Abstract
The enzyme sn-glycerol-1-phosphate dehydrogenase (Gro1PDH, EC 1.1.1.261) is key to the formation of the enantiomeric configuration of the glycerophosphate backbone (sn-glycerol-1-phosphate) of archaeal ether lipids. This enzyme catalyzes the reversible conversion between dihydroxyacetone phosphate and glycerol-1-phosphate. To date, no information about the active site and catalytic mechanism of this enzyme has been reported. Using the sequence and structural information for glycerol dehydrogenase, we constructed six mutants (D144N, D144A, D191N, H271A, H287A and D191N/H271A) of Gro1PDH from Aeropyrum pernix K1 and examined their characteristics to clarify the active site of this enzyme. The enzyme was found to be a zinc-dependent metalloenzyme, containing one zinc ion for every monomer protein that was essential for activity. Site-directed mutagenesis of D144 increased the activity of the enzyme. Mutants D144N and D144A exhibited low affinity for the substrates and higher activity than the wild type, but their affinity for the zinc ion was the same as that of the wild type. Mutants D191N, H271A and H287A had a low affinity for the zinc ion and a low activity compared with the wild type. The double mutation, D191N/ H271A, had no enzyme activity and bound no zinc. From these results, it was clarified that residues D191, H271 and H287 participate in the catalytic activity of the enzyme by binding the zinc ion, and that D144 has an effect on substrate binding. The structure of the active site of Gro1PDH from A. pernix K1 seems to be similar to that of glycerol dehydrogenase, despite the differences in substrate specificity and biological role.
Keywords: glycerophosphate backbone, metalloenzyme, zinc
Introduction
The Archaea comprise a phylogenetically distinct group (domain) that diverged from the Bacteria and Eukarya at an early stage of evolution (Woese et al. 1990, Nelson et al. 2000). Aeropyrum pernix K1 (JCM no. 9820) is an aerobic hyperthermophilic archaeon for which the complete genome sequence has been determined (Faguy and Doolittle 1999, Kawarabayasi et al. 1999). Aeropyrum pernix is the only hyperthermophile known to obtain energy exclusively through aerobic respiration of complex organic matter (Sako et al. 1996). One characteristic feature of archaea is that their cellular membrane has an ether linkage between the glycerol backbone and the hydrocarbon residues. The core lipid of the phospholipids and glycolipids in archaeal cells is sn-2,3-dialkylglycerol, which has a polar head group in the sn-1 position. In contrast, the major lipids of eukaryotic and eubacterial cells consist of sn-1,2-diacylglycerol, which has a polar head group in the sn-C-3 position (Zhang and Poulter 1993). The enzyme sn-glycerol-1-phosphate dehydrogenase (Gro1PDH) is key to the formation of the enantiomeric configuration of the glycerophosphate backbone (sn-glycerol-1-phosphate) of archaeal ether lipids, and catalyzes the reversible conversion between dihydroxyacetone phosphate (DHAP) and glycerol-1-phosphate (Gro1P) using either NADH or NADPH as a coenzyme (Zhang and Poulter 1993, Nishihara and Koga 1995). We have found and characterized a hyperthermostable Gro1PDH from A. pernix (Han et al. 2002) that exhibits maximum activity at 96 °C.
From the homology modeling of Gro1PDH (Daiyasu et al. 2002), aspartic acid (Asp or D) and histidine (His or H) residues are speculated to be located at the active site and related to zinc ion (Zn2+) binding. Until now, however, there has been no evidence concerning the roles of these residues. Information on the active site and catalytic mechanism of Gro1PDH will lead to an initial understanding of the formation mechanism for polar lipids in archaea, and of the difference between archaea and bacteria in evolution.
