Abstract
The genes encoding aromatic aminotransferase II (AroAT II) and aspartate aminotransferase (AspAT) from Pyrococcus furiosus have been identified, expressed in Escherichia coli and the recombinant proteins characterized. The AroAT II enzyme was specific for the transamination reaction of the aromatic amino acids, and uses α-ketoglutarate as the amino acceptor. Like the previously characterized AroAT I, AroAT II has highest efficiency for phenylalanine (kcat/Km = 923 s–1 mM–1). Northern blot analyses revealed that AroAT I was mainly expressed when tryptone was the primary carbon and energy source. Although the expression was significantly lower, a similar trend was observed for AroAT II. These observations suggest that both AroATs are involved in amino acid degradation. Although AspAT exhibited highest activity with aspartate and α-ketoglutarate (kcat ~105 s–1), it also showed significant activity with alanine, glutamate and the aromatic amino acids. With aspartate as the amino donor, AspAT catalyzed the amination of α-ketoglutarate, pyruvate and phenylpyruvate. No activity was detected with either branched-chain amino acids or α-keto acids. The AspAT gene (aspC) was expressed as a polycistronic message as part of the aro operon, with expression observed only when the aromatic amino acids were absent from the growth medium, indicating a role in the biosynthesis of the aromatic amino acids.
Keywords: phenylalanine, phenylpyruvate
Introduction
Aminotransferases are pyridoxal 5′-phosphate (PLP)-dependent enzymes that catalyze the reversible transfer of an amino group, usually the α-amino group of an amino acid, to an amino acceptor, usually an α-keto acid such as α-ketoglutarate. These enzymes have been divided into four subfamilies based on structural similarities (Mehta et al. 1993). Subfamily I comprises the aspartate, alanine, aromatic and histidinolphosphate aminotransferases. The best characterized enzyme of this subfamily is aspartate aminotransferase (AspAT), which catalyzes the conversion of the dicarboxylic α-amino acids, L-aspartate and L-glutamate, with the corresponding α-keto acids, α-ketoglutarate and oxaloacetate (Christen and Metzler 1985). The AspAT enzyme is a homodimer comprising two 45 kDa subunits, each subunit having one PLP covalently bound to a lysine residue through a Schiff base linkage. The crystal structure has been solved for AspATs from numerous sources and the active site residues identified (McPhalen et al. 1992, Okamoto et al. 1994, Nobe et al. 1998). Based on structural homologies, the AspATs are divided into two subgroups: one including AspATs from bacteria and eukaryotes, and the other including AspATs from bacteria and archaea (Jensen and Gu 1996).
Pyrococcus furiosus is a hyperthermophilic archaeon that grows optimally at 100 °C by the fermentation of various carbohydrates, including starch, glycogen, β-glucans, cellobiose and maltose, and by the breakdown of proteins and peptides (Fiala and Stetter 1986, Kengen et al. 1996). Although much is known about the fermentation of oligosaccharides such as cellobiose and maltose, there is a contrasting lack of insight concerning proteolytic fermentation. For example, the metabolism of the individual amino acids of P. furiosus has not been investigated in detail. It has been implied that the aromatic amino acids could serve as a source of ATP via substrate-level phosphorylation in P. furiosus (Mai and Adams 1994). It has been proposed that this occurs by the coordinated actions of an aromatic aminotransferase (AroAT), the indolepyruvate ferredoxin oxidoreductase (IOR) and an ADP-dependent acetyl-CoA synthetase, thereby generating the free acids and ATP (Mai and Adams 1996). Pyrococcus furiosus is capable of synthesizing all three aromatic amino acids, distinguishing it from Pyrococcus abyssi, which appears to be auxotrophic for Phe and Tyr (Watrin et al. 1995), and Pyrococcus horikoshii, which requires Phe, Tyr and Trp (Gonzalez et al. 1998). It is anticipated that, in P. furiosus, specific aminotransferases are involved in the catalysis of the last step in the biosynthesis of Phe and Tyr. Two distinct aromatic aminotransferases have been identified in P. furiosus, of which one, AroAT I, has been biochemically characterized in detail (Andreotti et al. 1995). Recently, Matsui et al. (2000) isolated and solved the crystal structure of a second aromatic aminotransferase, AroAT II, from P. horikoshii. Similar to AspATs and AroATs from mesophilic sources, the P. horikoshii AroAT II is a homodimer with one PLP bound per subunit.
