP19, SRP14 and SRP9) and a 7S RNA, the eukaryal SRP recognizes and binds to the signal sequence of an emerging nascent polypeptide via the SRP54 subunit (Walter and Johnson 1994, Lütcke 1995, Keenan et al. 2001). Then, relying on the interaction between the SRP9/14 heterodimer and the translating ribosome, the SRP arrests further elongation of the nascent chain. Subsequently, the ribosome–nascent polypeptide–SRP complex is targeted to the membrane of the endoplasmic reticulum via the affinities of SRP for its membrane-bound receptor (SR) and of the ribosome for the translocon, the complex responsible for protein translocation into the lumenal space of the endoplasmic reticulum (Kalies et al. 1994, Young et al. 1995). The interaction of the SRP with its receptor initiates a synchronized series of GTP hydrolysis events in which three GTP-hydrolyzing proteins (SRP54 and the SR a and b subunits) regulate the release of SRP from the ribosome–nascent protein–SRP complex, leading to the resumption of protein translation directly into the translocon pore (Miller et al. 1993, Powers and Walter 1995, Bacher et al. 1999).
In Bacteria (i.e., Escherichia coli), a much simpler version of the SRP is present (Herskovits et al. 2000). Consisting solely of the SRP54 homolog Ffh and a 4.5S RNA, the bacterial SRP, acting together with the bacterial SRP receptor protein, FtsY, plays a role in the insertion of certain plasma membrane proteins (Seluanov and Bibi 1997, Ulbrandt et al. 1997) and may also be involved in protein secretion (Phillips and Silhavy 1992).
At first glance, the archaeal SRP pathway appears to correspond to a hybrid of the eukaryal and bacterial systems (Table 1, Figure 1). Consisting of an SRP RNA that is strikingly similar in secondary structure to its eukaryal counterpart (Figure 2), as well as SRP54 and SRP19 subunits, the archaeal SRP can be considered a precursor of the SRP of higher eukaryotes. The mode of SRP-mediated membrane targeting in Archaea is, however, apparently similar to that in Bacteria, relying on archaeal versions of bacterial FtsY rather than homologs of the eukaryal SR a and b subunits. However, despite these general similarities between the archaeal and eukaryal or bacterial SRP pathways, closer examination reveals Archaea-specific SRP characteristics.
Table 1.
Components of the signal recognition particle (SRP)-mediated pathway in the three domains of life.
| Bacteria | Archaea | Eukarya | |
| SRP components | 4.5S RNA | SRP RNA | SRP RNA |
| Ffh (SRP54) | SRP54 | SRP54 | |
| SRP19 | SRP19 | ||
| SRP9 | |||
| SRP14 (mammals) | |||
| SRP21 (yeast) | |||
| SRP68 | |||
| SRP72 | |||
| SRP receptor | FtsY | FtsY | SRα |
| SRβ | |||
Figure 1.
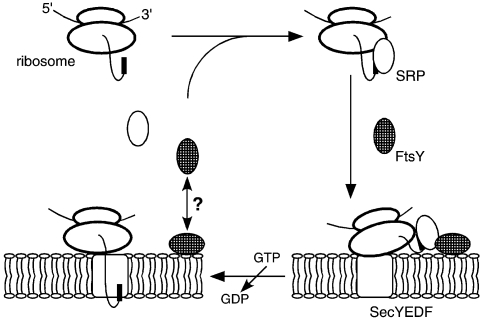
The hypothetical signal recognition particle (SRP) cycle of Archaea. Proteins destined to cross the archaeal plasma membrane are synthesized with a cleavable N-terminal extension (the signal sequence, shown as a bold black line), although exceptions to this rule exist. The signal sequence is recognized by the SRP54 subunit of the SRP. Although the diagram depicts the interaction of SRP54 and the signal sequence occurring soon after the onset of protein synthesis, it remains unclear at which step this interaction takes place. The SRP–polypeptide complex is then delivered to the membrane, presumably through the affinity of the SRP for FtsY (hatched), the archaeal SRP receptor. It is presently unknown whether a soluble form of FtsY binds the complex and only then interacts with the membrane or whether a membrane-associated form of FtsY is employed. The nature of the FtsY–membrane interaction also remains to be determined. Although the role played by GTP in the archaeal SRP cycle is currently undefined, it is likely that GTP hydrolysis events mediated by SRP54 or FtsY lead to the release of SRP and FtsY from the complex and the transfer of the ribosome to the membrane-embedded SecYEDF translocation machinery, possibly with the bulk of the nascent polypeptide chain still being translated. The Sec complex, presumably responsible for both protein translocation and insertion in Archaea, may contain additional Archaea-specific subunits.
Figure 2.