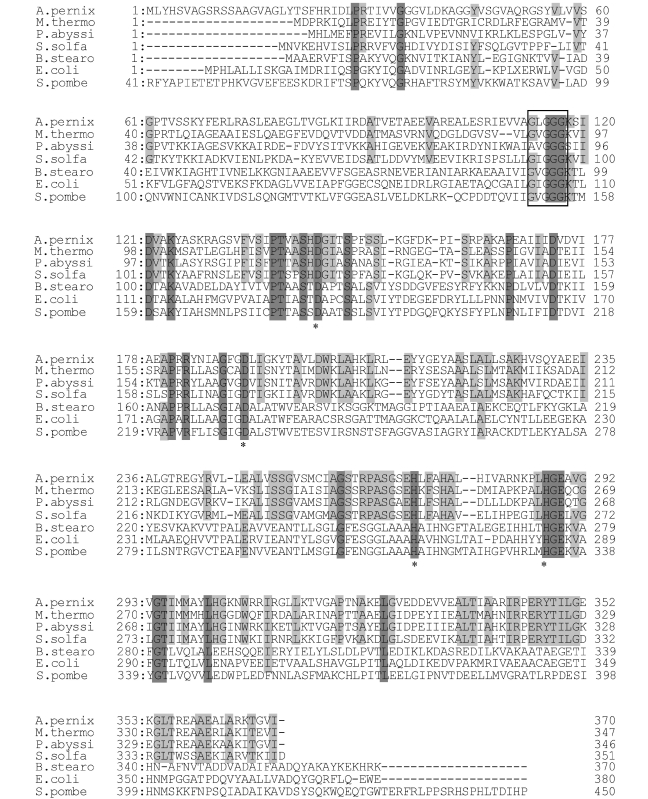
According to the results of sequence alignment, the Gro1PDH isolated from A. pernix seems to have a different three-dimensional structure to that of glycerol-3-phosphate dehydrogenases, which have a role in bacteria corresponding to that of Gro1PDH in archaea (Nishihara and Koga 1995, Han et al. 2002). Glycerol dehydrogenases (GroDH), which allow the use of glycerol as a carbon source (May and Sloan 1981), have been isolated from several different organisms (Scharschmidt et al. 1983, Spencer et al. 1989). Despite its different biological role and substrate specificity, GrolPDH exhibits a sequence similarity to GroDH. Therefore, the design of the GrolPDH mutants was performed on the basis of the sequence homology between GrolPDH and GroDH. Analysis of the crystal structures of family III metal-dependent GroDHs showed that residues D123, D173, H256 and H274 are important in the orientation of the glycerol that forms ligands to the coordinated Zn2+ (Ruzheinikov et al. 2001). These amino acids are conserved in the GrolPDH from A. pernix, and correspond to D144, D191, H271 and H287, respectively. It has also been reported that GrolPDH from Methanobacterium thermoautotrophicum contains Zn2+ (Koga et al. 2003). In this study, we prepared mutant enzymes by site-directed mutagenesis and examined their characteristics to clarify the active site of GrolPDH.
Materials and methods
Construction of mutated Gro1PDH
Site-directed mutation of Gro1PDH was performed by the polymerase chain reaction (PCR) method using the wild-type Gro1PDH gene (Mullis et al. 1986). Synthetic oligonucleotides were designed to produce the desired point mutations: D144N (Asp144→asparagine (Asn or N)), 5′-ACCGTGGCTAGCCACAACGGGATAACATCG-3′; D144A (Asp144→ alanine (Ala or A)), 5′-ACCGTGGCTAGCCACGCCGGGATAACATCG-3′; D191N, 5′-TCGCTGGCTTCGGAAACCTGATAGGCAAG-3′; H271A, 5′-GCAAGCGGCAGTGAAGCCCTCTTCGCCCAC-3′ and H287A, 5′-GATGGCCTACCTCGCCGGTAAGAACTGGAG-3′. The mutated positions are indicated in bold. These substituted codons are used frequently in Escherichia coli. The D191N/H271A double mutant was constructed from a single mutant plasmid (D191N) as a template. All mutants were transformed with the pET11a vector system into the host, BL21 (DE3) (Han et al. 2002). The transformant cells were grown in 2YT medium containing ampicillin (100 µg ml–1) at 37 °C. After incubation with shaking at 37 °C until the optical density at 600 nm reached 0.6–1.0, induction of the protein was carried out by adding isopropyl β-D-thiogalactopyranoside to a final concentration of 1 mM, and shaking for 4 h at 37 °C. The complete DNA sequence of each mutated gene was verified with an automated DNA sequencing system (ABI PRISM 310, Applied Biosystems, Foster City, CA), and only the expected differences from the wild type at the mutation sites were observed.
Preparation and purification of mutated Gro1PDH
Each Gro1PDH mutant was expressed and purified by the same methods as previously reported for the wild-type Gro1PDH (Han et al. 2002). Cells were harvested by centrifugation, frozen at –80 °C, and disrupted by sonication in 50 mM Tris-HCl buffer (pH 8.0). The homogenate was heated at 85 °C for 30 min and centrifuged. The supernatant was dialyzed against 50 mM Tris-HCl buffer (pH 8.0), and the dialyzed sample was purified by chromatography on a HiTrap Q column (Pharmacia, Uppsala, Sweden), a HiLoad Phenyl Sepharose column (Pharmacia) and then a HiLoad Superdex column (Pharmacia). The homogeneity of each Gro1PDH mutant was confirmed by SDS-PAGE (12%). The protein concentration was determined with Coomassie protein assay reagent (Pierce Chemical Company, Rockford, IL) with bovine serum albumin as the standard.