We report on the identification and functional expression of the P. furiosus AspAT in Escherichia coli and its biochemical characterization. The AroAT II was also expressed and characterized in further detail. In addition, transcriptional analyses were performed in P. furiosus showing that the three aminotransferases, AroAT I, AroAT II and AspAT, are each regulated differently at the level of transcription. The combined biochemical and transcriptional results provide insight into the potential role of the distinct pyrococcal aminotransferases in both the catabolism and biosynthesis of aromatic amino acids.
Materials and methods
Growth of microorganisms Pyrococcus furiosus
(DSM 3638) was grown either in a sea salts medium (SSM) (Ward et al. 2000) or a chemically defined medium (CDM). The CDM medium was made as described by Kengen et al. (1993) except that yeast extract was replaced by the 20 amino acids at a final concentration of 0.25 mM. Cellobiose (10 mM), maltose (10 mM), pyruvate (40 mM) or tryptone (5 g l–1) were included as the primary carbon source. Escherichia coli strains BL21(lDE3) and XL-1 were grown at 37 °C in Luria-Bertani medium, and when required, kanamycin (50 µg ml–1) was included.
Cloning and expression of the genes encoding aromatic aminotransferase II and aspartate aminotransferase
The genes coding for AroAT II (PF1187250) and AspAT (PF1582969) were amplified from P. furiosus genomic DNA with the oligonucleotides BG550 (5′-CCATGGCACTTAGCGACAAGC) and BG551 (5′-CGCGGGATCCCTCAGATGAGCTTTTTCTCC) for AroAT II, and BG810 (5′-CGCGCCATGGCCTTCAACGTTTATGAGCTATTTAATAAG) and BG783 (5′-CGCGGGATCCTTATTGTGATTCGCATTCTGCTACCTCC) for AspAT. The N-terminus of the AroAT II was based on the N-terminal sequence of the Thermococcus litoralis AroAT II (Andreotti et al. 1994), the presence and proper spacing of a ribosomal binding site (RBS) and the annotation from the genome sequence. The N-terminus of the AspAT was identified based on the presence and proper spacing of the RBS, annotation from the genome sequence and its agreement with the AspAT identified from Thermococcus kodakaraensis (Accession Number BAA75925). The oligonucleotides BG550 and BG810 contain NcoI sites, whereas BG551 and BG783 contain BamHI sites. The PCR amplification was carried out with Pfu polymerase (Stratagene) and the resulting 1.2 kb (AroAT II) and 1.0 kb (AspAT) PCR products were cloned into pET-24d using the NcoI and BamHI sites. The recombinant plasmids, pLUW771 (AroAT II) and pLUW790 (AspAT), were transformed into Escherichia coli BL21(lDE3). For expression of the recombinant enzymes, E. coli BL21(lDE3), harboring the appropriate plasmid, was grown in 1 l of medium at 37 °C for 16–18 h, at which point the cells were harvested for purification of the recombinant enzyme.
Purification of the recombinant aminotransferases
The recombinant AroAT II (rAroAT II) was purified by a 2-step method. A 1.0-l overnight culture of E. coli BL21 (lDE3), harboring pLUW771, was harvested and resuspended in 10 ml of 20 mM Tris-Cl (pH 7.8), 0.1 mM pyridoxal 5¢-phosphate and 2 mM a-ketoglutarate. The cells were lyzed by sonication and the cellular debris removed by centrifugation. The supernatant was incubated at 80 °C for 15 min and the denatured E. coli proteins were removed by centrifugation (13,000 g for 30 min). The supernatant (12 ml) was applied to a Mono-Q column equilibrated with Tris buffer and eluted with a linear gradient of 0 to 1.0 M NaCl at a flow rate of 0.5 ml min–1. The rAroAT II eluted between 0.05 and 0.12 M NaCl. The recombinant protein was stored at 4 °C. The rAspAT was purified using a similar procedure and eluted from the MonoQ at NaCl concentrations between 0.35 and 0.42 M.