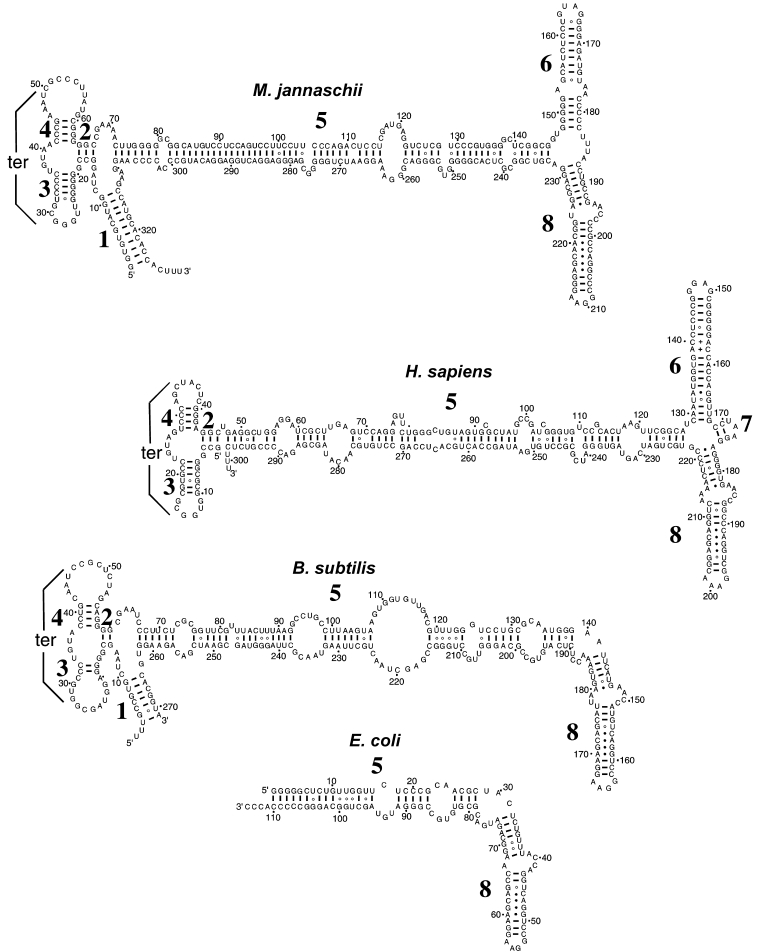
Secondary structures of signal recognition particle (SRP) RNA from Methanococcus jannaschii, Homo sapiens, Bacillus subtilis and Escherichia coli, with base pairings supported by comparative sequence analysis (Fox and Woese 1975). The 5′ and 3′ ends of RNA molecules are labeled, and helices are numbered 1 to 8 according to the nomenclature of Larsen and Zwieb (1991). One A –C and two A –G pairings in Helix 8 that exist in a homologous structure (Batey et al. 2000) are connected by dots. The “+” symbols in Helix 6 of human SRP RNA indicate interactions in the crystal structure of a 28-mer duplex (Wild et al. 1999). Residues are numbered in increments of 50 and marked in increments of 10 nucleotides. Tertiary interactions (“ter”) between the loops of Helices 3 and 4 are indicated.
The SRP of Archaea
SRP RNA
Like its eukaryal counterpart, the archaeal SRP contains a 7S RNA molecule instead of a smaller SRP RNA like those found in most bacteria (Figure 2). In contrast to the variability in size associated with bacterial SRP RNA, archaeal SRP RNAs that have been identified to date contain about 300 nucleotides, much like human SRP RNA (Kaine 1990, Althoff et al. 1994). Eukaryal and archaeal SRP RNA are both composed of seven helices. In Eukarya, Helices 2, 3 and 4 bind the SRP9/14 heterodimer; most of Helix 5 binds the SRP68 /72 heterodimer; Helices 6 and 8 bind SRP 19; and Helix 8 binds the SRP54 protein (Siegel and Walter 1988, Strub and Walter 1990, Larsen and Zwieb 1991). In terms of primary structure, only the proximal and distal bulges of Helix 8 and the tetranucleotide loop of Helix 6 exhibit striking sequence conservation in all three domains of life (Larsen and Zwieb 1991, Althoff et al. 1994, Batey et al. 2000). Despite the overall lack of sequence conservation, archaeal SRP RNA can be folded into a secondary structure virtually identical to that of human SRP RNA, differing only in the presence of archaeal Helix 1, formed upon pairing of the 5′ and 3′ ends (Kaine 1990), and the absence of the uniquely eukaryotic Helix 7 (Larsen and Zwieb 1991). Although not found in eukaryotes, Helix 1 is present in the SRP RNA of the gram-positive bacterium Bacillus subtilis (Struck et al. 1988; see Figure 2). Examination of archaeal SRP RNA sequences available to date reveals that despite their phylogenetic and phenotypic diversity, the various archaeal SRP RNA molecules display striking similarities in even fine details of secondary structure, such as the positions and sizes of internal loops in Helix 5, the major backbone of the molecule (Figure 2). Finally, archaeal SRP RNA may fulfill the roles of the SRP 68 /72 and 9/14 heterodimers; although binding sites for these proteins may exist on the archaeal SRP RNA molecule (Bhuiyan et al. 2000), genomic analyses have failed to reveal the existence of archaeal homologs of these eukaryal proteins.
