Abstract
A phylogenetic analysis of the genes encoding enzymes in the pentose phosphate pathway (PPP), the ribulose monophosphate (RuMP) pathway, and the chorismate pathway of aromatic amino acid biosynthesis, employing data from 13 complete archaeal genomes, provides a potential explanation for the enigmatic phylogenetic patterns of the PPP genes in archaea. Genomic and biochemical evidence suggests that three archaeal species (Methanocaldococcus jannaschii, Thermoplasma acidophilum and Thermoplasma volcanium) produce ribose-5-phosphate via the nonoxidative PPP (NOPPP), whereas nine species apparently lack an NOPPP but may employ a reverse RuMP pathway for pentose synthesis. One species (Halobacterium sp. NRC-1) lacks both the NOPPP and the RuMP pathway but may possess a modified oxidative PPP (OPPP), the details of which are not yet known. The presence of transketolase in several archaeal species that are missing the other two NOPPP genes can be explained by the existence of differing requirements for erythrose-4-phosphate (E4P) among archaea: six species use transketolase to make E4P as a precursor to aromatic amino acids, six species apparently have an alternate biosynthetic pathway and may not require the ability to make E4P, and one species (Pyrococcus horikoshii) probably does not synthesize aromatic amino acids at all.
Keywords: aromatic amino acid biosynthesis, chorismate, genomic analysis, nucleotide biosynthesis, pentose phosphate pathway, ribulose-5-phosphate, transketolase
Introduction
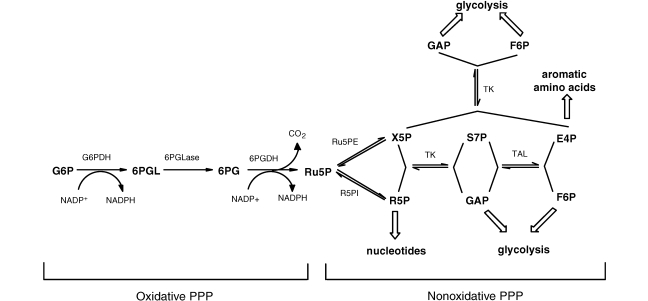
The pentose phosphate pathway (PPP) is a ubiquitous metabolic pathway in bacteria and eukaryotes. The function of the PPP is threefold: to generate NADPH for reducing power in reductive biosynthesis, to convert three- and six-carbon sugar phosphates from the glycolytic pathway into ribose-5-phosphate (R5P) for nucleotide biosynthesis, and to provide erythrose-4-phosphate (E4P), a precursor for aromatic amino acid biosynthesis. The classical PPP is divided into two phases (Figure 1). In the oxidative PPP (OPPP), glucose-6-phosphate (G6P) undergoes oxidative decarboxylation to form ribulose-5-phosphate (Ru5P) with the concomitant reduction of two molecules of NADP+ to NADPH. In the nonoxidative phase (NOPPP), R5P isomerase converts Ru5P to R5P, while the enzymes Ru5P 3-epimerase, transketolase and transaldolase act in concert to shuttle excess Ru5P back into three- and six-carbon glycolytic intermediates, with E4P formed in the process.
Figure 1.
The pentose phosphate pathway (PPP). Abbreviations: G6P = glucose-6-phosphate; G6PDH = glucose-6-phosphate dehydrogenase; 6PGL = 6-phosphoglucono-δ-lactone; 6PGLase = 6-phosphoglucono-δ-lactonase; 6PG = 6-phosphogluconate; 6PGDH = 6-phosphogluconate dehydrogenase; Ru5P = ribulose-5-phosphate; Ru5PE = ribulose-5-phosphate-3-epimerase; R5PI = ribose-5-phosphate isomerase; R5P = ribose-5-phosphate; X5P = xylulose-5-phosphate; TK = transketolase; S7P = sedoheptulose-7-phosphate; GAP = glyceraldehyde-3-phosphate; TAL = transaldolase; E4P = erythrose-4-phosphate; and F6P = fructose-6-phosphate.