Enzyme assays
Aspartate aminotransferase activity was routinely assayed at 50 °C, because of the instability of oxaloacetate at elevated temperatures. The amount of aspartate converted to oxaloacetate was measured indirectly by the NADH-dependent conversion of oxaloacetate to malate catalyzed by malate dehydrogenase (MDH). The AspAT reaction mixture (1 ml) contained 20 mM Tris (pH 7.8), 50 µM pyridoxal 5′-phosphate, 20 mM a-ketoglutarate, 50 mM L-aspartate, 0.2 mM NADH and 3 U MDH from beef muscle. The reaction was started by the addition of the AspAT, and enzyme activity was monitored by the decrease in absorbance at 340 nm. One unit of AspAT or MDH is defined as the amount of enzyme that catalyzes the oxidation of 1 µmol NADH min–1. The activity with glutamate and oxaloacetate as substrates was determined indirectly by the NADH-dependent conversion of a-ketoglutarate to glutamate, catalyzed by glutamate dehydrogenase (GDH). The reaction mixture (1 ml) contained 20 mM Tris (pH 7.8), 50 µM pyridoxal 5′-phosphate, 20 mM oxaloacetate, 50 mM L-glutamate, 0.2 mM NADH, 50 mM NH4Cl and 15 U GDH.
Alanine aminotransferase activity was assayed at 80 °C as described previously by Ward et al. (2000). The amount of alanine converted to pyruvate was measured by the conversion of NADH in a separate lactate dehydrogenase (LDH) assay at 30 °C. The AlaAT reaction mixture (1 ml) contained 50 mM MOPS (pH 7.2), 100 mM KCl, 50 µM pyridoxal 5′-phosphate, 20 mM a-ketoglutarate and 50 mM L-alanine. The reaction mixture was pre-incubated at 80 °C for 5 min and the reaction started by the addition of enzyme. The reaction was stopped by snap freezing in an ice/ethanol bath. Samples were taken (5–100 µl) and added to the LDH assay, which contained 100 mM potassium phosphate (pH = 7.0), 0.2 mM NADH and 3 µl LDH from beef muscle. One unit of AlaAT or LDH is defined as the amount of enzyme that catalyzes the oxidation of 1 µmol NADH min–1.
Aromatic aminotransferase activity was determined as described by Andreotti et al. (1994). The assay involved the detection of an arsenate-catalyzed formation of aromatic α-ketoacid-enol-borate complexes. The assays were carried out in a volume of 800 µl, containing 50 mM MOPS (pH 7.2), 20 mM phenylalanine and 5 mM α-ketoglutarate. The reaction mixture was pre-incubated at 80 °C for 5 min and the reaction started by the addition of enzyme. The reaction was stopped by the addition of 20% (w/v) trichloroacetic acid, to a final concentration of 4% (w/v), rapidly cooled on ice and centrifuged. A 100-µl sample of the supernatant was diluted with 500 µl of 1 M boric acid (pH 6.5) and 1 M sodium arsenate and incubated for 25 min at room temperature. The amount of phenylpyruvate-enol-borate was measured at 300 nm and estimated based on an extinction coefficient of 7100 M–1 cm–1.