SRP19
To date, 10 archaeal SRP19 sequences have been identified (Table 2). Recent SRP reconstitution studies have shown that, as in mammals, archaeal SRP19 plays a role in SRP assembly, interacting with SRP RNA and thereby facilitating the binding of SRP54 to the SRP RNA molecule (Bhuiyan et al. 2000, Diener and Wilson 2000). Unlike in Eukarya, however, the interaction between SRP RNA and SRP54 is not entirely SRP19-dependent, and significant amounts of binding can occur in the absence of SRP19 (Moll et al. 1999, Bhuiyan et al. 2000). Therefore, examination of SRP19 binding to the RNA molecule in Archaea provides an opportunity to assess the contribution of SRP19 to SRP assembly. Accordingly, SRP19 of Archaeoglobus fulgidus has been shown to bind a truncated SRP RNA fragment containing only Helices 6 and 8 (Bhuiyan et al. 2001). It is thus likely that archaeal SRP19, like human SRP19 (Rose and Weeks 2001), binds to the tetraloop of SRP RNA Helix 6 and the distal section of Helix 8, possibly leading to conformational changes at the asymmetric bulge in Helix 8. The SRP19-induced change in the position or folding of Helix 8 would result from the interaction of SRP19 with the more rigid Helix 6 (Bhuiyan et al. 2000, Diener and Wilson 2000). The structural changes realized in Helix 8 are, in turn, thought to promote SRP54 binding (Diener and Wilson 2000, Rose and Weeks 2001), as also proposed for the 4.5S SRP RNA and the Ffh protein in E. coli (Batey et al. 2000).
Table 2.
Identified components of the signal recognition particle (SRP) pathway in Archaea, arranged according to NCBI taxonomy. Bolded accession numbers represent sequences obtained from completed genomes, indicating that the failure to detect a given sequence in a particular strain may be meaningful. GSP refers to a genome sequencing project with sequences that have not yet been submitted to GenBank. The Pyrococcus furiosus sequences were provided at http://www.genome.utah.edu/Pfu102000.gb. The listed sequences are accessible from the SRPDB at http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html and its European mirror site at http://bio.lundberg.gu.se/dbs/SRPDB/SRPDB.htm
| Subdomain | Order | Species | SRP RNA | SRP19 | SRP54 | SRP receptor |
| Crenarchaeota | Desulfurococcales | Pyrodictium occultum | M21985 | |||
| Aeropyrum pernix | AP000058 | BAA79750 | BAA80736 | BAA80066 | ||
| Sulfolobales | Acidianus ambivalens | Y08735 | X95989 | |||
| Sulfolobus solfataricus | X17239 | AAK40510 | AAK4124 | X58538 | ||
| AE006647 | ||||||
| Sulfolobus acidocaldarius | Y12702 | X77509 | ||||
| Euryarchaeota | Archaeoglobales | Archaeoglobus fulgidus | X17237 | AF1258 | 3334342 | 2648469 |
| Halobacteriales | Halobacterium sp. NRC-1 | X01698 | Q9HQ21 | AAG20536 | AAG20538 | |
| AE005009 | ||||||
| Methanobacteriales | Methanothermus fervidus | M32222 | ||||
| Methanobacterium | ||||||
| thermoautotrophicum | X15364 | AE000804 | 3334341 | 2622733 | ||
| AE000940 | ||||||
| Methanococcales | Methanococcus jannaschii | U67510 | U67546 | U67467 | MJU67484 | |
| Methanococcus voltae | M22560 | |||||
| Methanosarcinales | Methanosarcina acetivorans | X17238 | ||||
| Thermococcales | Thermococcus sp. AN1 | TSU95207 | ||||
| Thermococcus zilligii | 2130562 | |||||
| Thermococcus celer | X17240 | |||||
| Pyrococcus horikoshii | AB009466 | AP000007 | 3258124 | D1031724 | ||
| Pyrococcus furiosus | GSP | GSP | GSP | GSP | ||
| Pyrococcus abyssi | AJ248283 | CAB49350 | CAB49401 | CAB49424 | ||
| Thermoplasmales | Thermoplasma volcanium | NC002689 | 13540831 | BAB60164 | BAB59674 | |
| Thermoplasma acidophilum | AL445063 | CAC11655 | CAC12205 | |||
Sequence alignments of available SRP19 sequences have revealed that the archaeal SRP19 proteins are generally shorter than their eukaryal homologs, partly as a result of the absence of a connecting region that separates conserved regions II and III of the protein (Bhuiyan et al. 2001). This missing region, which would be expected to lie between Residues 72 and 73 of the A. fulgidus sequence (Figure 3), may be involved in the binding of SRP19 to Helix 7, which is absent in archaeal SRP RNA but present in the eukaryal molecule (Figure 2). Examination of the recently released structure of A. fulgidus SRP19 at the atomic level (Pakhomova et al. 2001) should shed light on structure–function properties of the protein.