The role of the PPP in archaea has been an open question for more than a decade. Before the advent of whole genome sequencing projects, it was reported that activities of NOPPP enzymes could be detected in the methanogenic archeaon Methanococcus maripaludis, but OPPP enzyme activities were not observed (Yu et al. 1994). The authors proposed that R5P synthesis in methanococci occurs solely via the NOPPP enzymes acting in the ‘reverse’ direction (F6P and GAP to Ru5P), and not by oxidative decarboxylation of G6P. This hypothesis was further supported by studies with Methanococcus voltae (Choquet et al. 1994) and M. maripaludis (Tumbula et al. 1997), in which 13C labeling patterns were consistent with R5P and E4P synthesis via the NOPPP. Further genomic analysis indicated that Methanocaldococcus jannaschii has a complete NOPPP but is missing the OPPP entirely (Selkov et al. 1997). Conversely, 13C labeling studies with other methanogenic archaea including Methanobacterium thermoautotrophicum (Ekiel et al. 1983, Eisenreich et al. 1991, Choquet et al. 1994) showed ribose labeling patterns consistent with the synthesis of R5P by the OPPP. The same studies also revealed labeling patterns in phenylalanine and tyrosine that were inconsistent with the biosynthesis of these aromatic amino acids starting from E4P, calling into question the role of E4P and the NOPPP in these species.
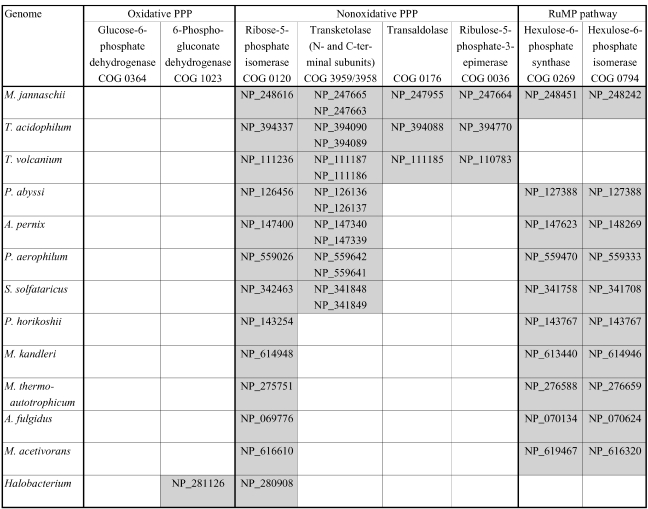
Recent progress in the sequencing of archaeal genomes has revealed a confusing picture of the PPP in the archaeal domain (Figure T1). Among the thirteen completed archaeal genomes included in the Clusters of Orthologous Groups (COG) database (Tatusov et al. 1997, 2001; http://www.ncbi.nlm.nih.gov/ COG/), orthologs of genes encoding enzymes of the OPPP are virtually nonexistent: only one species, Halobacterium sp. NRC-1, has a recognizable ortholog of 6-phosphogluconate dehydrogenase (6PGDH), whereas glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate lactonase (6PGL) orthologs are missing in all 13 genomes. Nonoxidative PPP orthologs are present to a varying extent in archaea: all 13 genomes contain R5P isomerase orthologs, but only three species (M. jannaschii, Thermoplasma volcanium and Thermoplasma acidophilum) have a complete set of recognizable NOPPP genes. Transketolase orthologs are present in an additional four archaeal genomes, but these four are missing genes for transaldolase and Ru5P 3-epimerase. Only two archaeal PPP enzymes have been biochemically characterized to date: R5P isomerase from Pyrococcus horikoshii (Ishikawa et al. 2002) and transaldolase from M. jannaschii (Soderberg and Alver 2004).
Figure T1.
Phylogenetic patterns of archaeal gene orthologs from the pentose phosphate pathway (PPP) and the ribulose monophosphate (RuMP) pathway. GenBank accession numbers are listed for each ortholog.
The biochemical and genomic data raise important questions about how R5P and E4P are synthesized in the 10 archaeal species that apparently lack a complete NOPPP. A possible answer could involve the existence of novel, as-yet unrecognized PPP genes in archaea. Another possibility is the existence of alternate pathways. Aromatic amino acid biosynthesis in some archaeal species appears to occur via an alternate route that does not use E4P (Tumbula et al. 1997, White 2004). An alternate source of pentose biosynthesis, which to our knowledge has not yet been recognized as such, is the ribulose monophosphate (RuMP) pathway.
