Abstract
The Z-curve is a three-dimensional curve that constitutes a unique representation of a DNA sequence, i.e., both the Z-curve and the given DNA sequence can be uniquely reconstructed from the other. We employed Z-curve analysis to identify one replication origin in the Methanocaldococcus jannaschii genome, two replication origins in the Halobacterium species NRC-1 genome and one replication origin in the Methanosarcina mazei genome. One of the predicted replication origins of Halobacterium species NRC-1 is the same as a replication origin later identified by in vivo experiments. The Z-curve analysis of the Sulfolobus solfataricus P2 genome suggested the existence of three replication origins, which is also consistent with later experimental results. This review aims to summarize applications of the Z-curve in identifying replication origins of archaeal genomes, and to provide clues about the locations of as yet unidentified replication origins of the Aeropyrum pernix K1, Methanococcus maripaludis S2, Picrophilus torridus DSM 9790 and Pyrobaculum aerophilum str. IM2 genomes.
Keywords: Halobacterium, Methanocaldococcus jannaschii, Methanosarcina mazei
Introduction
The Archaea are a group of prokaryotes that were recognized in 1977 as an independent monophyletic domain of life (Woese and Fox 1977). The evolutionary relationships among the Archaea and the other domains of life, the Bacteria and the Eukarya, are uncertain. However, based on similarities in the proteins involved, the process of replication in archaea appears to be more closely related to that in eukarya than in bacteria (Edgell and Doolittle 1997, Tye 2000, MacNeill 2001, Giraldo 2003, Kelman and Hurwitz 2003). Our understanding of archaeal replication mechanisms has advanced dramatically in the past few years (Bernander 2000, 2003, Kelman 2000, Tye 2000, Bohlke et al. 2002, Grabowski and Kelman 2003, Kelman and Kelman 2003), and it appears that archaea have a simplified version of the eukaryotic replication apparatus. Clarification of the archaeal replication mechanism is therefore important not only to the understanding of archaeal replication, but also for the insight it may provide into the replication mechanisms of eukarya.
Replication initiates bidirectionally at a specific locus called the origin of replication. Knowing the positions and sequences of replication origins is critical to understanding the initiation phase of replication. Replication origins have currently been identified in vivo for only four of the 19 available archaeal genomes (Myllykallio et al. 2000, Maisnier-Patin et al. 2002, Berquist and DasSarma 2003, Matsunaga et al. 2003, Lundgren et al. 2004, Robinson et al. 2004). The experimental methods for identifying replication origins in vivo are reliable, but time-consuming and labor-intensive. In silico analysis, however, is fast and suitable for handling a large number of genomes. In addition, in some experimental methods, e.g., as used to identify the replication origin of Halobacterium species NRC-1 (Berquist and DasSarma 2003), the replication origin must first be located approximately in a known sequence.
With the advent of the post-genomic era, genomic data are accumulating exponentially. High-throughput methods for genome annotations, e.g., replication origin identification, are thus needed to meet the challenge of interpreting this information. The identification of replication origins based on in silico analysis has been the subject of intensive study during the past few years. The GC skew method was first proposed to detect nucleotide composition asymmetry around the replication origin (Lobry 1996a). Other algorithms were later proposed to tackle the same task (Grigoriev 1998, McLean et al. 1998, Mrazek and Karlin 1998, Salzberg et al. 1998, Rocha et al. 1999).
The Z-curve is a three-dimensional curve that constitutes a unique representation of a DNA sequence, i.e., for the Z-curve and the given DNA sequence, each can be uniquely reconstructed from the other (Zhang and Zhang 1991, 1994). We have used Z-curve analysis to identify one replication origin in the Methanocaldococcus jannaschii genome (Zhang and Zhang 2004b), two replication origins in the Halobacterium species NRC-1 genome (Zhang and Zhang 2003c) and one replication origin in the Methanosarcina mazei genome (Zhang and Zhang 2002). One predicted replication origin of Halobacterium species NRC-1 is the same as the replication origin later identified by in vivo experiments (Berquist and DasSarma 2003). The Z-curve analysis suggested the existence of three replication origins in the Sulfolobus solfataricus P2 genome, and indicated their approximate locations (Zhang and Zhang 2003c), the results being consistent with the results of subsequent in vivo studies (Lundgren et al. 2004, Robinson et al. 2004).
This review summarizes past applications of the Z-curve in identifying replication origins in archaeal genomes, and applies the same technique in the search for clues about the locations of as yet unidentified archaeal replication origins.
The Z-curve representation of genome sequences
The Z-curve is a three-dimensional curve that provides a unique representation of a DNA sequence in that the DNA sequence and the Z-curve can each be uniquely reconstructed from the other. Therefore, the Z-curve contains all the information that the corresponding DNA sequence carries. The resulting curve has a zigzag shape, hence the name Z-curve. A DNA sequence can be analyzed by studying the corresponding Z-curve. One of the advantages of the Z-curve is its intuitiveness; the entire Z-curve of a genome can be viewed on a computer screen or on paper, regardless of genome length, thus allowing both global and local compositional features of genomes to be easily grasped. By combining use of the Z-curve with statistical analysis, better results may be obtained.
The Z-curve is composed of a series of nodes, P0, P1, P2, ..., PN, with coordinates xn, yn and zn (n = 0, 1, 2, …, N, where N is the length of the DNA sequence), which are uniquely determined by the Z-transform of a DNA sequence (Zhang and Zhang 1991, 1994, Zhang et al. 2003):
where An, Cn, Gnand Tn are the cumulative occurrence numbers of A, C, G and T, respectively, in the subsequence from the first base to the nth base in the sequence. We define A0 = C0 = G0 = T0 = 0, and therefore, x0 = y0 = z0 = 0. Here R, Y, M, K, W and S represent the purine, pyrimidine, amino, keto, weak hydrogen (H) bond and strong H bond bases, respectively, according to the Recommendation 1984 by the NC-IUB (Cornish-Bowden 1985). The Z-curve is defined as the sequential connection of the nodes P0, P1, P2, ..., PN with straight lines. Note that the Z-curve always starts from the origin of the three-dimensional coordinate system. Once the coordinates xn, yn and zn (n = 1, 2, …, N) of a Z-curve are given, the corresponding DNA sequence can be reconstructed from the so-called inverse Z-transform:
where An + Cn + Gn+ Tn = n.