Figure 3.
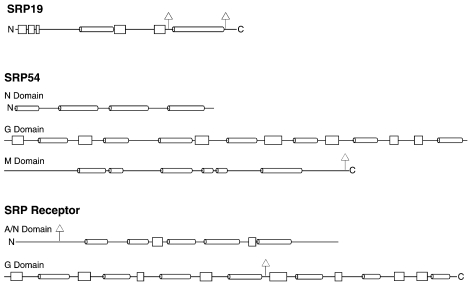
Protein secondary structures of archaeal SRP19, SRP54 and the SRP receptor. Alpha helices are drawn as cylinders, beta conformations as rectangles. The N- and C-termini are indicated. Only the secondary structure of Archaeoglobus fulgidus SRP19 has been determined experimentally (Pakhomova et al. 2001). For SRP54, structural information was obtained by comparison with the NG region of SRP54 of Acidianus ambivalens (Montoya et al. 2000) and the human SRP54 M domain (Clemons et al. 1999). Delta sign posts indicate regions that are absent in Archaea but present in Bacteria, or Eukarya or both.
Surprisingly, after examination of the recently completed genome of Thermoplasma acidophilum (Ruepp et al. 2000), we have failed to detect an SRP19 subunit. Moreover, examination of the SRP19 binding site on Helix 6 of the T. acidophilum SRP RNA reveals a G to A modification in the first tetraloop position. The same modification has been detected in Helix 6 of the SRP RNA of Thermoplasma volcanium, which contains a gene encoding SRP19 (Kawashima et al. 2000). The T. volcanium SRP19 is, however, the smallest known archaeal version of SRP19 (84 amino acid residues) and lacks residues that are expected to be required for RNA binding. Thermoplasma volcanium SRP19, if expressed, may therefore not interact with SRP RNA. Thus, it is possible that the SRP of the genus Thermoplasma represents a version of SRP intermediate between that of Archaea and Bacteria. Indeed, Thermoplasma possesses traits reminiscent of gram-positive bacteria (Gupta 2000). Another element of the protein translocation apparatus of Thermoplasma, i.e., signal peptidase (the enzyme responsible for removing signal sequences following the completion of translocation), also possesses more bacteria-like properties than do other archaeal versions of the protein (Eichler 2002). Alternatively, SRP19 may not be essential for the protein targeting function of archaeal SRP.
SRP54
Responsible for binding the emerging signal sequence of a nascent polypeptide chain and interacting with the SRP receptor, SRP54 (or its bacterial homolog, Ffh) is an essential component of the SRP pathway in all three domains of life. The protein (Figure 3) can be divided functionally into the C-terminal M region, which is involved in signal sequence recognition and SRP RNA binding, and the NG region, which is responsible for binding of guanidine nucleotides through five well-conserved sequence elements, G1 –G5 (Bernstein et al. 1989, Römisch et al. 1989).
The recent determination of the crystal structure of the NG region of Acidianus ambivalens SRP54 offers an understanding of archaeal SRP54 nucleotide binding behavior at the molecular level (Montoya et al. 2000). In the eukaryal SRP54 protein, a conserved threonine residue within the G2 sequence element is essential for GTP hydrolysis, because of its interaction with the Mg2+ ion of Mg2+-GTP (Bourne et al. 1991). In the A. ambivalens SRP54 NG region, this threonine is substituted with a hydrophobic valine residue (V136). Therefore, the role normally played by the absent threonine residue must be assumed by a neighboring amino acid. Alignment of available archaeal SRP54 sequences also reveals a similar threonine–valine substitution in T. acidophilum and T. volcanium, whereas in Sulfolobus acidocaldarius, the threonine residue is replaced by an isoleucine residue. The absence of threonine in the SRP54 nucleotide binding site suggests that some archaeal strains may employ a mechanism of GTP hydrolysis that differs from that found in eukaryal SRP54 proteins. This modified mechanism may be similar to that employed by bacteria, where hydrophobic valine, isoleucine or leucine residues often replace the G2 threonine residue.