RNA isolation, northern blot analysis and primer extension
Isolation of total RNA from P. furiosus and northern blot analyses were carried out as described by Ward et al. (2000). The 5′ end of the aspC, tyrB and tyrC genes were mapped using the oligonucleotides 5′-TCTCCCCTTTTTAGAGAATTTATGG, 5′-TTCTGAGGCTTTCATCTTTAGAGC and 5′-AAAGTCGGGCTCTCCAATTCCC. The oligonucleotides were labeled with the fluorescent label IRD800 by the manufacturer (MWG-BIOTECH AG, Ebersberg, Germany). The primer extension reaction was carried out using the Reverse Transcription System (Promega) according to the manufacturer’s instructions with the following modifications: 2.0 pmol of oligonucleotide was hybridized with 20 µg of total RNA in 10 µl of the supplied buffer by heating at 70 °C for 10 min, before allowing to cool to room temperature. The reaction was started by the addition of dNTPs (1 mM), MgCl2 (5 mM), RNAsin (20 U) and AMV-RT (22.5 U) in a final volume of 20 µl. The reaction was incubated at 45 °C for 30 min, at which point the volume was adjusted to 50 µl with 10 mM Tris-HCl (pH 8.5), 1 µl of RNase A (5 mg ml–1) was added, and the mixture incubated at 37 °C for 10 min. The RNA were precipitated with ethanol and the pellets dissolved in 5 µl of loading buffer; 1 µl was subjected to electrophoresis on a sequencing gel in parallel with a sequencing reaction obtained with the same oligonucleotide. The following oligonucleotide pairs were used to generate probes for northern blot analyses: for the AroAT I, BG720 (5′-CAAAAGTTGACATAATGATTACA) and BG721 (5′-GTCAACACTCTGCTTTGCAATCTC), and for the AroAT II, BG722 (5¢-GATCCCAAGTCCAATGTTCGT GAG) and BG723 (5′-CAGCTTCTCTCGTCTCTTAGTGC). Oligonucleotides BG846 (5′-GGTAATTGTTGGGCCTGG AGCC) and BG847 (5′-CCCAGCCCTTTGAACGAAAGGA GG) were used for AspAT analysis.
Results
Comparison of the genome organization and primary structure of AspAT and AroATs in Pyrococcus
Based on the N-terminal sequence previously determined by Andreotti et al. (1995), the gene encoding the AroAT I (tyrB) was identified and contains the gene identifier PF0121. A BLAST search with the pyrococcal AlaAT (Ward et al. 2000) of the P. furiosus genome (http://www.genome.utah.edu) revealed two putative paralogs (PF1253 and PF1702). To study their function, the latter genes were cloned, overexpressed, and the corresponding enzymes were characterized (see below) and identified as AroAT II (tyrC) and AspAT (aspC).
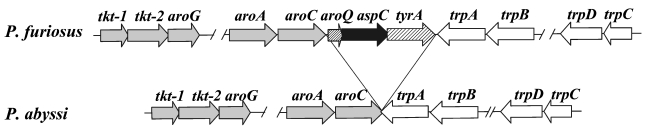
The aspC gene is located in the 13 kb aro gene cluster in P. furiosus, and the trp gene cluster is located immediately downstream and is transcribed in the opposite orientation (Figure 1). Directly upstream of aspC is a gene that encodes a putative monofunctional chorismate mutase (aroQ) and downstream is a gene that encodes a putative prephenate dehydrogenase (tyrA). The AroQ was 30% identical to the monofunctional chorismate mutase identified and characterized in Methanococcus jannaschii (MacBeath et al. 1998), whereas the TyrA exhibited significant identity (24–34%) to prephenate dehydrogenases from both mesophilic and thermophilic sources. Comparison to the P. abyssi genome reveals that the aroQ, aspC and tyrA genes are missing from the aro gene cluster. Moreover, homologs to aroQ, aspC or tyrA could not be identified on the chromosome of P. abyssi. The G + C content (38%) and codon usage of aroQ, aspC and tyrA are in good agreement with the rest of the P. furiosus genome, and suggest a deletion rather than an insertion. Analysis of the surrounding sequence revealed no elements that might be involved in a deletion or insertion event, such as inverted repeats or IS-elements. Pyrococcus horikoshii lacks both the aro and trp operons, including the aroQ, aspC and tyrA cluster. Similar to P. abyssi, an aspC homolog was not identified elsewhere on the chromosome. In an effort to better understand the distribution in the Thermococcales, southern blot analysis was carried out with aspC as a probe. The aspC was identified in single copy in both P. furious and P. woesei, but was absent from P. abyssi, P. horikoshii, T. profundus, T. peptonophilus, T. barophilus and T. stetteri (data not shown). However, an aspC homolog is present in T. kodakaraensis (Accession Number BAA75925). Neither tyrB nor tyrC appeared to be organized in an operon-like structure. Homologs of tyrB and tyrC were easily identified in both P. abyssi (PAB2227 and PAB0525) and P. horikoshii (PH0207 and PH1371).