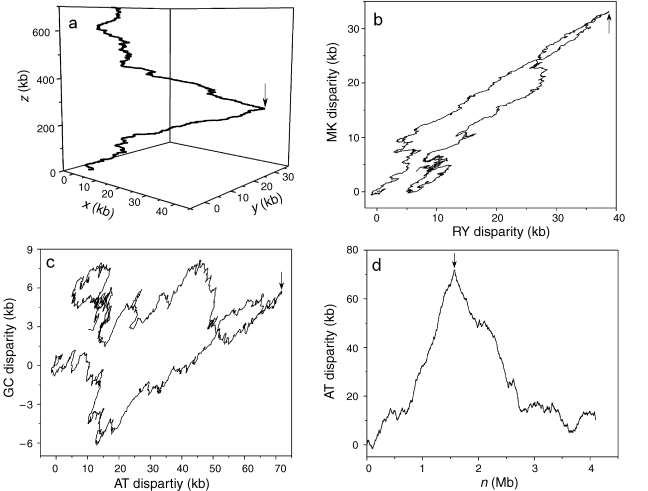
The three components of the Z-curve, xn, yn and zn, represent three independent distributions that completely describe the DNA sequence being studied. The components xn, yn and zn display the distributions of purine versus pyrimidine (R vs. Y), amino versus keto (M vs. K) and strong H-bond versus weak H-bond (S vs. W) bases along the sequence, respectively. In the subsequence constituted from the first base to the nth base of the sequence, when purine bases (A and G) are in excess of pyrimidine bases (C and T), xn > 0, otherwise, xn < 0, and when the numbers of purine and pyrimidine bases are identical, xn = 0. Similarly, when amino bases (A and C) are in excess of keto bases (G and T), yn > 0, otherwise, yn < 0, and when the numbers of amino and keto bases are identical, yn = 0. Finally, when weak H-bond bases (A and T) are in excess of strong H-bond bases (G and C), zn > 0, otherwise, zn < 0, and when the numbers of weak and strong H-bond bases are identical, zn = 0. The xn and yn components are termed RY and MK disparity curves, respectively. The AT and GC disparity curves are defined by (xn + yn)/2 and (xn – yn)/2, which shows the excess of A over T and G over C, respectively, along the genome. The RY and MK disparity curves, as well as AT and GC disparity curves, can be used to predict replication origins. Figure 1 shows an example of the Z-curves for the M. mazei genome. The Z-curve for a genome is a three-dimensional (3-D) curve (Figure 1a). To facilitate the use of the Z-curve, it can be plotted as two-dimensional (2-D) curves. Figure 1b is a plot based on RY and MK disparities, whereas Figure 1c is a plot based on AT and GC disparities. The most convenient method, however, is to plot one of the Z-curve components, i.e., RY, MK, AT or GC disparities, along the chromosome. Figure 1d shows an AT disparity curve and Figure 2d shows RY and MK disparity curves for the M. mazei genome. Arrows indicate the position of cdc6 genes, and also the putative replication origin. Therefore, in the case of M. mazei, all 3-D, 2-D and various disparity curves (RY, MK, AT and GC) show a peak at the position of the putative replication origin.
Figure 1.
The Z-curves for the Methanosarcina mazei genome. (a) The 3-D Z-curve, (b) the 2-D Z-curve based on RY and MK disparity, (c) the 2-D Z-curve based on AT and GC disparity and (d) the AT disparity curves. Arrows indicate the positions of the cdc6 gene, which is also the position of the predicted replication origin.
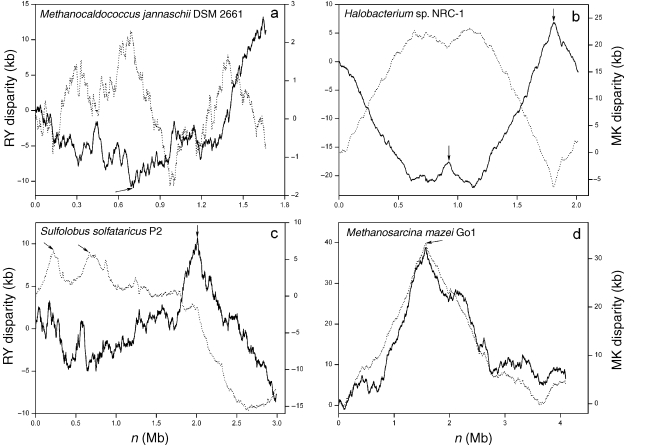
Figure 2.
The Z-curves for the genomes of (a) Methanocaldococcus jannaschii DSM 2661, (b) Halobacterium sp. NRC-1, (c) Sulfolobus solfataricus P2 and (d) Methanosarcina mazei Go1. Unbroken lines denote RY disparity curves, and broken lines denote MK disparity curves. Arrows indicate the positions of cdc6 genes, which are also the positions of predicted replication origins. In the Halobacterium sp. NRC-1 genome, Berquist and DasSarma (2003) have identified a chromosomal autonomously replicating sequence element, which is at the location of the cdc6-3 (arrow at about 1.8 Mb). Robinson et al. (2004) have identified two replication origins in the S. solfataricus genome in vivo. The two replication origins, oriC1 and oriC2, are close to cdc6-1 and cdc6-3, respectively (the positions of the first and third arrows).
Replication origin identification in the Methanocaldococcus jannaschii genome
Methanocaldococcus jannaschii is an autotroph that grows at pressures greater than 20 MPa and at temperatures up to 94 °C (Jones et al. 1983). As the first completely sequenced archaeon (Bult et al. 1996), M. jannaschii is notorious for the difficulty it presents to those seeking to identify its replication origins. Despite extensive efforts, the locations of the replication origins of this species remain elusive 8 years after the publication of its complete genome sequence. Ambiguous results were obtained in identifying the replication origins of M. jannaschii based on all in silico genome analyses, which usually assess biases in nucleotide, codon and oligomer usages (Salzberg et al. 1998, Lopez et al. 1999, Rocha et al. 1999). Recently, a technique called marker frequency analysis was successfully applied in vivo to identify the location of the replication origin of the archaeon Archaeoglobus fulgidus. It failed, however, in the case of M. jannaschii (Maisnier-Patin et al. 2002). Distinguishing it from other archaea, the genome of M. jannaschii was generally thought to lack a clear cdc6 homologue (Bernander 2000).
The RY disparity curve for the M. jannaschii genome shows a global minimum at the position of about 695 kb, indicating that the genome changes from CT-rich to AG-rich at this site (Figure 2a). Therefore, the site around 695 kb may contain a replication origin. We scanned the region around the minimum for a potential cdc6 gene. Surprisingly, we found that an open reading frame (ORF), MJ0774, is highly similar to the cdc6 gene (Zhang and Zhang 2004b). The ORF MJ0774 encodes a 409 amino-acid-long polypeptide, and is annotated as a hypothetical protein. We searched the amino acid sequence against the NCBI Conserved Domain Database (Marchler-Bauer et al. 2003), and a Cdc6 protein was assigned to MJ0774, from amino acids 13 to 404. The alignment of the MJ0774 (13–404) with the consensus sequence of Cdc6 proteins (12–355) showed that MJ0774 is a homologue of the Cdc6 protein. In addition, a helix-turn-helix domain was found at the region from residues 327–403, and this domain is believed to be involved in the DNA binding (Liu et al. 2000).