Thermal denaturation measurement by circular dichroic spectroscopy
Circular dichroism (CD) spectra were recorded with a J-820 spectrometer (Jasco, Tokyo, Japan). Wavelength scans were collected in 0.1 nm increments from 250 to 200 nm at 20 nm min–1 and 25 °C, and temperature scans were collected in 0.2 °C increments from 20 to 95 °C with a 12 s per point mean time at 222 nm. Baseline buffer spectra were subtracted from sample spectra.
Enzyme assay
Enzyme activity was measured spectrophotometrically with a J-520 spectrophotometer (Jasco) by monitoring the decrease in the NAD(P)H concentration in 50 mM Tris-HCl buffer (pH 8.0) containing 70 mM KCl, 2.1 mM DHAP and 0.32 mM NADH (or 0.32 mM NADPH) at 65 °C. The reaction was initiated by the addition of 10 µl of enzyme solution. The total volume of the reaction mixture was 1.2 ml. The kinetic constants of Gro1PDH from A. pernix were obtained by activity measurements in the direction of DHAP reduction, with substrate concentrations ranging from 0.1 to 10 Km. Each individual rate measurement was performed in triplicate and the kinetic mechanism was determined by the damped nonlinear least-squares method (Marquardt-Levenberg Method) according to the following equation (Menke 1989, Press et al. 1992, Han et al. 2002):
where A is the coenzyme (NAD(P)H), B is DHAP, Kia is the dissociation constant for NAD(P)H, Ka is Km for NAD(P)H, and Kb is Km for DHAP.
Analysis of bound metal ions
The bound metals of the enzyme were analyzed by inductively coupled plasma atomic emission spectroscopy (IRIS AP; TJA Solutions, Franklin, MA). The wild-type enzyme and double mutant D191N/H271A were dialyzed against 50 mM Tris-HCl buffer (pH 8.0) containing 0.15 M NaCl. The dialyzed enzyme, which contained 0.35 mg ml–1 protein, was used for the analysis, and the amount of zinc was calculated with a zinc standard solution.
Materials
The cloned Gro1PDH gene from A. pernix was used as a template for constructing mutant products (Han et al. 2002). The DNA primers were synthesized by Hokkaido System Science (Sapporo, Japan). All other chemicals and reagents were of commercially available analytic grade.
Results
Selection of mutation sites
There was no information about the structure and active site residues of Gro1PDH. Sequence alignment (see Figure 1) suggested a similar three-dimensional structure between Gro1PDH and GroDH despite their different substrate specificities (Ruzheinikov et al. 2001). The structure of GroDH has been solved (Ruzheinikov et al. 2001), and most of the active site residues are conserved in Gro1PDH (Han et al. 2002). The crystal structures of family III metal-dependent GroDHs showed that the amino acids D123, D173, H256 and H274 are important in the orientation of the glycerol that forms ligands to the enzyme-bound Zn2+ at the active site (Ruzheinikov et al. 2001). These amino acids are conserved in Gro1PDH from A. pernix, corresponding to D144, D191, H271 and H287, respectively (see Figure 1). Six mutant Gro1PDHs (D144N, D144A, D191N, H271A, H287A and D191N/H271A) were constructed by site-directed mutagenesis.
Figure 1.
Amino acid alignment of several glycerol dehydrogenases (GroDHs) together with sn-glycerol-1-phosphate dehydrogenase (Gro1PDH) of Aeropyrum pernix K1. Archaeal Gro1PDH: Methanobacterium thermoautotrophicum (M. thermo; 370 aa); Pyrococcus abyssi (P. abyssi; 346 aa); and Sulfolobus solfataricus (S. solfa; 351 aa). Bacterial or eukaryotic GroDH: Bacillus stearothermophilus (B. stearo; 370 aa); Escherichia coli (E. coli; 380 aa); and Schizosaccharomyces pombe (S. pombe; 450 aa). Amino acid residues that are identical in all sequences are shaded in dark gray, and residues that are identical in all Gro1PDHs are shaded in light gray. Asterisks indicate amino acid residues changed by site-directed mutagenesis. The box indicates the NAD(P)H binding site.