Figure 1.
Molecular organization of the aro gene cluster and the AspAT (aspC) in P. furiosus and P. abyssi: aroQ, putative monofunctional chorismate mutase; aspC, aspartate aminotransferase; tyrA, putative prephenate dehydrogenase.
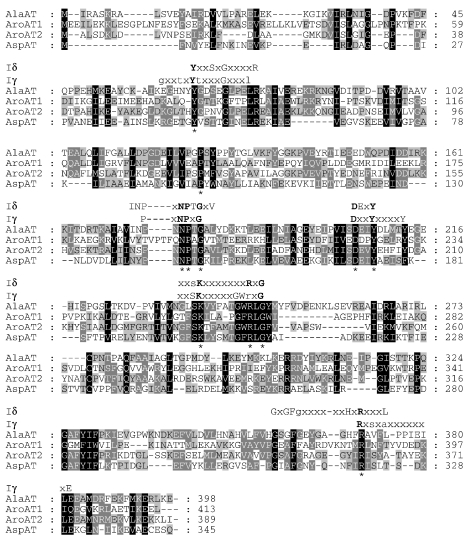
The structural tyrC gene encoding the AroAT II consists of 1203 bp and encodes a protein of 400 residues with a predicted molecular mass of 45.5 kDa. The structural aspC gene encoding the AspAT consists of 1041 bp and encodes a protein of 346 residues with a predicted molecular mass of 39 kDa. The AroAT I, AroAT II and AspAT all share similarities to the subfamily Ig and Id aminotransferases (Mehta et al. 1993, Jensen and Gu 1996), which is in contrast to the previously characterized pyrococcal AlaAT, which clearly belongs to the subfamily Id (Figure 2). BLAST and TFASTA analyses of the AspAT and AroATs revealed the highest degree of identity to the thermophilic AspATs of the subfamily Ig, and for this reason all three have been classified within the subfamily Ig. The 11 invariant residues involved in binding of the substrate or the cofactor pyridoxal 5′-phosphate were conserved in all three aminotransferases.
Figure 2.
Multiple sequence alignment of P. furiosus AlaAT, AroAT I, AroAT II and AspAT. The alignment was performed with Clustal W. Invariant residues found within the subclass I aminotransferases are designated with an asterisk. The motifs associated with subfamilies Id and Ig aminotransferases are also shown.
Purification and characterization of rAspAT and rAroAT II
The rAspAT was purified 19-fold with a yield of 65% and a specific activity of 158 U mg–1 with aspartate and α-ketoglutarate as substrates (Table 1). The rAspAT had a subunit molecular weight of 42 kDa and the molecular mass of the native enzyme was 80 kDa as determined by SDS-PAGE and gel filtration, respectively, suggesting that the active form of the enzyme is a homodimer similar to AspATs from both mesophilic and thermophilic sources. The kinetic parameters were determined at 50 °C and the results are summarized in Table 1. The substrate specificity of the rAspAT was determined with saturating amounts of the amino acids (20 mM) and α-ketoglutarate (10 mM) as the amino acceptor. The enzyme exhibited broad substrate specificity and was active with aspartate, alanine and all three aromatic amino acids (Table 2). As with all AspATs, the enzyme was able to use glutamate and oxaloacetate as substrates, and had a specific activity of 46 U mg–1. Using aspartate as the amino donor, the enzyme was also capable of utilizing α-ketoglutarate, pyruvate and phenylpyruvate as amino acceptors (Table 2). No activity was observed with the branched-chain α-keto acids α-ketoisocaproate or α-keto-β-methylvalerate.
Table 1.