A closer look at the region revealed that an intergenic region of about 700 bp between the cdc6 homologue and an adjacent gene has many characteristics of a replication origin. This intergenic region is between the ORF MJ0773 and MJ0774, from 694,540–695,226 bp of the genome. The region is 687 bp in length and is highly AT-rich (80%). In addition, there are multiple copies of direct repeat elements and AT stretches. This region contains almost all the features of known replication origins and is, therefore, very likely a true replication origin, which has been designated oriC1 (Zhang and Zhang 2004b).
Recently, marker frequency analysis was successfully applied in vivo to identify the location of a replication origin of A. fulgidus. However, M. jannaschii displayed a complex pattern of marker frequency distributions with multiple peaks and valleys. An intriguing explanation proposed for this pattern is that it reflects the presence of multiple replication origins (Maisnier-Patin et al. 2002). The features of the MK disparity curve for M. jannaschii are consistent with this hypothesis.
The MK disparity curve for M. jannaschii shows four extremes, including one probable replication origin associated with the oriC1 (Figure 2a). The locations of these maxima and minima are 695 (oriC1) and 1388 kb, and 127 and 986 kb, respectively. Studying the positions of the four extremes suggests the possibility that the maximum at 1388 kb is associated with another replication origin, whereas the minima at 127 and 986 kb correspond to replication termini. Supporting this hypothesis, the distances between the maximum at 1388 kb and the two predicted replication termini are exactly the same (402 kb), which is consistent with the characteristics of most identified replication origins, i.e., in genomes with a single replication origin, oriC and terC divide the genome into parts of similar length. However, we also noticed that the distances between the oriC1 and the two predicted replication termini are different. It is known that some horizontally transferred elements are present in the genome of M. jannaschii (Bult et al. 1996). Although the exact amount of horizontally transferred DNA is unclear, these horizontal transfer events could explain why the two replichores have different sizes, i.e., the horizontally transferred DNA increased the length of one of the replichores. In addition, a gene coding for replication factor C (MJ1422) is situated at the position of the maximum associated with the putative oriC2. However, there is no evidence to suggest that the gene coding for replication factor C is close to replication origins. Nevertheless, some archaeal replication origins are indeed situated in the regions close to some replication factors, such as DNA polymerases and helicases (Salzberg et al. 1998).
Replication origin identification in the Halobacterium species NRC-1 and Sulfolobus solfataricus genomes
Halobacterium NRC-1 belongs to the obligatorily halophilic Halobacterium species, and is an experimental model among archaea. The exact locations of all replication origins have not been identified, although the possibility of multiple replication origins was suggested based on the GC-skew analysis (Ng et al. 2000, Kennedy et al. 2001).
The RY and MK disparity curves show two relatively sharp and two relatively broad peaks. Interestingly, two of the three cdc6 genes are located at the positions of the two sharp peaks (Figure 2b). Furthermore, two intergenic regions immediately beside the corresponding cdc6 genes show many features of replication origins. Therefore, the two intergenic regions were assigned as putative replication origins oriC1 and oriC2 (Zhang and Zhang 2003c).
The putative replication origin oriC1 is at the intergenic region close to the cdc6-1 gene, which is from 921,863–922,014 bp. The oriC1 contains two long direct repeats. The putative replication origin oriC2 is at the intergenic region close to the cdc6-3 gene, which is from 1,806,444–1,807,229 bp. In addition, two helicase genes were located about 20 kb away from these two regions, respectively (Zhang and Zhang 2003c). Soon afterwards, a replication origin of Halobacterium NRC-1 was identified in vivo by Berquist and DasSarma (2003). These authors found that sequences located up to 750 bp upstream of the orc7 gene (cdc6-3) translational start, plus the orc7 gene and 50 bp downstream, are sufficient to endow the plasmid with replication ability. Further, they found that the sequence within the 750-bp region upstream of orc7 contains a nearly perfect inverted repeat of 31 bp, which flanks an extremely AT-rich stretch of 189 bp. The region containing these inverted repeats and AT-rich stretch is within the predicted oriC2, 1,806,444–1,807,229 bp (Zhang and Zhang 2003c).
A breakthrough in the study of archaeal replication origins was the demonstration that S. solfataricus has multiple replication origins. This is the first archaeon found to have multiple replication origins, referred to as oriC1 and oriC2, according to the nomenclature of Lundgren et al. (2004) and Robinson et al. (2004). The replication origins oriC1 and oriC2 are located at sites close to cdc6-1 and cdc6-3, respectively (Robinson et al. 2004). Interestingly, the RY disparity curve for the archaeon S. solfataricus shows a global maximum around the position of the cdc6-3 genes, whereas the MK disparity curve shows a maximum at the position of cdc6-1 (Figure 2c) (Zhang and Zhang 2003c).
Replication origin identification in the Methanosarcina mazei genome
The archaeon Methanosarcina mazei and related species have great ecological importance, because they are the only organisms that ferment acetate, methylamines and methanol to methane, carbon dioxide and ammonia. Since acetate is the precursor of 60% of the methane produced on Earth, these organisms contribute significantly to the production of this greenhouse gas (Deppenmeier et al. 2002).
Both RY and MK disparity curves for M. mazei show a global maximum at about 1600 kb and a minimum at about 3600 kb (Figure 2d). The maximum and minimum correspond to a sharp peak and relatively broad peak, respectively. The cdc6 gene is located exactly at the global maximum. Based on the known behaviors of the Z-curves for archaea whose replication origins have been identified, we hypothesize that the replication origin and termination sites in M. mazei correspond to the positions of the sharp and broad peaks, respectively. We have located an intergenic region that is between the cdc6 gene (MM1314) and the adjacent gene (MM1315), which shows many characteristics of known replication origins. This region is highly AT-rich (74%), and contains multiple copies of consecutive repeats. Our results strongly suggest that the single replication origin of M. mazei is situated at the intergenic region between the cdc6 gene and the adjacent gene, from 1,564,657 to 1,566,241 bp of the genome (Zhang and Zhang 2002).
Common features of archaeal replication origins
So far, replication origins of four archaea have been identified in vivo. Two replication origins have been identified in the S. solfataricus P2 genome by 2-D gel analysis (Robinson et al. 2004) and the approximate location of the third was suggested by marker frequency analysis (Lundgren et al. 2004). One replication origin has been identified in Pyrococcus abyssi GE5 based on oligomer skew analysis, which was later confirmed in vivo (Lopez et al. 1999, Myllykallio et al. 2000, Matsunaga et al. 2003). An autonomously replicating sequence element has been identified in Halobacterium sp. NRC-1 (Berquist and DasSarma 2003). The marker frequency analysis showed a candidate region of a replication origin in A. fulgidus; however, the exact location of the replication origin has not been determined (Maisnier-Patin et al. 2002).