Expression of the mutants
The mutant enzymes were constructed, expressed and purified as described under Materials and methods. The amount of the expressed enzyme was unaffected by the mutations. The chromatographic patterns during purification and the yields of the expressed mutants were similar to those of the wild-type Gro1PDH. The purified mutant enzymes each migrated as a single band on SDS-PAGE, with an apparent molecular mass of 38 kDa, the same as that of the wild type.
Thermal stability of the mutants
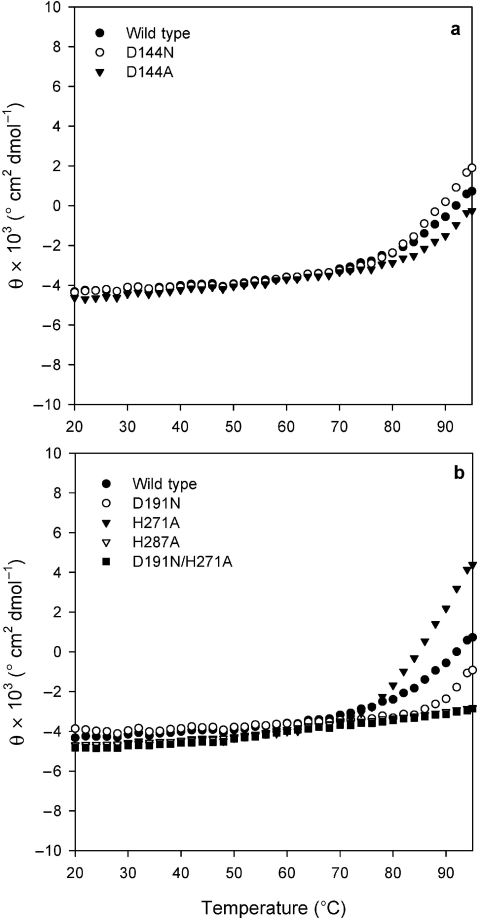
The CD spectrum of the wild type in the far-UV region was examined from 20 to 95 °C. The thermal stabilities of the wild type and mutants were compared by CD. Circular dichroism at 222 nm of the mutants was measured from 20–95 °C (see Figures 2a and 2b). Some mutants exhibited a slight decrease in ellipticity at 220 nm at temperatures greater than 70 °C. These results indicate that major conformational changes of the wild type and mutants were unaffected by temperatures between 20 and 70 °C.
Figure 2.
Thermal unfolding curves for the wild type and mutant sn-glycerol-1-phosphate dehydrogenase (Gro1PDH) (0.21–0.34 mg ml–1) at 222 nm of CD. Abbreviation: θ = ellipticity.
Characteristics of the mutants
All enzyme assays were performed at 65 °C because of the thermostability of the substrates and enzymes. Residues D144 and D191 were changed to their amidated forms (D144N and D191N), and H271 and H278 were changed to alanine (H271A and H278A) by site-directed mutagenesis. Mutant D144N exhibited a 13-fold increase in activity on NADH-dependent DHAP reduction. The activity of D144A also increased. The activity of D191N, H271A and H287A decreased. It has been reported that the catalytic reaction of Gro1PDH from A. pernix follows an ordered bi-bi mechanism (Han et al. 2002). Analysis of DHAP reduction was performed by bi-substrate kinetic analysis with NAD(P)H and DHAP because of the low Gro1P oxidation activity (Nishihara and Koga 1997). The experimental data for the mutants could be fitted to the equation for an ordered bi-bi mechanism. The kinetic constants Ka (Km for NAD(P)H), Kia (dissociation constant for NAD(P)H), Kb (Km for DHAP) and kcat were determined (see Tables 1 and 2). The kinetic parameters for NADH and DHAP of D144N differed from those of the wild type. The kcat value increased 12-fold compared with that of the wild type and the Ka value was also increased. Based on the kcat/Kavalue, D144N was more effective than the wild type. The dissociation constant for NADH was decreased greatly by this mutation. A threefold increase in the kcat value was observed for the D144A mutant, but this mutant was less effective than the wild type when the kcat/Kavalues were compared.
Table 1.