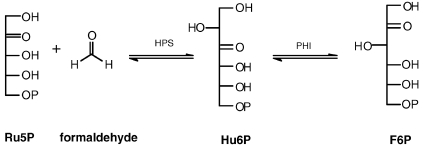
In the RuMP pathway (Figure 2), Ru5P condenses with formaldehyde to form 3-hexulose-6-phosphate (D-aribino-3-ketohexulose 6-phosphate, Hu6P) in an aldol reaction catalyzed by 3-hexulose-6-phosphate synthase (HPS, encoded by hps). 3-Hexulose-6-phosphate is then converted to F6P by 3-hexulose-6-phosphate isomerase (PHI, encoded by phi). Until recently, it was believed that the RuMP pathway was present exclusively in methylotrophic bacteria for formaldehyde fixation. However, hps and phi orthologs have recently been cloned from Bacillus subtilis and shown to encode HPS and PHI enzymes (Yasueda et al. 1999). Further sequence analysis indicates that hps and phi orthologs are widely distributed among bacterial and archaeal genomes (Reizer et al. 1997). The function of the phi ortholog in the archaeon M. jannaschii has been biochemically confirmed (Martinez-Cruz et al. 2002), and in the same study HPS activity was indirectly observed in a Pyrococcus horikoshii extract. However, the function of an archaeal hps ortholog has yet to be experimentally confirmed. It has been proposed that the role of the RuMP pathway in nonmethylotrophic bacteria could be xylose metabolism (Reizer et al. 1997), methylamine metabolism or formaldehyde detoxification (Yasueda et al. 1999). However, all of these functions require the presence of a complete NOPPP in order to supply Ru5P, a requirement that is met by bacteria but not by most archaea. This suggests that the RuMP pathway enzymes have another function in archaea, possibly related to pentose synthesis.
Figure 2.
The reactions of the ribulose monophosphate (RuMP) pathway. Abbreviations: Ru5P = ribulose-5-phosphate; HPS = 3-hexulose-6-phosphate synthase; Hu6P = 3-hexulose-6-phosphate; PHI = 3-hexulose-6-phosphate isomerase; and F6P = fructose-6-phosphate.
The availability of a growing database of complete microbial genome sequences and the development of new software tools to analyze phylogenetic patterns of inheritance provide a novel opportunity to better understand how various species have evolved different metabolic strategies. This report gives the results of a genomic analysis study that suggests an explanation for the enigmatic phylogenetic patterns of the PPP genes in archaeal genomes, and discusses the implications for nucleotide and aromatic amino acid biosynthesis in archaea.
Materials and methods
Included in this analysis are the 13 complete archaeal genomes in the COG database at the National Center for Biotechnology Information (NCBI) (Tatusov et al. 1997, 2000; http://www. ncbi.nlm.nih.gov/COG/). The STRING resource (Snel et al. 2000, von Mering et al. 2003; http://string.embl.de/) was used to compare phylogenetic profiles and gene position. Homology searches were performed with the BLASTP program on the NCBI website.
Results and discussion
Biosynthesis of ribose-5-phosphate
The genomic evidence strongly suggests that in archaea, as in bacteria and eukaryotes, Ru5P is the original source of the pentose moiety in nucleotides. Orthologs of R5P isomerase (COG 0120), which catalyzes the reversible interconversion of R5P and Ru5P, are present in all 13 archaeal genomes compared in this study (Figure T1). Also ubiquitous in archaeal genomes are most of the genes for nucleotide biosynthesis, including putative genes for phosphoribosylpyrophosphate (PRPP) synthase (COG 0462), orotate phosphoribosyltransferase (COG 0461) and glutamine phosphoribosyl amidotransferase (COG 0034), the key enzymes responsible for incorporating R5P into nucleotides. The question thus becomes how Ru5P is synthesized by those archaea that lack a complete set of NOPPP orthologs.
The RuMP pathway, operating in the ‘reverse’ direction, is a possible alternate source of Ru5P. The two reactions of the RuMP pathway—an aldol condensation and a 3-ketose to 2-ketose isomerization—are thermodynamically reversible. Furthermore, it appears that in archaeal genomes, the phylogenetic profiles of the RuMP pathway and the NOPPP are for the most part complementary. Orthologs for hps and phi (COG 0269 and COG 0794, respectively), are present in 10 of the 13 genomes (Figure T1), and all but one of these (M. jannaschii) lack a complete NOPPP, which is the source of Ru5P in the ‘forward’ RuMP pathway of methylotrophic bacteria. This indicates that the role of the RuMP pathway in these archaeal species may be reversed: that is, its function may be to produce Ru5P from F6P.