Kinetic parameters of Pyrococcus furiosus rAroAT II and rAspA T.1
| Enzyme | Substrate | Apparent Km (mM) | Specific activity (µmol min–1 mg–1) | kcat (s–1) | kcat/Km (s–1m M–1) |
| rAroAT II | Phenylalanine | 1.3 | 1600 | 1200 | 923 |
| Tyrosine | 2.6 | 1560 | 1130 | 434 | |
| Tryptophan | 3.4 | 1960 | 1420 | 418 | |
| α-Ketoglutarate | 0.9 | ||||
| rAspAT | Aspartate | 4.2 | 158 | 105 | 25 |
| α-Ketoglutarate | 0.46 | ||||
1 For the amino acids, 5 mM α-ketoglutarate was used as the amino acceptor. For α-ketoglutarate, 20 mM phenylalanine or aspartate was used as the amino donor. The AspAt was assayed at 50 °C and the AroAT II was assayed at 80 °C.
Table 2.
Substrate specificity of rAspAT from P. furiosus. The amino acids (20 mM) were assayed with 5 mM α-ketoglutarate as the amino acceptor, and the α-ketoacids (5 mM) were assayed with 20 mM aspartate as the amino donor.
| Substrate | Relative activity (%) |
| Aspartate | 100 |
| Glutamate | 30 |
| Alanine | 1.2 |
| Phenylalanine | 1.0 |
| Tyrosine | 0.6 |
| Tryptophan | 1.0 |
| α-Ketoglutarate | 100 |
| Pyruvate | 8 |
| Phenylpyruvate | 3 |
| α-Ketoisovalerate | < 0.01 |
| α-Keto-α-methylvalerate | < 0.01 |
The rAroAT II was purified 27-fold with a yield of 74% and a specific activity of 1600 U mg–1 with phenylalanine and α-ketoglutarate as substrates (Table 1). The purified rAroAT II migrated as a single band in an SDS-PAGE with an apparent molecular mass of 46 kDa. The molecular mass of the native enzyme was 95.4 kDa as determined by gel filtration, suggesting that the active form of the enzyme exists as a dimer of identical subunits. This resembles the AroATs from E. coli (Hayashi et al. 1993), P. denitrificans (Oue et al. 1997) and T. litoralis (Andreotti et al. 1994), and the AroAT I from P. furiosus (Andreotti et al. 1995), which exist as homodimers. The kinetic parameters of the AroAT II were determined at 80 °C and the results are summarized in Table 1. There was no indication of substrate inhibition as was found with the AroAT from Klebsiella aerogenes (Paris and Magasanik 1981). Similar to the AroATs from M. aeolicus (Xing and Whitman 1992) and T. litoralis (Andreotti et al. 1994) and the AroAT I from P. furiosus (Andreotti et al. 1995), AroAT II was active with all three aromatic amino acids and displayed low activity with the amino acids alanine and aspartate (Table 3). Furthermore, the AroAT II from P. furiosus was almost identical to that from T. litoralis.
Table 3.
Substrate specificity of AroATs from Pyrococcus furiosus and Thermococcus litoralis, expressed as relative maximal activity (%). Abbreviation: nd = not determined.
| Substrate | AroAT I1 | AroAT II | AroAT I2 | AroAT II2 |
| Alanine | nd | 1.0 | 0.8 | 0.2 |
| Aspartate | nd | 0.1 | < 0.1 | 0.1 |
| Phenylalanine | 100 | 84 | 100 | 82 |
| Tyrosine | 28 | 79 | 44 | 56 |
| Tryptophan | 24 | 100 | 29 | 100 |
1 Values for the P. furiosus AroAT I are from Andreotti et al. (1995). 2 Values for the T. litoralis AroATs are from Andreotti et al. (1994).