More pronounced than these structural differences is the presence of a dipeptide (GY) insertion between Residues 193 and 194 of the A. ambivalens sequence, immediately downstream of the highly conserved G3 sequence element. Examination of available SRP54 sequences reveals that this insertion is detected only in crenarchaeal proteins for which sequences are available (S. acidocaldarius, Sulfolobus solfataricus and Aeropyrum pernix, where the tyrosine residue is replaced by histidine) yet is absent from euryarchaeal, bacterial and eukaryal SRP54 proteins. The hydroxyl group of Y194 is connected to residues within the G1 and G4 sequence elements through a network of hydrogen bonds or via water molecules. The functional significance of this insertion is unclear, although it has been proposed that it could function as a sensor of nucleotide occupancy of the active site, communicating such information between the G1 and G4 sequence elements, and to the neighboring I-box, an α-β-α insertion within the GTPase domain of the protein, which is thought to play a role in nucleotide exchange (Moser et al. 1997).
As noted above, reconstitution studies involving SRP RNA and SRP54 from homogeneous or heterogeneous archaeal systems have revealed the ability of these two components to interact in the absence of SRP19. For instance, A. fulgidus SRP54 was shown to bind not only to A. fulgidus SRP RNA, but also to Methanococcus jannaschii (and human) SRP RNA (Bhuiyan et al. 2000). Similarly, A. ambivalens SRP54 was shown to bind specifically to the SRP RNA of the related crenarchaeon S. solfataricus (Moll et al. 1999). The ability of SRP RNA and SRP54 to interact in the absence of SRP19 could reflect the requirement for a more stable SRP in Archaea, given the various environmental challenges with which these microorganisms must cope (Diener and Wilson 2000).
The SRP receptor, FtsY
Delivery of SRP–nascent chain complexes to the archaeal plasma membrane is likely to be mediated by FtsY, the SRP receptor (Miller et al. 1994). Having arisen along with SRP54 as the result of a gene duplication event (Althoff et al. 1994), FtsY also contains the G1–G5 sequence elements involved in GTP binding. Indeed, comparison of archaeal SRP54 and FtsY sequences lends support to the hypotheses that the tree of life bifurcated between the bacterial and archaea l/eukaryal branches (Gribaldo and Cammarano 1998), and that average protein evolutionary rates in Archaea are much slower than in Bacteria or Eukarya (Kollman and Doolittle 2000).
The FtsY protein can be functionally divided into the N-terminal A region and the C-terminal NG region (Figure 3). Responsible for GTP binding (Montoya et al. 1997), the NG region is well conserved in Archaea, Bacteria and Eukarya (Zwieb and Samuelsson 2000). As in Bacteria, the archaeal FtsY A region is believed to be responsible for binding the protein to the membrane. The manner of this membrane–protein interaction remains unclear, however, as does the significance of substantial cytoplasmic FtsY pools. Although archaeal FtsY A regions differ significantly in length and amino acid composition, clusters of lysine or arginine residues are found in the extreme N-terminal region in all cases. In the absence of either transmembrane sequences in FtsY or identified proteinaceous membrane receptors, it is thought that these positively charged regions represent a site that binds archaeal FtsY proteins to anionic phospholipids of the plasma membrane, as proposed for the N-terminal region of the bacterial protein (de Leeuw et al. 1997, Powers and Walter 1997, Zelazny et al. 1997). A possible exception to this hypothesis may be the FtsY protein of Halobacterium sp. NRC-1, in which only three positively charged residues are found within the 46 N-terminal residues (as opposed to 8–13 positively charged residues in other archaeal FtsY proteins). This exception could represent an adaptation to the saline environment encountered by this archaeon, or could reflect an alternative mode of FtsY–membrane interaction. Indeed, it is entirely plausible that archaeal FtsY proteins rely on a novel mode of phospholipid interaction, given the ether-based lipids that make up the archaeal membrane.
The role of SRP in archaeal protein translocation
Despite the presence of SRP-mediated translocation pathway elements in Archaea (Althoff et al. 1994, Eichler and Moll 2001), the in vitro reconstitution of an archaeal SRP particle from purified components (Bhuiyan et al. 2000, Diener and Wilson 2000) and the reported ability of archaeal SRP54 to interact with a signal sequence (Bhuiyan et al. 2000), the involvement of SRP in archaeal protein translocation has yet to be conclusively demonstrated. To date, the only experiments addressing this point have focused on the biosynthesis of the membrane protein bacterioopsin in Halobacterium salinarium (Gropp et al. 1992). These studies revealed the co-sedimentation of 7S RNA and bacterioopsin mRNA with membrane-bound polysomes, puromycin-mediated release of 7S RNA from polysomes and a correlation of amount of ribosome-bound 7S RNA with bacterioopsin expression. Subsequent studies have reported the co-translational nature of bacterioopsin insertion (Dale et al. 2000). Collectively, these results can be interpreted as support for the existence of an SRP-dependent mode of protein translocation in Archaea, although it has been suggested that a dedicated system may be responsible for purple membrane biogenesis (Ortenberg and Mevarech 2000).