The operation of a reverse RuMP pathway for the production of Ru5P implies the concurrent production of formaldehyde, which is toxic to cells. Some mechanism must therefore exist to remove formaldehyde in all species that synthesize RuMP in this manner. Five of the 13 genomes analyzed in this study (the four methanogens plus the sulfate reducer Archaeoglobus fulgidus) contain members of COG 1795, a formaldehyde-activating enzyme first characterized in the methylotroph Methylobacterium extorquens (Vorholt et al. 2000). It is significant that in four of these five species this gene is fused to hps, possibly encoding a bifunctional enzyme that activates formaldehyde for oxidation as soon as it is produced as a by-product of Ru5P synthesis. Also present in nine of the 13 genomes are members of COG 2414, a tungsten-dependent formaldehyde ferredoxin oxidoreductase that has been characterized in Pyrococcus furiosus (Roy et al. 1999). Among the nine archaeal genomes that contain the RuMP pathway genes, only Sulfolobus solfataricus is missing both COG 1795 and COG 2414. This genome does, however, contain an open reading frame (GenBank accession no. AAK40795) that appears to be an ortholog of a glutathione-independent formaldehyde dehydrogenase characterized from Pseudomonas putida (Ogushi et al. 1984, Tanaka et al. 2002).
Experimental confirmation of the biochemical function of the protein encoded by hps in archaea will be critical to confirming or denying the role of the RuMP pathway in ribose synthesis. The homology between some archaeal hps orthologs and the biochemically characterized hps gene in B. subtilis is relatively low, with expected values ranging from 10–43 for the Pyrococcus abyssi gene to 10–14 for the M. jannaschii gene. In addition, hps orthologs are fused in several archaeal genomes to members of COG 0684, which encodes demethylmenaquinone methyltransferase. This fusion event further clouds predictions of the function of hps in archaea.
An alternate interpretation of results from previous isotopic labeling studies can be provided in light of the genomic data now available. Previous reports (Eisenreich et al. 1991, Choquet et al. 1994) stated that 13C labeling patterns in several methanogenic archaea (but not in methanococci) pointed to the action of OPPP enzymes in the conversion of F6P to Ru5P. However, the same labeling pattern observed in these studies would also be expected if the F6P to Ru5P conversion was carried out instead by a reverse RuMP pathway, a more likely scenario given the lack of OPPP orthologs in methanogenic genomes.
Although both T. volcanium and T. acidophilum are missing the RuMP pathway genes, these species do have a complete NOPPP that could provide a source of Ru5P. Methanocaldococcus jannaschii has both a complete NOPPP and an RuMP pathway, therefore Ru5P could derive from either or both of these pathways. However, labeling studies (Choquet et al. 1994, Tumbula et al. 1997) suggest that the NOPPP alone is responsible for ribose biosynthesis in methanococci.
Only Halobacterium lacks both an NOPPP and an RuMP pathway. However, Halobacterium is also the only archaeal genome that contains a recognizable OPPP gene (the ortholog of genes encoding 6PGDH (COG 1023)), which suggests that pentose synthesis in Halobacterium may take place via a modified OPPP. Although it is not yet clear how this could occur, there are several reasonable possibilities. Halobacterium may have a novel G6PDH that is not recognizable by sequence homology to known enzymes. 6-Phosphogluconate could be produced by oxidation of glucose to gluconate by glucose dehydrogenase (COG 1063, GenBank accession no. AAG18991), followed by phosphorylation of gluconate by an as yet uncharacterized putative sugar kinase (e.g., COG 0524, GenBank accession no. AAG20057). Alternatively, the oxidative carboxylation of gluconate to ribulose could be followed by phosphorylation to Ru5P by an as yet unidentified ribulokinase, if the predicted 6PGDH enzyme in Halobacterium is capable of oxidizing a nonphosphorylated substrate. Biochemical characterization of the predicted 6PGDH enzyme in Halobacterium, as well as experimental confirmation of the presence or absence of G6PDH, gluconate-6-kinase and ribulokinase activities in cell extracts, will be necessary to address the questions surrounding the origin of Ru5P in Halobacterium.