Common features of archaeal replication origins can be summarized based on what is known about replication origins identified in vivo. Except that of A. fulgidus, all identified replication origins are associated with an extreme in one of the components of the Z-curve. In addition, the extremes associated with replication origins are relatively sharp compared with those associated with replication termini, probably because termination sometimes occurs at multiple loci. These replication origins are located immediately beside a cdc6 gene. This is similar to the case in bacteria, where a gene coding for DnaA is frequently close to the oriC (Mackiewicz et al. 2004). Replication origins are highly rich in AT content. The identified replication origins have AT stretches, as well as multiple copies of direct or inverted repeat elements. Furthermore, some replication origins, e.g., those of S. solfataricus, contain conserved Cdc6 binding elements.
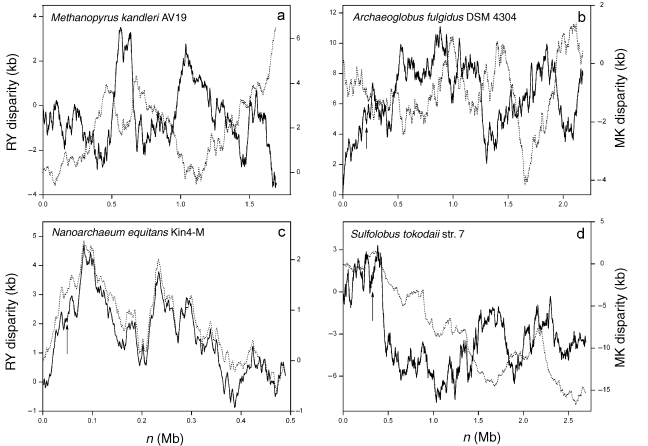
Based on the above conserved features, some putative replication origins have been identified by in silico analysis, but have yet to be confirmed in vivo. These include a replication origin of Methanothermobacter thermautotrophicus str. Delta H (Lopez et al. 1999), a replication origin of Methanosarcina acetivorans C2A (Galagan et al. 2002), one of the two putative replication origins in Halobacterium sp. NRC-1 (Zhang and Zhang 2003c), a replication origin in the M. mazei genome (Zhang and Zhang 2002) and a replication origin in the M. jannaschii genome (Zhang and Zhang 2004b). A replication origin of Pyrococcus furiosus DSM 3638 and a replication origin of Pyrococcus horikoshii OT3 were identified based on homologue analysis with Pyrococcus abyssi (Lopez et al. 1999). In addition, a replication origin of Thermoplasma acidophilum DSM 1728 was predicted based on different nucleotide skews; however, other conserved features of archaeal replication origins, e.g., the close proximity to a cdc6 gene and the presence of repeat elements, were not mentioned (Ruepp et al. 2000). Furthermore, one replication origin of Methanopyrus kandleri AV19 was predicted based on the GC-skew analysis; however, the figure of GC-skew provided by the authors does not seem to have a clear minimum or maximum at the site of predicted replication origin (Slesarev et al. 2002). Furthermore, various components of the Z-curve show a complex pattern in the case of M. kandleri (Figure 3a). The current status of replication origin identification in the 19 available archaeal genomes is listed in Table 1.
Figure 3.
The Z-curves for the genomes of (a) Methanopyrus kandleri AV19, (b) Archaeoglobus fulgidus DSM 4304, (c) Nanoarchaeum equitans Kin4-M and (d) Sulfolobus tokodaii str. 7. Among the 19 available archaeal genomes, the Z-curves for these four genomes show a complex pattern, with no clear global minima or maxima. Unbroken lines denote RY disparity curves, and broken lines denote MK disparity curves. Arrows indicate the positions of cdc6 genes in the A. fulgidus, N. equitans and S. tokodaii genomes. The approximate location of the replication origin of A. fulgidus was suggested to be at about the middle of the chromosome based on marker frequency analysis.
Table 1.
Status of replication origin identification in the currently available archaeal genomes.
| Name (reference) | Order | ID | Length (bp) | Status of replication origin identification (reference) | Z-curve extremes | Position of Cdc6 binding element (kb) (Robinson et al. 2004) | |
| 1 | Aeropyrum pernix K1 (Kawarabayasi et al. 1999) | Crenarchaeota | NC_000854 | 1,669,695 | Unknown | Yes | 445 |
| 2 | Archaeoglobus fulgidus | Euryarchaeota | NC_000917 | 2,178,400 | Approximate location is known based | ||
| DSM 4304 (Klenk et al. 1997) | on marker frequency analysis | No | 1430 | ||||
| (Maisnier-Patin et al. 2002). | |||||||
| 3 | Halobacterium sp. NRC-1 | Euryarchaeota | NC_002607 | 2,014,239 | Two replication origins have been | Yes | 1806 |
| (Ng et al. 2000) | predicted based on the Z-curve and | ||||||
| GC skew analysis (Kennedy et al. 2001, | |||||||
| Zhang and Zhang 2003c). One replication | |||||||
| origin has been identified in vivo | |||||||
| (Berquist and DasSarma 2003). | |||||||
| 4 | Methanocaldococcus jannaschii | Euryarchaeota | NC_000909 | 1,664,970 | One replication origin has been identified | Yes | |
| DSM 2661 (Bult et al. 1996) | based on the Z-curve analysis (Zhang and | ||||||
| Zhang 2004b). | |||||||
| 5 | Methanococcus maripaludis S2 | Euryarchaeota | NC_005791 | 1,661,137 | Unknown | Yes | |
| (Unpublished) | |||||||
| 6 | Methanopyrus kandleri AV19 | Euryarchaeota | NC_003551 | 1,694,969 | See footnote 1 | No | |
| (Slesarev et al. 2002) | |||||||
| 7 | Methanosarcina acetivorans | Euryarchaeota | NC_003552 | 5,751,492 | One replication origin has been identified | Yes | |
| C2A (Galagan et al. 2002) | based on the GC-skew analysis. | ||||||
| 8 | Methanosarcina mazei Go1 | Euryarchaeota | NC_003901 | 4,096,345 | One replication origin has been identified | Yes | |
| (Deppenmeier et al. 2002) | based on the Z-curve analysis (Zhang and | ||||||
| Zhang 2002). | |||||||
| 9 | Methanothermobacter | Euryarchaeota | NC_000916 | 1,751,377 | One replication origin has been identified | Yes | |
| thermautotrophicus str. Delta H | based on the oligomer-skew analysis | ||||||
| (Smith et al. 1997) | (Lopez et al. 1999). | ||||||
| 10 | Nanoarchaeum equitans Kin4-M | Nanoarchaeota | NC_005213 | 490,885 | Unknown | No | |
| (Waters et al. 2003) | |||||||
| 11 | Picrophilus torridus DSM 9790 | Euryarchaeota | NC_005877 | 1,545,895 | Unknown | Yes | |
| (Futterer et al. 2004) | |||||||
| 12 | Pyrobaculum aerophilum str. IM2 | Crenarchaeota | NC_003364 | 2,222,430 | Unknown | Yes | |
| (Fitz-Gibbon et al. 2002) | |||||||
| 13 | Pyrococcus abyssi GE5 | Euryarchaeota | NC_000868 | 1,765,118 | One replication origin has been identified | Yes | 123 |
| (Lecompte et al. 2001, | by 2-D gel analysis (Myllykallio et al. | ||||||