Kinetic constants for the wild-type and sn-glycerol-1-phosphate dehydrogenase (Gro1PDH) mutants in dihydroxyacetone phosphate (DHAP) reduction with NADH as the coenzyme. These parameters were estimated by the Marquardt-Levenberg method (Menke 1989, Press et al. 1992) in experiments in which a fixed concentration of substrate or coenzyme and appropriate ranges of concentrations of the other reactants were used. Abbreviations: kcat = turnover number; Ka = Km for NADH; Kb = Km for DHAP; Kia = dissociation constant for NADH; and nd = not detected.
| Kinetic parameter | Wild type | D144N | D144A | D191N | H271A | H287A | D191N/H271A |
| kcat (min–1) | 299.9 ± 23.4 | 3628.2 ± 380.5 | 1126.5 ± 158.6 | 196.2 ± 38.4 | 29.58 ± 16.6 | 176.7 ± 4.99 | nd |
| Ka (mM) | 0.0477 ± 0.02 | 0.415 ± 0.06 | 0.213 ± 0.06 | 0.296 ± 0.10 | 0.321 ± 0.31 | 0.036 ± 0.004 | nd |
| Kb (mM) | 0.177 ± 0.01 | 1.12 ± 0.20 | 1.12 ± 0.34 | 0.633 ± 0.32 | 1.09 ± 0.13 | 0.259 ± 0.026 | nd |
| Kia (mM) | 0.180 ± 0.17 | 0.0017 ± 0.00 | 0.245 ± 0.07 | 0.227 ± 0.14 | 0.042 ± 0.02 | 0.0025 ± 0.00 | nd |
| % Fit | 98.6 | 99.7 | 99.8 | 99.1 | 98.57 | 99.8 | |
| SD | 0.562 | 1.04 | 0.294 | 0.108 | 0.027 | 0.125 | |
Table 2.
Kinetic parameters for the wild-type, D144N and D144A sn-glycerol-1-phosphate dehydrogenases (Gro1PDH) in dihydroxyacetone phosphate (DHAP) reduction with NADPH as the coenzyme. These parameters were estimated with the Marquardt-Levenberg method (Menke 1989, Press et al. 1992) in experiments in which a fixed concentration of substrate or coenzyme and appropriate ranges of concentrations of the other reactant were used. Abbreviations: kcat = turnover number; Ka = Km for NADPH; Kb = Km for DHAP; and Kia = dissociation constant for NADPH.
| Kinetic parameter | Wild type | D144N | D144A |
| kcat (min–1) | 90.42 ± 3.01 | 284.1 ± 50.8 | 210.4 ± 7.91 |
| Ka (mM) | 0.086 ± 0.01 | 0.165 ± 0.06 | 0.039 ± 0.006 |
| Kb (mM) | 0.050 ± 0.02 | 0.360 ± 0.03 | 0.218 ± 0.030 |
| Kia (mM) | 0.257 ± 0.02 | 0.022 ± 0.01 | 0.013 ± 0.002 |
| % Fit | 99.8 | 99.7 | 99.7 |
| SD | 0.06 | 0.19 | 0.18 |
The D191N, H271A and H287A mutations decreased kcat. The kcat value for H271A decreased markedly, to about one tenth that of the wild type. The kinetic constants for H287A were similar to those for the wild type, although the kcat value was lower for the mutant enzyme, and there was a 140-fold decrease in the dissociation constant Kia. Comparison with the active site structure of GroDH (Ruzheinikov et al. 2001) suggests that residues D191 and H271 are located in the vicinity of the Zn2+ ion in the active site of Gro1PDH. Therefore, we designed the D191N/H271A double mutant and found it had no enzyme activity.
The kinetic constants for D144N and D144A were also determined with NADPH as the coenzyme (see Table 2). The kcat and Kb values for D144N and D144A increased significantly when determined with NADH as the coenzyme (see Table 1).