Biosynthesis of erythrose-4-phosphate and aromatic amino acids
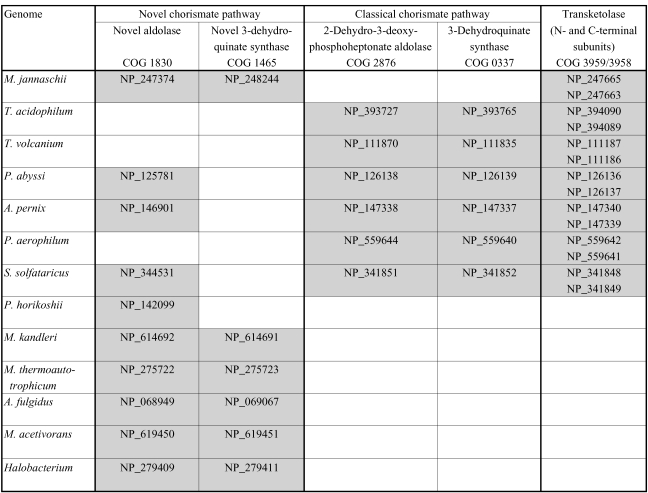
Another critical function of the NOPPP in bacteria and eukaryotes is the generation of E4P as a precursor for aromatic amino acid biosynthesis. The curious NOPPP phylogeny in archaeal genomes can be explained by the presence in some archaea of an alternate biosynthetic pathway that does not use E4P. The first phase of the classical aromatic amino acid biosynthesis pathway involves the seven-step synthesis of chorismate, beginning with the condensation of E4P and phosphoenolpyruvate (PEP) to form 2-dehydro-3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) and the subsequent cyclization to 3-dehydroquinate (DHQ). Most of this pathway is present in 12 of the 13 archaeal genomes analyzed here, although it is entirely missing in Pyrococcus horikoshii, which requires at least one aromatic amino acid (tryptophan) for growth (Gonzalez et al. 1998). For the fifth enzymatic step, archaea possess a novel shikimate kinase (COG 1685) that has only recently been identified and characterized (Daugherty et al. 2001). Seven of the genomes are missing orthologs from COG 2876 and COG 0337, the first two biosynthetic steps responsible for the formation of DHQ from E4P and PEP (Figure T2). Significantly, these are the same genomes (with the addition of M. jannaschii) that are also missing transketolase, and thus do not possess a recognizable means of making E4P.
Figure T2.
Phylogenetic distribution of the novel archaeal chorismate biosynthesis pathway genes (White 2004) compared with the first two steps of classical chorismate biosynthesis and transketolase. GenBank accession numbers are listed for each ortholog.
The hypothesis that these archaeal species possess an alternate beginning phase of the aromatic amino acid biosynthesis pathway that does not involve E4P as a precursor (Tumbula et al. 1997) was recently confirmed (White 2004). In M. jannaschii, two genes were identified that catalyze the formation of DHQ from L-aspartate semialdehyde and 6-deoxy-5-ketohexulose, rather than from E4P and PEP. Although the novel aldolase identified in that study is widespread among archaea, including several species that have genes for the classical (E4P- and PEP-utilizing) chorismate synthesis pathway, the phylogenetic pattern of the novel DHQ synthase is highly complementary to the patterns observed both for the classical chorismate pathway genes and also for transketolase (Figure T2).
It is notable that four of the 13 genomes (Pyrococcus abyssi, Aeropyrum pernix, Pyrobaculum aerophilum and Sulfolobus solfataricus) have an incomplete NOPPP: orthologs of transketolase and R5P isomerase are present but not orthologs of epimerase or transaldolase (Figure T1). Apparently, these four species use the RuMP pathway for Ru5P synthesis and thus do not require a complete NOPPP, but do require transketolase for generation of E4P from F6P and GAP. All four genomes contain orthologs for DAHP synthase, the E4P-utilizing step in aromatic amino acid biosynthesis. Furthermore, in three of these four genomes, the transketolase genes are located in operon-like clusters along with aromatic amino acid biosynthesis genes.
Conclusions
The archaeal phylogeny of the PPP, when taken in isolation, is an enigmatic picture of partial pathways and seemingly inexplicable patterns of gene inheritance. When combined with genomic information about the RuMP pathway and the chorismate pathway of aromatic amino acid biosynthesis, however, a more coherent picture begins to emerge. In most archaea, the ribose biosynthesis function of the OPPP is apparently carried out either by the enzymes of the NOPPP or by a reverse RuMP pathway. Those species that require E4P for aromatic amino acid biosynthesis produce it with transketolase, whereas those species with an alternate early phase of the chorismate pathway do not require transketolase. Finally, pentose synthesis in Halobacterium may occur via a modified OPPP, although the details of this putative pathway await experimental determination. The development of this picture of pentose and erythrose synthesis in archaea is an example of how the analysis of phylogenetic patterns within multiple genome sequences can yield valuable insights into diverse metabolic pathways. It will be interesting to see if the patterns observed in this study hold true as more complete genome sequences become available, and as the predicted biochemical functions of more genes are confirmed experimentally.