| Cohen et al. 2003) | 2000, Matsunaga et al. 2003). | ||||||
| 14 | Pyrococcus furiosus DSM 3638 | Euryarchaeota | NC_003413 | 1,908,256 | One replication origin has been identified | Yes | 15 |
| (Robb et al. 2001) | based on homologue analysis with Pyro- | ||||||
| coccus abyssi GE5 (Lopez et al. 1999). | |||||||
| 15 | Pyrococcus horikoshii OT3 | Euryarchaeota | NC_000961 | 1,738,505 | One replication origin has been identified | Yes | 111 |
| (Kawarabayasi et al. 1998) | based on homologue analysis with Pyro- | ||||||
| coccus abyssi GE5 (Lopez et al. 1999). | |||||||
| 16 | Sulfolobus solfataricus P2 | Crenarchaeota | NC_002754 | 2,992,245 | Two replication origins have been identified | Yes | 222 |
| (She et al. 2001) | in vivo (Robinson et al. 2004). The location | ||||||
| of the third replication origin was suggested | |||||||
| by microarray-based marker frequency | |||||||
| analysis (Lundgren et al. 2004). | |||||||
| 17 | Sulfolobus tokodaii str. 7 | Crenarchaeota | NC_003106 | 2,694,756 | Unknown | No | 323 |
| (Kawarabayasi et al. 2001) | |||||||
| 18 | Thermoplasma acidophilum | Euryarchaeota | NC_002578 | 1,564,906 | One replication origin has been identified | Yes | |
| DSM 1728 (Ruepp et al. 2000) | based on GC skew analysis (Ruepp et al. 2000). | ||||||
| 19 | Thermoplasma volcanium GSS1 | Euryarchaeota | NC_002689 | 1,584,804 | Unknown | Yes | |
| (Kawashima et al. 2000) | |||||||
1 It was reported that one replication origin of Methanopyrus kandleri AV19 was predicted based on the GC-skew analysis, however, the figure of GC-skew provided by the authors does not seem to have a clear minimum or maximum at the site of predicted replication origin (Slesarev et al. 2002). Various components of the Z-curve for M. kandleri also show a complex pattern, suggesting that the replication origin predicted by Slesarev et al. (2002) is questionable.
Besides the above common features observed among replication origins, there are some differences. For instance, sometimes all disparity curves (MK, RY, AT and GC) show a global maximum or minimum for a given origin, whereas in other cases, only one or a subset of curves shows significant peaks. In addition, in the A. fulgidus genome, although an approximate region of replication origin was suggested by marker frequency analysis, both Z-curve (Figure 3b) and oligomer skew (Lopez et al. 1999) show no extremes at the site of the replication origin. Furthermore, some replication origins are not associated with cdc6 genes, e.g., it was suggested that the third replication origin of S. solfataricus is about 80 kb away from the nearby cdc6 gene (Lundgren et al. 2004), but the MK disparity curve shows a maximum at the position of the cdc6 gene (Figure 2c). It is interesting that although the three replication origins are within the same chromosome, only two of them are close to cdc6 genes. This may suggest different mechanisms of replication from the three origins. One reviewer of this manuscript noticed that for S. solfataricus and M. jannaschii, different DNA asymmetry is associated with replication origins. For instance, one replication origin of S. solfataricus corresponds to the global maximum of the RY disparity curve, whereas another replication origin corresponds to a maximum of the MK disparity curve. The different behaviors of the Z-curve for different replication origins are consistent with the hypothesis that the three replication origins have different replication mechanisms. The close proximity of the cdc6 gene and replication origin may serve to ensure that the proteins can associate with the origin as soon as they are synthesized (Kelman and Kelman 2003). It is unclear why the third replication origin of S. solfataricus is not adjacent to a cdc6 gene. Lundgren et al. (2004) proposed that one of the three initiation sites might act as the master regulator, with the other two origins being subordinate and therefore different in sequence or organization, or both (Lundgren et al. 2004). Taken together, different Z-curve behaviors of the three replication origins of S. solfataricus are consistent with the hypothesis that the three replication origins have different replication mechanisms. The absence of a Z-curve extreme or a cdc6 gene cannot exclude the possibility of a replication origin at a certain position of a chromosome.
A reasonable procedure for identifying replication origins by the Z-curve method appears to be: (1) generate RY, MK, AT and GC disparity curves for the available genomes; and (2) if there is a minimum or maximum in any of the curves, investigate the regions around each extreme for some replication origin specific features such as the presence of cdc6 genes or AT- rich intergenic regions that contain repeats.
Z-curve analysis of archaeal genomes with unknown replication origins
In seven out of the 19 available archaeal genomes, replication origins have yet to be identified, and clues to some of their locations have not been found. These seven genomes are Aeropyrum pernix K1, Methanococcus maripaludis S2, Nanoarchaeum equitans Kin4-M, Picrophilus torridus DSM 9790, Pyrobaculum aerophilum str. IM2, Sulfolobus tokodaii str. 7 and Thermoplasma volcanium GSS1. Among these seven genomes, the Z-curves for N. equitans Kin 4-M and S. tokodaii str. 7 have a complex pattern, i.e., no global minima or maxima (Figures 3c and 3d).
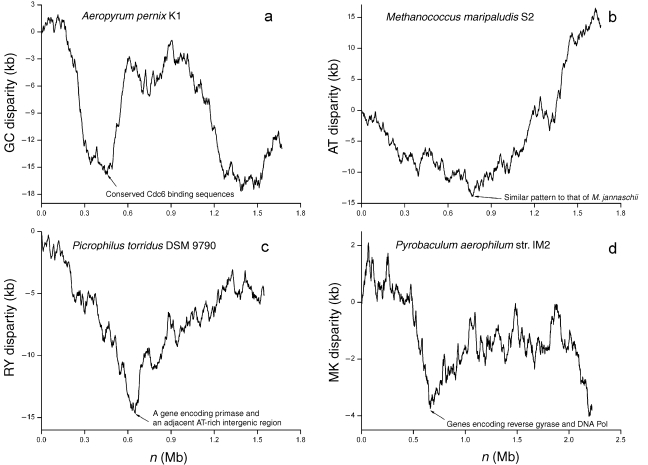
The RY and MK disparity curves for T. volcanium GSS1 show a similar pattern to that of T. acidophilum DSM 1728 and have a global minimum and maximum (data not shown), suggesting the presence of a single replication origin. However, no replication origin specific features, such as the presence of a cdc6 gene, could be found around the Z-curve extremes. The Z-curves for the remaining four genomes, A. pernix K1, M. maripaludis S2, P. torridus DSM 9790 and P. aerophilum str. IM2 show some replication origin-specific features at the extremes, and thus provide additional clues to regions that may contain replication origins.