The evolution of SRP
Comparison of the archaeal SRP with its eukaryal and bacterial counterparts has led to the hypothesis that the archaeal complex, comprising SRP RNA, SRP54 and SRP19, represents the precursor of the SRPs found in the other domains (Eichler and Moll 2001). It is possible that with the development of alternative protein targeting pathways, such as the SecB or Tat systems (Fekkes and Driessen 1999, Berks et al. 2000), during evolution, the bacterial system became simplified and may have lost the need for the SRP19 subunit found in the primitive particle. Concomitant with such changes, an enhanced ability of SRP54 to bind SRP RNA could have evolved (Diener and Wilson 2000). In contrast, with the development of various intracellular organelles, the eukaryal SRP became more complex in structure, incorporating additional subunits. Such complexity is reflected in the spatial and temporal separation of SRP assembly in Eukarya (Politz et al. 2000). In the apparent absence of these additional subunits, it is possible that archaeal SRP RNA may have assumed novel functions. For instance, it has been proposed that the Alu domain of archaeal SRP RNA could fulfill the roles of the SRP9/14 heterodimer (Bovia and Strub 1996) responsible for translational arrest in mammals (Chang et al. 1997). Figure 4 depicts how, in the absence of the SRP9/14 heterodimer, additional RNA found in the archaeal molecule leads to the formation of a molecular fold in the RNA molecule similar to that which is assumed to form in the presence of the heterodimer, presumably to fulfill the functions of the absent proteins. The greater importance of RNA as opposed to protein in the archaeal SRP may also be reflected in the occurrence of several small deletions in the three SRP-associated proteins, as indicated in Figure 3. Alternatively, it is possible that the archaeal SRP also includes SRP9/14- and SRP68 /72-like proteins that share little sequence homology with their eukaryal homologs, given the failure of genome searches to detect such components in Archaea. Finally, considering the intriguing possibility that SRP RNA could interact directly with signal sequences (Batey et al. 2000), it is also conceivable that the primitive SRP contained only RNA, and later evolved to incorporate the SRP54 subunit. With time, other subunits were added as the need arose. The SRP19 protein began to appear in some, but not all Archaea. Eukarya developed a more complex SRP, adding additional subunits with specialized functions and compartmentalizing SRP assembly.
Figure 4.
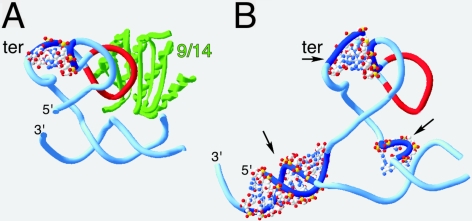
Small SRP RNA domains. (A) Structure of the human SRP Alu domain (Weichenrieder et al. 2000). The RNA backbone is shown as a ribbon, with backbone and side-chain residues drawn to emphasize the tertiary interaction (marked “ter”). The SRP9/14 protein heterodimer, present only in the human SRP, is labeled “9/14.” The conserved RNA loop that serves as a protein binding site for SRP9/14 is colored in red. (B) Three-dimensional homology model of the Methanococcus jannaschii domain incorporating the conserved features of the human structure and using comparative sequence analysis (Larsen and Zwieb 1991) for other structural features. Backbone and side-chain residues are drawn to show the more extensive tertiary interaction. Three regions containing “extra” RNA are labeled with arrows. The model was generated with the program ERNA-3D (Mueller et al. 1995) on a Silicon Graphics Octane workstation. The figure was generated with the SwissPdbViewer program available at http://www.expasy.ch/spdbv/.
Conclusions and future directions
The near future promises advances in our understanding of the archaeal SRP pathway. With the upcoming release of additional completed archaeal genomes, a larger sample set of archaeal SRP components will become available for comparative analysis. Given the ability of archaeal proteins to remain properly folded in the face of a variety of harsh conditions, archaeal components could become preferred substrates for protein crystallization and other structural studies of the SRP pathway. As improvements are made to SRP reconstitution efforts, questions such as the respective roles of the RNA and protein components of the SRP can be addressed. With recent advances in the establishment of archaeal in vitro translation and translocation systems (Sanchez and Amils 1995, Ring and Eichler 2001), it will soon be possible to directly address the role of the SRP in archaeal protein translocation as well as the interplay between protein translation, translocation and SRP. Information obtained from such studies will not only further our understanding of protein translocation, but will also serve to elucidate the evolutionary relationship among the three domains of life.
Acknowledgments
C.Z. is supported by NIH grant GM -49034. J.E. is supported by the Israel Ministry of Absorption and the Israel Science Foundation (Grant #291/99) and is the incumbent of the Murray Shusterman Career Development Chair in Microbiology.