References
- R1.Choquet C.G., Richards J.C, Sprott G.D. Ribose synthesis in methanogenic archaea. Arch. Microbiol. 1994;161:481–488. [Google Scholar]
- R2.Daugherty M., Vonstein V., Overbeek R., Osterman A. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 2001;183:292–300. doi: 10.1128/JB.183.1.292-300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Eisenreich W., Schwarzkopf B., Bacher A. Biosynthesis of nucleotides, flavins, and deazaflavins in Methanobacterium thermoautotrophicum . J. Biol. Chem. 1991;266:9622–9631. [PubMed] [Google Scholar]
- R4.Ekiel I., Smith I.C.P., Sprott G.D. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 1983;156:316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5.Gonzalez J.M., Masuchi Y., Robb F.T., Ammerman J.W., Maeder D.L., Yanagibayashi J., Tamaoka J., Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- R6.Ishikawa K., Matsui I., Payan F., Cambillau C., Ishida H., Kawarabayasi Y., Kikuchi H., Roussel A. A hyperthermostable D-ribose-5-phosphate isomerase from Pyrococcus horikoshii: characterization and three-dimensional structure. Structure. 2002;10:877–886. doi: 10.1016/s0969-2126(02)00779-7. [DOI] [PubMed] [Google Scholar]
- R7.Martinez-Cruz L.A., Dreyer M.K., Boisvert D.C., Yokota H., Martinez-Chanter M.L., Kim R., Kim S.H. Crystal structure of MJ1247 protein from M. jannaschii at 2.0 Å resolution infers a molecular function of 3-hexulose-6-phosphate isomerase. Structure. 2002;10:195–204. doi: 10.1016/s0969-2126(02)00701-3. [DOI] [PubMed] [Google Scholar]
- R8.Ogushi S., Ando M., Tsuru D. Formaldehyde dehydrogenase from Pseudomonas putida: a zinc metalloenzyme. J. Biochem. 1984;96:1587–1591. doi: 10.1093/oxfordjournals.jbchem.a134988. [DOI] [PubMed] [Google Scholar]
- R9.Reizer J., Reizer A., Saier M.H. Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology. 1997;143:2519–2520. doi: 10.1099/00221287-143-8-2519. [DOI] [PubMed] [Google Scholar]
- R10.Roy R., Swarnalatha M., Schut G.J., Dunn D.M., Weiss R., Adams M.W.W. Purification and characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: the third of a putative five-member tungstoenzyme family. J. Bacteriol. 1999;181:1171–1180. doi: 10.1128/jb.181.4.1171-1180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Selkov E., Maltsev N., Olsen G.J., Overbeek R., Whitman W.B. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- R12.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Soderberg T., Alver R.C. Transaldolase of Methanocaldococcus jannaschii . Archaea. 2004;1:255–262. doi: 10.1155/2004/608428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Tanaka N., Kusakabe Y., Ito K., Yoshimoto T., Nakamura K.T. Crystal structure of formaldehyde dehydrogenase from Pseudomonas putida: the structural origin of the tightly bound cofactor in nicotinoprotein dehydrogenases. J. Mol. Biol. 2002;324:519–533. doi: 10.1016/s0022-2836(02)01066-5. [DOI] [PubMed] [Google Scholar]
- R15.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- R16.Tatusov R.L., Natale D.A., Garkavtsev I.V., et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Tumbula D.L., Teng Q., Bartlett M.G., Whitman W.B. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis . J. Bacteriol. 1997;179:6010–6013. doi: 10.1128/jb.179.19.6010-6013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Vorholt J.A., Marx C.J., Lidstrom M.E., Thauer R.K. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 2000;182:6645–6650. doi: 10.1128/jb.182.23.6645-6650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.White R.H. L-Aspartate semialdehyde and a 6-deoxy-5-ketohexose-1-phosphate are the precursors to the aromatic amino acids in Methanocaldococcus jannaschii . Biochemistry. 2004;43:7618–7627. doi: 10.1021/bi0495127. [DOI] [PubMed] [Google Scholar]
- R21.Yasueda H., Kawahara Y., Sugimoto S. Bacillus subtilusyckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 1999;181:7154–7160. doi: 10.1128/jb.181.23.7154-7160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Yu J.P., Ladapo J., Whitman W.B. Pathway of glycogen metabolism in Methanococcus maripaludis . J. Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]