Robinson et al. (2004) found some conserved Cdc6 binding elements across archaeal genomes. In the A. pernix K1 genome, such an element is located at 445 kb of the genome (Robinson et al. 2004). At 445 kb, the GC disparity curve shows a minimum, implying that the nucleotide composition changes around this site (Figure 4a). These lines of evidence suggest the presence of a replication origin around this site.
Figure 4.
The Z-curve analysis for the genomes of Aeropyrum pernix K1, Methanococcus maripaludis S2, Picrophilus torridus DSM 9790 and Pyrobaculum aerophilum str. IM2, in which replication origins are unknown. (a) The GC disparity curve for the A. pernix K1 genome. Some conserved Cdc6 binding sequences are located at a minimum. (b) The AT disparity curve for the M. maripaludis S2 genome. The AT disparity curve shows a global minimum, suggesting the existence of a replication origin around this site. In addition, the overall pattern of the AT disparity curve is similar to the RY disparity curve of the M. jannaschii genome. Compare Figure 4b with Figure 2a. (c) The RY disparity curve for the P. torridus DSM 9790 genome. A DNA primase gene (PTO0617) is located at the site of the global minimum. In addition, immediately beside this primase gene, a 174 bp intergenic sequence between the ORF PTO0617 and PTO0616 is highly rich in AT content (81.1%). (d) The MK disparity curve for the P. aerophilum str. IM2 genome. Genes coding for reverse gyrase and DNA polymerase are located at a minimum. The presence of Cdc6 binding elements, AT-rich intergenic sequence, or replication-associated genes at one of the Z-curve extremes provides additional clues for potential candidate regions that may contain replication origins.
A putative replication origin has been assigned in the M. jannaschii DSM 2661 genome (Zhang and Zhang 2004b). A relative of M. jannaschii DSM 2661, M. maripaludis S2, has been sequenced recently. The AT disparity curve for M. maripaludis S2 shows a global minimum, suggesting the presence of a replication origin around this site. In addition, the pattern of the AT disparity curve for M. maripaludis is similar to the RY disparity curve of M. jannaschii (compare Figures 4b and 2a). However, we could not detect a cdc6 homologue around the global minimum of the AT disparity curve of the M. maripaludis genome. Nevertheless, the conserved pattern of the AT disparity curve suggests the region around the global minimum needs further investigation.
The RY disparity curve for the P. torridus DSM 9790 genome shows a global minimum at the position 650 kb (Figure 4c), and a DNA primase gene (PTO0617) is located at the site of the extreme. In addition, immediately beside the primase gene, a 174 bp intergenic sequence between the ORF PTO0617 and PTO0616 has high AT content (81.1%). The MK disparity curve for P. aerophilum str. IM2 genome shows a minimum at 662 kb (Figure 4d). Two replication associated genes, a reverse gyrase gene (PAE1108) and a DNA polymerase gene (PAE1113) are all situated around the position of the minimum. In addition to cdc6, several replication-related genes are close to archaeal replication origins, e.g., genes encoding DNA polymerases in M. thermautotrophicus and Pyrococcus species(Lopez et al. 1999, Myllykallio et al. 2000), genes encoding replication factor C and helicases in Pyrococcus species (Myllykallio et al. 2000), and a gene encoding radA in S. solfataricus (Robinson et al. 2004). Thus, sequences around the 650 kb of the P. torridus DSM 9790 genome and the 662 kb of the P. aerophilum str. IM2 genome are good candidate regions that may contain replication origins.
Among the 19 available archaeal genomes, the Z-curves for the genomes of four species show a complex pattern, with no clear global minima or maxima: M. kandleri AV19, A. fulgidus DSM 4304, N. equitans Kin4-M and S. tokodaii str. 7 (Figure 3). Methanococcus kandleri has a high evolutionary rate and a surprisingly large number of specific insertions and deletions (Brochier et al. 2004). Nanoarchaeum equitans is an obligate symbiont with a small genome (490,885 bp), and is currently the only member of the archaeal kingdom Nanoarchaeota whose genome has been sequenced (Waters et al. 2003). Because of its small size and parasitic reduction, the genome of N. equitans may also be fast evolving. In the S. tokodaii genome, it was proposed that plasmid integration, rearrangement of genomic structure and duplication of genomic regions have increased the genome size (Kawarabayasi et al. 2001). Furthermore, extensive gene duplications have been found in the A. fulgidus genome (Klenk et al. 1997). Therefore, horizontal gene transfer, genome reduction, genome rearrangement and extensive gene duplication may explain the complex pattern of the Z-curves for these four genomes. Another possible explanation for the complex pattern is the presence of multiple replication origins in the genomes, or some of the above factors may act together, resulting in the complex pattern of the Z-curves.
Comparison of the Z-curve method with others
Various methods for the graphical representation of DNA sequences have been proposed, such as the H curve (Hamori and Ruskin 1983), the game representation (Jeffrey 1990), color DNA tetragram (Pickover 1992) and the two-dimensional DNA walk (Gates 1986, Lobry 1996b). It was shown that most are special cases of the Z-curve, and an extensive comparison between the Z-curve and other methods proposed before 1994 was detailed in Zhang and Zhang (1994). It is noteworthy that the so-called purine excess and keto excess (Freeman et al. 1998) are identical to the x and y components of the Z-curve, which was proposed 4 years earlier (Zhang and Zhang 1994).
Traditionally, the GC skew analysis is often used to assess the nucleotide compositional asymmetry around the replication origin. The GC skew is defined as (C – G)/(C + G), where C and G are the number of C and G residues in a sliding window (Lobry 1996a). Later, a method of cumulative GC skew without sliding windows was proposed, which is thought to give better resolution (Grigoriev 1998). Because the Z-curve provides a unique representation of a DNA sequence, it contains all the information that the DNA sequence carries. Therefore, the Z-curve is not any DNA walk, but almost all DNA walks are special cases of the Z-curve or functions of xn, yn and zn. For instance, the cumulative GC skew is equal to (yn – xn)/(n – zn) (see Equation 1). Indeed, almost all the replication origins that were identified based on the GC skew, including those of bacteria, viruses and mitochondria, are indicated by a change in polarity in the Z-curve (Zhang et al. 2003). However, for some genomes, e.g., that of S. solfataricus, GC skew failed to show the compositional asymmetry around the replication origins that is detected with the Z-curve (Zhang and Zhang 2003c).