Metal ions bound to Gro1PDH
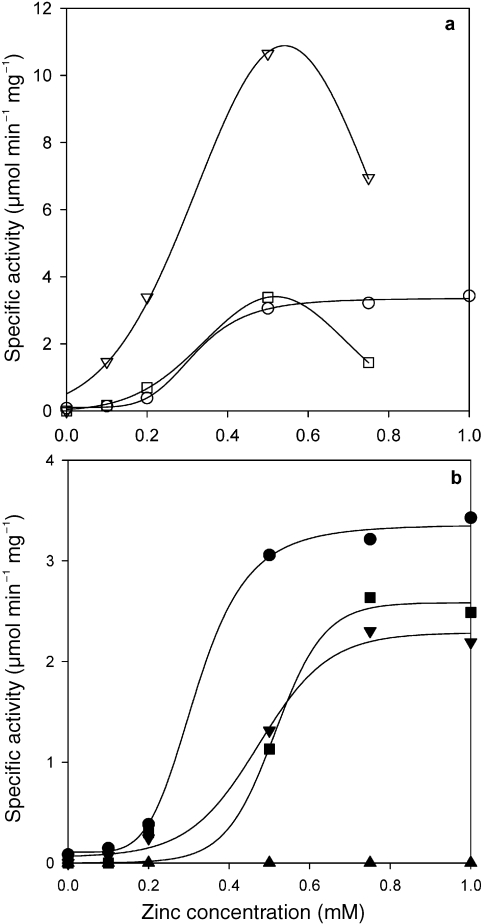
Atomic absorption analysis showed that Gro1PDH contained 0.81 mol of Zn2+ per monomer enzyme protein, and small amounts (less than 0.05 mol of metal ion per monomer enzyme protein) of magnesium and manganese. The enzyme did not contain other divalent cations, such as calcium, cobalt, molybdenum, nickel or copper. The effect of the metal-chelating reagent EDTA on the activity of Gro1PDH was examined after the enzymes had been dialyzed against 50 mM EDTA in Tris-HCl buffer (pH 8.0) for 12 h at 4 °C and then against 50 mM Tris-HCl buffer (pH 8.0). Exposure to EDTA led to a total loss of activity and a loss of Zn2+ to less than 0.05 mol Zn2+ per monomer protein for these enzymes. Activity was restored by incubation in 50 mM Tris-HCl buffer (pH 8.0) containing ZnCl2 at 4 °C for 1 h (Figure 3), whereas activity was not restored by incubation in MnCl2 solutions (data not shown). Figures 3a and 3b show the restored activity of the wild-type and mutant forms of Gro1PDH following incubation in a ZnCl2 solution. The zinc reactivation curves for the wild type and all mutants were sigmoid, and the effect depended on the zinc concentration. The specific activity abruptly increased until 0.5 mM ZnCl2 was added, and it was suppressed in the wild type when the ZnCl2 concentration exceeded 1.0 mM. Enzyme activity increased in D144N and D144A until 0.5 mM ZnCl2 was added, similar to the response of the wild type, indicating that this mutation had no effect on the zinc binding affinity of this enzyme. However, the specific activities of the D191N and H287A mutations increased until 0.75 mM ZnCl2 had been added, with lower maximum specific activities. The curves for both mutants are shifted to the right compared with those for the wild type, indicating that these mutations decrease the apparent affinity of the enzyme for zinc. For the H271A mutant, activity was not restored by incubation in a ZnCl2 solution up to 2.0 mM (see Figure 3). Atomic absorption analysis showed that the D191N/H271A double mutant contained less than 12% Zn2+ (0.09 mol Zn2+ per monomer enzyme protein) compared with the wild type.
Figure 3.
Restoration of activity by zinc ions in sn-glycerol-1-phosphate (Gro1PDH). The Gro1PDH was dialyzed against 50 mM EDTA (pH 8.0) for 24 h at 4 °C, and then against 50 mM Tris-HCl buffer/0.15 M NaCl (pH 8.0). Activity was measured after incubation in 50 mM Tris-HCl buffer/0.15 M NaCl (pH 8.0) containing ZnCl2 at 4 °C for 1 h. (a) Wild type (○), D144N (▽) and D144A (□). (b) Wild type (●), D191N (▼), H271A (▲) and H287A (■).
Discussion
The results of metal analysis and restoration of activity on addition of a zinc ion to the EDTA-treated enzyme indicate that the binding of one zinc ion to the enzyme plays a decisive role in determining Gro1PDH activity. This enzyme also contains small numbers of magnesium and manganese ions that do not affect its activity. Therefore, GrolPDH is a zinc-dependent metalloenzyme.
The thermostability of the enzyme at 20–70 °C was unaffected by the mutations (Figure 2), indicating that the mutations did not affect the overall structure and folding of the enzyme. By changing residue D144 to Asn or Ala, the activity of the enzyme was increased with NADH and NADPH as the coenzyme. From the kinetic data (see Tables 1 and 2), it is clear that the DHAP binding affinity was decreased and the kcat value was increased by these mutations. These results indicate that D144 is not the catalytic residue, although it is likely to be close to the DHAP binding site, and its carboxyl side chain may contribute to the affinity of the enzyme for DHAP.