References
- R1.Althoff S., Selinger D., Wise J.A. Molecular evolution of SRP cycle components: functional implications. Nucleic Acids Res. 1994;22:1933–1947. doi: 10.1093/nar/22.11.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Bacher G., Pool M., Dobberstein B. The ribosome regulates the GTPase of the beta-subunit of the signal recognition particle receptor. J. Cell Biol. 1999;23:723–730. doi: 10.1083/jcb.146.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Batey R.T., Rambo R.P., Lucast L., Rha B., Doudna J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- R4.Berks B.C., Sargent F., Palmer T. The Tat protein export pathway. Mol. Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- R5.Bernstein H.D., Poritz M.A., Strub K., Hoben P.J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- R6.Bhuiyan S.H., Gowda K., Hotokezaka H., Zwieb C. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 2000;15:1365–1373. doi: 10.1093/nar/28.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Bhuiyan S.H., Pakhomova O.N., Hinck A.P., Zwieb C. Complexes with truncated RNAs from the large domain of Archaeoglobus fulgidus signal recognition particle. FEMS Microbiol. Lett. 2001;198:105–110. doi: 10.1111/j.1574-6968.2001.tb10626.x. [DOI] [PubMed] [Google Scholar]
- R8.Bourne H.R., Sanders D.A., Mccormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- R9.Bovia F., Strub K. The signal recognition particle and related small cytoplasmic ribonucleoprotein particles. J. Cell Sci. 1996;109:2601–2608. doi: 10.1242/jcs.109.11.2601. [DOI] [PubMed] [Google Scholar]
- R10.Chang D.Y., Newitt J.A., Hsu K., Bernstein H.D., Maraia R.J. A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res. 1997;25:1117–1122. doi: 10.1093/nar/25.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Clemons W.J., Gowda K., Black S., Zwieb C., Ramakrishnan V. Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 Å resolution: Evidence for the mechanism of signal peptide binding. J. Mol. Biol. 1999;292:697–705. doi: 10.1006/jmbi.1999.3090. [DOI] [PubMed] [Google Scholar]
- R12.Dale H., Angevine C.M., Krebs M.P. Ordered membrane insertion of an archaeal opsin in vivo. Proc. Natl. Acad. Sci. USA. 2000;97:7847–7852. doi: 10.1073/pnas.140216497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.de Leeuw E., Poland D., Mol O., Sinning I., Ten Hagen-Jongman C.M., Oudega B., Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- R14.Diener J.L., Wilson C. Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry. 2000;39:12862–12874. doi: 10.1021/bi001180s. [DOI] [PubMed] [Google Scholar]
- R15.Eichler J. Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. Evol. 2002 doi: 10.1007/s00239-001-0035-8. In press. [DOI] [PubMed] [Google Scholar]
- R16.Eichler J., Moll R. The signal recognition particle of archaea. Trends Microbiol. 2001;9:130–136. doi: 10.1016/s0966-842x(01)01954-0. [DOI] [PubMed] [Google Scholar]
- R17.Fekkes P., Driessen A.J. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Fox G.E., Woese C.R. 5S RNA secondary structure. Nature. 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- R19.Gribaldo S., Cammarano P. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. J. Mol. Evol. 1998;47:508–516. doi: 10.1007/pl00006407. [DOI] [PubMed] [Google Scholar]
- R20.Gropp R., Gropp F., Betlach M.C. Association of the halobacterial 7S RNA to the polysome correlates with expression of the membrane protein bacterioopsin. Proc. Natl. Acad. Sci. USA. 1992;89:1204–1208. doi: 10.1073/pnas.89.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Gupta R.S. The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 2000;26:111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- R22.Herskovits A.A., Bochkareva E.S., Bibi E. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 2000;38:927–939. doi: 10.1046/j.1365-2958.2000.02198.x. [DOI] [PubMed] [Google Scholar]
- R23.Kaine B.P. Structure of the archaeabacterial 7S RNA molecule. Mol. Gen. Genet. 1990;221:315–321. doi: 10.1007/BF00259394. [DOI] [PubMed] [Google Scholar]
- R24.Kalies K.U., Görlich D., Rapoport T.A. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Kawashima T., Amano N., Koike H., et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium . Proc. Natl. Acad. Sci. USA. 2000;97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Keenan R.J., Freymann D.M., Stroud R.M., Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- R27.Kollman J.M., Doolittle R.F. Determining the relative rates of change for prokaryotic and eukaryotic proteins with anciently duplicated paralogs. J. Mol. Evol. 2000;51:173–181. doi: 10.1007/s002390010078. [DOI] [PubMed] [Google Scholar]