Availability of the Z-curve drawing software
Software has been developed to facilitate the use of the Z-curve. The software, Zplotter online, draws and manipulates the Z-curve online, based on a user’s input sequence. With this software, RY, MK, AT and GC disparity curves can be shown for a user’s DNA sequence in the forward (5′ to 3′) and inverted (3′ to 5′) directions and for their complementary strands. The resolution of any local parts of each curve can be arbitrarily adjusted with the built-in zoom function. The Z-curve coordinates can also be shown by putting the cursor at the site of interest. In addition, a user can download the local version of the Zplotter program and run it on their own computer. This software is freely available from the Z-curve database (Zhang et al. 2003) at http://tubic.tju.edu.cn/zcurve/.
Perspective
In bacteria, replication initiates at a unique site, whereas in eukarya, replication occurs at multiple sites along the genome. A recent breakthrough was the demonstration that the archaeon S. solfataricus has at least two replication origins—the first example of the presence of multiple replication origins in archaea (Robinson et al. 2004). Eukaryotic genomes, such as the human genome, have thousands of replication origins, thus complicating the study of replication. In this respect, the simplified version of eukaryotic replication, i.e., archaeal replication that utilizes two or three replication origins, is an excellent model, especially for the study of how the cell coordinates replications occuring at multiple origins. The Z-curve analysis for the Halobacterium species NRC-1 and M. jannaschii shows the possibility that these genomes also have multiple replication origins, and some candidate sites are suggested, e.g., the second replication origin of Halobacterium species NRC-1 is suggested to be 921,863–922,014 bp of the genome (Zhang and Zhang 2003c, 2004b). It is hoped that further in vivo studies will confirm the multiple replication origins in the Halobacterium species NRC-1 and M. jannaschii genomes.
The Z-curve is a powerful tool for in silico identification of archaeal and bacterial replication origins. Because the Z-curve contains all the information that the corresponding DNA sequence carries, the DNA sequence can be studied by geometrical methods with the Z-curve, which is nicely complementary to widely used mathematical methods. Consequently, the Z-curve has been used for many purposes in addition to the identification of replication origins. For instance, algorithms based on the Z-curve have been used to recognize protein-coding genes in both prokaryotic (Guo et al. 2003) and eukaryotic genomes (Zhang and Wang 2000). Furthermore, it has been shown that the algorithm based on the Z-curve is among the best available for gene recognition (Gao and Zhang 2004). The Z-curve has also been used in isochore identification (Zhang and Zhang 2003a, 2004a), detection of horizontally transferred genomic islands (Zhang and Zhang 2004c), comparative genomics (Zhang and Zhang 2003b), and in studying the distribution of nucleotide composition (Ou et al. 2003). With the availability of an increasing number of complete genome sequences, it is hoped that the Z-curve may play a more and more important role in genome research.
Acknowledgments
The present study was supported in part by the 973 Project of China (Grant 1999075606).
References
- R1.Bernander R. Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol. 2000;8:278–283. doi: 10.1016/s0966-842x(00)01760-1. [DOI] [PubMed] [Google Scholar]
- R2.Bernander R. The archaeal cell cycle: current issues. Mol. Microbiol. 2003;48:599–604. doi: 10.1046/j.1365-2958.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- R3.Berquist B.R., DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Bohlke K., Pisani F.M., Rossi M., Antranikian G. Archaeal DNA replication: spotlight on a rapidly moving field. Extremophiles. 2002;6:1–14. doi: 10.1007/s007920100222. [DOI] [PubMed] [Google Scholar]
- R5.Brochier C., Forterre P., Gribaldo S. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004;5:R17. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Bult C.J., White O., Olsen G.J., et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii . Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- R7.Cohen G.N., Barbe V., Flament D., et al. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi . Mol. Microbiol. 2003;47:1495–1512. doi: 10.1046/j.1365-2958.2003.03381.x. [DOI] [PubMed] [Google Scholar]
- R8.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Deppenmeier U., Johann A., Hartsch T., et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- R10.Edgell D.R., Doolittle W.F. Archaea and the origin(s) of DNA replication proteins. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- R11.Fitz-Gibbon S.T., Ladner H., Kim U.J., Stetter K.O., Simon M.I., Miller J.H. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum . Proc. Natl. Acad. Sci. USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Freeman J.M., Plasterer T.N., Smith T.F., Mohr S.C. Patterns of genome organization in bacteria. Science. 1998;279:1827. [Google Scholar]
- R13.Futterer O., Angelov A., Liesegang H., Gottschalk G., Schleper C., Schepers B., Dock C., Antranikian G., Liebl W. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. USA. 2004;101:9091–9096. doi: 10.1073/pnas.0401356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Galagan J.E., Nusbaum C., Roy A., et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Gao F., Zhang C.T. Comparison of various algorithms for recognizing short coding sequences of human genes. Bioinformatics. 2004;20:673–681. doi: 10.1093/bioinformatics/btg467. [DOI] [PubMed] [Google Scholar]
- R16.Gates M.A. A simple way to look at DNA. J. Theor. Biol. 1986;119:319–328. doi: 10.1016/s0022-5193(86)80144-8. [DOI] [PubMed] [Google Scholar]
- R17.Giraldo R. Common domains in the initiators of DNA replication in bacteria, archaea and eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 2003;26:533–554. doi: 10.1111/j.1574-6976.2003.tb00629.x. [DOI] [PubMed] [Google Scholar]
- R18.Grabowski B., Kelman Z. Archaeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- R19.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Guo F.B., Ou H.Y., Zhang C.T. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31:1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Hamori E., Ruskin J. H curves, a novel method of representation of nucleotide series especially suited for long DNA sequences. J. Biol. Chem. 1983;258:1318–1327. [PubMed] [Google Scholar]
- R22.Jeffrey H.J. Chaos game representation of gene structure. Nucleic Acids Res. 1990;18:2163–2170. doi: 10.1093/nar/18.8.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.Jones W.J., Leigh J.A., Mayer F., Woese C.R., Wolfe R.S. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 1983;136:254–261. [Google Scholar]
- R24.Kawarabayasi Y., Sawada M., Horikawa H., et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- R25.Kawarabayasi Y., Hino Y., Horikawa H., et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101, 145–152. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- R26.Kawarabayasi Y., Hino Y., Horikawa H., et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- R27.Kawashima T., Amano N., Koike H., et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium . Proc. Natl. Acad. Sci. USA. 2000;97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Kelman Z. The replication origin of archaea is finally revealed. Trends Biochem. Sci. 2000;25:521–523. doi: 10.1016/s0968-0004(00)01687-x. [DOI] [PubMed] [Google Scholar]