The study for the homology modeling of Gro1PDH (Daiyasu et al. 2002) indicates that D144 forms hydrogen bonds with the carboxyl oxygen at C2 of DHAP. Furthermore, the affinity of the coenzymes was also influenced by these mutations. Replacement of the carboxyl side chain on D144 appears to stimulate activity by removing the negative charge that affects coenzyme binding. In addition, the similar zinc affinity of D144N and D144A compared with that of the wild type (Figure 3a) indicates that D144 does not take part in the metal–environment interaction of this enzyme. The decrease in activity by high zinc concentrations of the zinc-deplete D144N and D144A mutants (Figure 3a) suggests that these mutants have a secondary zinc ion binding site and that excess zinc ions decrease the enzyme’s activity (Larsen and Auld 1991, Gomez-Ortiz et al. 1997).
Glycerol dehydrogenase is a metalloenzyme containing a zinc or iron ion at the active center. It has been reported that Zn2+ is bound in a deep cleft, and is tetrahedrally coordinated through an ion–dipole interaction with amino acid residues D173, H256 and H274, and one water molecule (Ruzheinikov et al. 2001). Analysis of the conserved sequence in Gro1PDH and GroDH, and a study of homology modeling of Gro1PDH (Daiyasu et al. 2002), showed that the three amino acid residues that bind zinc are completely conserved in A. pernix Gro1PDH. These residues are D191, H271 and H287. The H271 (H271A) mutation decreased the affinity for a substrate and decreased activity, which strongly suggests that this mutation modified the environment of the active site. Although the D191 and H287 mutations decreased the activity, the H287A mutant exhibited a binding affinity for the substrates similar to that of the wild type. The zinc activation curves for D191N and H287A were shifted to the right compared with that of the wild type, indicating a small reduction in zinc binding. This indicates that the D191 and H287 mutations lower the apparent affinity of the enzyme for zinc, and D191 and H287 contribute ligands to the zinc ion. In addition, the simultaneous replacement of D191 and H271 (D191N/H271A) brought about a loss of both enzyme activity and the bound zinc ion. From the kinetic data obtained for D191N, H271A, H287A and D191N/H271A, and the restoration of their activity by Zn2+, it was apparent that the zinc ion is located at the active center and seems to coordinate with these three amino acid residues through an ion–dipole interaction.
The complete loss of enzyme activity and the zinc ion in the D191N/H271A mutant strongly indicates that these residues are not directly related to the catalytic mechanism, but that the coordinated Zn2+ plays an important catalytic role. Two of the three amino acid residues mutated in this study appear to be necessary for zinc binding; however, these residues are unrelated to substrate binding. The activity of H271A was not restored by Zn2+ (see Figure 3b), indicating that the environment of the zinc binding site of H271A is modified irreversibly on removal of the zinc ion, and that H271 plays an important role in stabilizing the conformation of the active site with the zinc ion.
These results for the mutant enzymes are not in conflict with the active site data predicted by the homology modeling of Gro1PDH (Daiyasu et al. 2002). Despite the differences in substrate specificity and biological roles of the two dehydrogenases, the results of these experiments indicate that the overall structures of the active sites and catalytic mechanisms of Gro1PDH and GroDH are similar, and that these enzymes are related to the divergent evolution of archaea and bacteria. The utilization of glycerol as a carbon source by GroDH seems to have evolved from the the formation of the glycerophosphate backbone of ether lipids by Gro1PDH.
Acknowledgments
This work was performed as part of the STA fellowship program, supported by the Japan Science and Technology Corporation. We thank H.-Y. Kim for great assistance with the experiments in this study.