- R28.Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991;19:209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Lütcke H. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem. 1995;228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- R30.Miller J.D., Wilhelm H., Gierasch L., Gilmore R., Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- R31.Miller J.D., Bernstein H.D., Walter P. Interaction of E. coli F fh/4.5S ribonucleotide and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657 –659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- R32.Moll R., Schmidtke S., Schäfer G. Domain structure, GTP-hydrolyzing activity and 7S RNA binding of Acidianus ambivalens Ffh-homologous protein suggest an SRP-like complex in archaea. Eur. J. Biochem. 1999;259:441–448. doi: 10.1046/j.1432-1327.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- R33.Montoya G., Svensson C., Luirink J., Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- R34.Montoya G., Te Kaat K., Moll R., Schäfer G., Sinning I. The crystal structure of the conserved GTPase of SRP54 from the archaeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP-SRP receptor complex. Structure. 2000;15:515–525. doi: 10.1016/s0969-2126(00)00131-3. [DOI] [PubMed] [Google Scholar]
- R35.Moser C., Mol O., Goody R.S., Sinning I. The signal recognition particle receptor of Escherichia coli (FtsY) has a nucleotide exchange factor built into the GTPase domain. Proc. Natl. Acad. Sci. USA. 1997;94:11339–11344. doi: 10.1073/pnas.94.21.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Mueller F., Doring T., Erdemir T., Greuer B., Junke N., Osswald M., Rinke-Appel J., Stade K., Thamm S., Brimacombe R. Getting closer to an understanding of the three-dimensional structure of ribosomal RNA. Biochem. Cell Biol. 1995;73:767–773. doi: 10.1139/o95-085. [DOI] [PubMed] [Google Scholar]
- R37.Ortenberg R., Mevarech M. Evidence for post-translational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii . J. Biol. Chem. 2000;275:22839–22846. doi: 10.1074/jbc.M908916199. [DOI] [PubMed] [Google Scholar]
- R38.Pakhomova O.N., Cui Y., Zwieb C., Hinck A.P. Sequence-specific 1H, 13C and 15N signal assignments and secondary structure of Archaeoglobus fulgidus SRP19. J. Biomol. NMR. 2001;20:187–188. doi: 10.1023/a:1011243402698. [DOI] [PubMed] [Google Scholar]
- R39.Phillips G.J., Silhavy T.J. The E. coliffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- R40.Politz J.C., Yarovoi S., Kilroy S.M., Gowda K., Zwieb C., Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Powers T., Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- R42.Powers T., Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R43.Ring G., Eichler J. Large-scale preparation and characterization of inverted membrane vesicles from the halophilic archaeon Haloferax volcanii . J. Membr. Biol. 2001;183:195–204. doi: 10.1007/s00232-001-0067-4. [DOI] [PubMed] [Google Scholar]
- R44.Rose M.A., Weeks K.M. Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol. 2001;8:515–520. doi: 10.1038/88577. [DOI] [PubMed] [Google Scholar]
- R45.Ruepp A., Graml W., Santos-Martinez M.L., Koretke K.K., Volker C., Mewes H.W., Frishman D., Stocker S., Lupas A.N., Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum . Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- R46.Römisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- R47.Sanchez E., Amils R. Absolute requirement of ammonium sulfate for reconstitution of active 70S ribosomes from the extreme halophilic archaeon Haloferax mediterranei . Eur. J. Biochem. 1995;233:809–814. doi: 10.1111/j.1432-1033.1995.809_3.x. [DOI] [PubMed] [Google Scholar]
- R48.Seluanov A., Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- R49.Siegel V., Walter P. Binding sites of the 19-kDa and 68 /72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined by protein-RNA “footprinting”. Proc. Natl. Acad. Sci. USA. 1988;85:1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R50.Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol. Cell. Biol. 1990;10:777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R51.Struck J.C., Vogel D.W., Ulbrich N., Erdmann V.A. The Bacillus subtilis scRNA is related to the 4.5S RNA from Escherichia coli. Nucleic Acids Res. 1988;16:2719. doi: 10.1093/nar/16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R52.Ulbrandt N.D., Newitt J.A., Bernstein H.D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- R53.Walter P., Johnson A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- R54.Weichenrieder O., Wild K., Strub K., Cusack S. Structure and assembly of the Alu domain of mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- R55.Wild K., Weichenrieder O., Leonard G.A., Cusack S. The 2 Å structure of helix 6 of the human signal recognition particle RNA. Structure. 1999;7:1345–1352. doi: 10.1016/s0969-2126(00)80024-6. [DOI] [PubMed] [Google Scholar]
- R56.Young J.C., Ursini J., Legate K.R., Miller J.D., Walter P., Andrews D.W. An amino-terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor a subunit to the b subunit on the endoplasmic reticulum membrane. J. Biol. Chem. 1995;270:15650–15657. doi: 10.1074/jbc.270.26.15650. [DOI] [PubMed] [Google Scholar]
- R57.Zelazny A., Seluanov A., Cooper A., Bibi E. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc. Natl. Acad. Sci. USA. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R58.Zwieb C., Samuelsson T. SRPDB (signal recognition particle database) Nucleic Acids Res. 2000;28:171–172. doi: 10.1093/nar/28.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]