- R29.Kelman Z., Hurwitz J. Structural lessons in DNA replication from the third domain of life. Nat. Struct. Biol. 2003;10:148–150. doi: 10.1038/nsb0303-148. [DOI] [PubMed] [Google Scholar]
- R30.Kelman L.M., Kelman Z. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 2003;48:605–615. doi: 10.1046/j.1365-2958.2003.03369.x. [DOI] [PubMed] [Google Scholar]
- R31.Kennedy S.P., Ng W.V., Salzberg S.L., Hood L., Dassarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Klenk H.P., Clayton R.A., Tomb J.F., et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus . Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- R33.Lecompte O., Ripp R., Puzos-Barbe V., Duprat S., Heilig R., Dietrich J., Thierry J.C., Poch O. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 2001;11:981–993. doi: 10.1101/gr.165301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Liu J., Smith C.L., Deryckere D., Deangelis K., Martin G.S., Berger J.M. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- R35.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- R36.Lobry J.R. A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie. 1996;78:323–326. doi: 10.1016/0300-9084(96)84764-x. [DOI] [PubMed] [Google Scholar]
- R37.Lopez P., Philippe H., Myllykallio H., Forterre P. Identification of putative chromosomal origins of replication in archaea. Mol. Microbiol. 1999;32:883–886. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- R38.Lundgren M., Andersson A., Chen L., Nilsson P., Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R39.Mackiewicz P., Zakrzewska-Czerwinska J., Zawilak A., Dudek M.R., Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R40.MacNeill S.A. Understanding the enzymology of archaeal DNA replication: progress in form and function. Mol. Microbiol. 2001;40:520–529. doi: 10.1046/j.1365-2958.2001.02390.x. [DOI] [PubMed] [Google Scholar]
- R41.Maisnier-Patin S., Malandrin L., Birkeland N.K., Bernander R. Chromosome replication patterns in the hyperthermophilic euryarchaea Archaeoglobus fulgidus and Methanocaldococcus (Methanococcus) jannaschii . Mol. Microbiol. 2002;45:1443–1450. doi: 10.1046/j.1365-2958.2002.03111.x. [DOI] [PubMed] [Google Scholar]
- R42.Marchler-Bauer A., Anderson J.B., Deweese-Scott C., et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R43.Matsunaga F., Norais C., Forterre P., Myllykallio H. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R44.McLean M.J., Wolfe K.H., Devine K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- R45.Mrazek J., Karlin S. Strand compositional asymmetry in bacterial and large viral genomes. Proc. Natl. Acad. Sci. USA. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R46.Myllykallio H., Lopez P., Lopez-Garcia P., Heilig R., Saurin W., Zivanovic Y., Philippe H., Forterre P. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- R47.Ng W.V., Kennedy S.P., Mahairas G.G., et al. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R48.Ou H.Y., Guo F.B., Zhang C.T. Analysis of nucleotide distribution in the genome of Streptomyces coelicolor A3(2) using the Z curve method. FEBS Lett. 2003;540:188–194. doi: 10.1016/s0014-5793(03)00263-1. [DOI] [PubMed] [Google Scholar]
- R49.Pickover C.A. DNA and protein tetragrams: biological sequences as tetrahedral movements. J. Mol. Graph. 1992;10:2–6, 17. doi: 10.1016/0263-7855(92)80001-t. [DOI] [PubMed] [Google Scholar]
- R50.Robb F.T., Maeder D.L., Brown J.R., Diruggiero J., Stump M.D., Yeh R.K., Weiss R.B., Dunn D.M. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- R51.Robinson N.P., Dionne I., Lundgren M., Marsh V.L., Bernander R., Bell S.D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus . Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- R52.Rocha E.P., Danchin A., Viari A. Universal replication biases in bacteria. Mol. Microbiol. 1999;32:11–16. doi: 10.1046/j.1365-2958.1999.01334.x. [DOI] [PubMed] [Google Scholar]
- R53.Ruepp A., Graml W., Santos-Martinez M.L., et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum . Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- R54.Salzberg S.L., Salzberg A.J., Kerlavage A.R., Tomb J.F. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- R55.She Q., Singh R.K., Confalonieri F., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R56.Slesarev A.I., Mezhevaya K.V., Makarova K.S., et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R57.Smith D.R., Doucette-Stamm L.A., Deloughery C., et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R58.Tye B.K. Insights into DNA replication from the third domain of life. Proc. Natl. Acad. Sci. USA. 2000;97:2399–2401. doi: 10.1073/pnas.97.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R59.Waters E., Hohn M.J., Ahel I., et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R60.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R61.Zhang C.T., Wang J. Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z curve. Nucleic Acids Res. 2000;28:2804–2814. doi: 10.1093/nar/28.14.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R62.Zhang C.T., Zhang R. Analysis of distribution of bases in the coding sequences by a diagrammatic technique. Nucleic Acids Res. 1991;19:6313–6317. doi: 10.1093/nar/19.22.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R63.Zhang R., Zhang C.T. Z curves, an intutive tool for visualizing and analyzing the DNA sequences. J. Biomol. Struct. Dyn. 1994;11:767–782. doi: 10.1080/07391102.1994.10508031. [DOI] [PubMed] [Google Scholar]
- R64.Zhang R., Zhang C.T. Single replication origin of the archaeon Methanosarcina mazei revealed by the Z curve method. Biochem. Biophys. Res. Commun. 2002;297:396–400. doi: 10.1016/s0006-291x(02)02214-3. [DOI] [PubMed] [Google Scholar]
- R65.Zhang C.T., Zhang R. An isochore map of the human genome based on the Z curve method. Gene. 2003;317:127–135. doi: 10.1016/s0378-1119(03)00665-6. [DOI] [PubMed] [Google Scholar]
- R66.Zhang R., Zhang C.T. Identification of genomic islands in the genome of Bacillus cereus by comparative analysis with Bacillus anthracis . Physiol. Genomics. 2003;16:19–23. doi: 10.1152/physiolgenomics.00170.2003. [DOI] [PubMed] [Google Scholar]
- R67.Zhang R., Zhang C.T. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 2003;302:728–734. doi: 10.1016/s0006-291x(03)00252-3. [DOI] [PubMed] [Google Scholar]
- R68.Zhang C.T., Zhang R. Isochore structures in the mouse genome. Genomics. 2004;83:384–394. doi: 10.1016/j.ygeno.2003.09.011. [DOI] [PubMed] [Google Scholar]
- R69.Zhang R., Zhang C.T. Identification of replication origins in the genome of the methanogenic archaeon, Methanocaldococcus jannaschii . Extremophiles. 2004;8:253–258. doi: 10.1007/s00792-004-0385-4. [DOI] [PubMed] [Google Scholar]
- R70.Zhang R., Zhang C.T. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics. 2004;20:612–622. doi: 10.1093/bioinformatics/btg453. [DOI] [PubMed] [Google Scholar]
- R71.Zhang C.T., Zhang R., Ou H.Y. The Z curve database: a graphic representation of genome sequences. Bioinformatics. 2003;19:593–599. doi: 10.1093/bioinformatics/btg041. [DOI] [PubMed] [Google Scholar]