References
- R1.Daiyasu H., Hiroike T., Koga Y., Toh H. Analysis of membrane stereochemistry with homology modeling of sn-glycerol-1- phosphate dehydrogenase. Protein Eng. 2002;15:987–995. doi: 10.1093/protein/15.12.987. [DOI] [PubMed] [Google Scholar]
- R2.Faguy D.M., Doolittle W.F. Genomics: lessons from the Aeropyrum pernix genome. Curr. Biol. 1999;9:R883–R886. doi: 10.1016/s0960-9822(00)80074-3. [DOI] [PubMed] [Google Scholar]
- R3.Gomez-Ortiz M., Gomis-Ruth F.X., Huber R., Aviles F.X. Inhibition of carboxypeptidase A by excess zinc: analysis of the structural determinant by X-ray crystallography. FEBS Lett. 1997;400:336–340. doi: 10.1016/s0014-5793(96)01412-3. [DOI] [PubMed] [Google Scholar]
- R4.Han J.-S., Kosugi Y., Ishida H., Ishikawa K. Kinetic study of sn-glycerol-1-phosphate dehydrogenase from the aerobic hyperthermophilic archaeon, Aeropyrum pernix K1. Eur. J. Biochem. 2002;269:969–976. doi: 10.1046/j.0014-2956.2001.02731.x. [DOI] [PubMed] [Google Scholar]
- R5.Kawarabayasi Y., Hiro Y., Horikawa H., et al. Complete genome sequence of an aerobic hyperthermophilic crenarchaeon, Aeropyrumpernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- R6.Koga Y., Sone N., Noguchi S., Morii H. Transfer of Pro-R hydrogen from NADH to dihydroxyacetonephosphate by sn-glycerol-1-phosphate dehydrogenase from the archaeon Methanobacterium thermoautotrophicum . Biosci. Biotechnol. Biochem. 2003;67:1605–1608. doi: 10.1271/bbb.67.1605. [DOI] [PubMed] [Google Scholar]
- R7.Larsen K.S., Auld S.D. Characterization of an inhibitory metal binding site in carboxypeptidase. Biochemistry. 1991;30:2613–2618. doi: 10.1021/bi00224a007. [DOI] [PubMed] [Google Scholar]
- R8.May J.W., Sloan J. Glycerol utilization by Schizosaccharomycespombe: dehydration as the initial step. J. Gen. Microbiol. 1981;123:183–185. [Google Scholar]
- R9.Menke W. Geophysical Data Analysis. New York: Revised Edn. Academic Press; 1989. Discrete inverse theory; pp. 143–160. [Google Scholar]
- R10.Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H.A. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- R11.Nelson K.H., Heidelberg J.F., Fraser C.M. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 2000;18:1049–1054. doi: 10.1038/80235. [DOI] [PubMed] [Google Scholar]
- R12.Nishihara M., Koga Y. sn-Glycerol-1-phosphate dehydrogenase in Methanobacterium thermoautotrophicum: key enzyme in biosynthesis of the enantiomeric glycerophosphate backbone of ether phospholipids of archaebacteria. J. Biochem. 1995;117:933–935. doi: 10.1093/oxfordjournals.jbchem.a124822. [DOI] [PubMed] [Google Scholar]
- R13.Nishihara M., Koga Y. Purification and properties of sn-glycerol-1-phosphate dehydrogenase from Methanobacterium thermoautotrophicum: characterization of the biosynthetic enzyme for the enatiomeric glycerophosphate backbone of ether polar lipids of archaea. J. Biochem. 1997;122:572–576. doi: 10.1093/oxfordjournals.jbchem.a021791. [DOI] [PubMed] [Google Scholar]
- R14.Press W.H., Teukolsky S.A., Vertterling W.T., Flannery B.P. Cambridge: 2nd Edn. Cambridge University Press; 1992. Numerical recipes in Fortran—the art of scientific computing; pp. 120–149. [Google Scholar]
- R15.Ruzheinikov S.N., Burke J., Sedelnikova S., Baker P.J., Taylor R., Bullough P.A., Muir N.M., Gore M.G., Rice D.W. Glycerol dehydrogenase: structure, specificity, and mechanism of family III polyol dehydrogenase. Structure. 2001;9:789–802. doi: 10.1016/s0969-2126(01)00645-1. [DOI] [PubMed] [Google Scholar]
- R16.Sako Y., Nomura N., Uchida A., Ishida Y., Morri H., Koga Y., Hoaki T., Maruyama T. Aeropyrum pernix gen. Nov., sp. Nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 °C. Int. J. Syst. Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- R17.Scharschmidt M., Pfleiderer G., Metz H., Brummer W. Isolation and characterization of the glycerol dehydrogenase from Bacillus megaterium . Hoppe-Seyler’s Z. Physiol. Chem. . 1983;364:911–921. [PubMed] [Google Scholar]
- R18.Spencer P., Brown K.J., Scawen M.D., Atkinson T., Gore M.G. Isolation and characterization of the glycerol dehydrogenase from Bacillus stearothermophilus . Biochim. Biophys. Acta. 1989;994:270–279. doi: 10.1016/0167-4838(89)90304-x. [DOI] [PubMed] [Google Scholar]
- R19.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eukarya. Proc. Natl. Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Zhang D., Poulter C.D. Biosynthesis of archaebacterial ether lipids. Formation of ether linkages by prenyltransferases. J. Am. Chem. Soc. 1993;115:1270–1277. [Google Scholar]