Regulation of AroAT I, AroAT II and AspAT
To elucidate the regulation of the individual aminotransferases, P. furiosus was grown with cellobiose (10 mM), pyruvate (40 mM) or tryptone (5 g l–1) as the primary carbon and energy source. Because AspAT is believed to play a role in the biosynthesis of aromatic amino acids, we used a chemically defined medium containing all the amino acids, except for the aromatic amino acids, and 10 mM cellobiose as the primary carbon source. The three aromatic amino acids (0.25 mM final) were added (C+) or left out (C–) of the culture. Early exponential-phase cells were used for assay of enzyme activities in crude extracts and for isolation of total RNA for transcriptional analyses. Internal fragments of the tyrB, tyrC and aspC genes were generated by PCR, labeled and used as probes for northern blot analyses. Both tyrB and tyrC were expressed as a single, monocistronic 1.7-kb mRNA species consisting of only thetyrB or tyrC genes (Figure 3A). Expression of the tyrB gene was 5-fold higher when P. furiosus was grown with tryptone as the primary carbon source compared with cellobiose as the carbon source. There was also a 2-fold increase in tyrB expression in pyruvate-grown cells compared with cellobiose-grown cells. Although the overall expression was lower, a similar trend was observed in the case of tyrC (Figure 3A). Similar results were observed for the catalytic activity of the AroATs in cell extracts with 3-fold higher total AroAT activity in tryptone-grown cells and 2-fold higher in pyruvate-grown cells compared with cellobiose-grown cells (Table 4). The presence or absence of the aromatic amino acids had no effect on the transcription of either tyrB or tyrC. This may indicate a global regulation of amino acid catabolism (like the leucine response in E. coli) rather than a specific induction of aromatic amino acids.
Figure 3.
(A) Northern blot analysis of the P. furiosus tyrB (AroAT I) and tyrC (AroAT II) transcripts. Pyrococcus furiosus was grown with the following compounds as the primary carbon and energy source: 10 mM cellobiose (C), 40 mM pyruvate (P), or 5 g l–1 tryptone (T).Pyrococcus furiosus was also grown in a chemically-defined medium with cellobiose as carbon source in the presence (C+) and absence (C–) of the aromatic amino acids. (B) Identification of the transcriptional start site for the tyrB and tyrC by primer extension analysis; arrows indicate major and minor primer extension product. (C) Sequence of the tyrB and tyrC promoter regions and alignment to the tyrB and tyrC promoter regions from P. abyssi and P. horikoshii. The transcriptional start site as determined by primer extension analysis is designated by an arrow. The TATA and BRE sequences are boxed. The putative ribosome binding site ( S/D) is shown and the final ATG of each sequence is the first codon of each gene.
Table 4.
Activities of AroATs and AspAT in crude extracts of P. furiosus grown on various carbon sources. Either 20 mM aspartate or phenylalanine, and 5 mM α-ketoglutarate were used as substrates.
| Carbon source | AspAT (U mg–1) | AroAT (U mg–1) |
| 10 mM Cellobiose | 0.5 | 2.9 |
| 40 mM Pyruvate | 0.6 | 5.7 |
| 5 g l–1Tryptone | 0.8 | 8.7 |
A single major, apparent mRNA 5′ end was identified upstream of tyrB by primer extension (Figure 3B). A minor 5′ end was also observed five base pairs downstream of the major start site. This minor start site was observed under all tested conditions, indicating that it was not caused by degradation. However, in all conditions tested it was also the minor promoter. Analysis of the upstream sequence identified features common to archaeal promoters. These include the TATA-box located 25 bp upstream of the transcription initiation site, and the TFB-responsive element or BRE element (Bell et al. 1999). Comparison of the tyrB promoter regions between P. furiosus, P. abyssi and P. horikoshii revealed 60% identity over the entire 120 bp and 100% identity at the TATA and BRE sites (Figure 3C). Furthermore, a 14-bp inverted repeat was identified upstream of the TATA and BRE sites that is also highly conserved in all three promoters. This high degree of conservation among the tyrB promoters suggests a similar mode of regulation in P. abyssi and P. horikoshii.
The AroAT II (tyrC) appears to be expressed constitutively at a low level regardless of the carbon source. The highest total AroAT activity observed in cell extracts was also found in tryptone. Because of the overlapping activities of AroAT I and AroAT II, it was impossible to determine if one enzyme was responsible for the increase in activity, or if both enzymes were involved. Based on northern blot analysis, we conclude that AroAT I is responsible for the observed increase in activity. For tyrC, two transcription start sites were identified by primer extension (Figure 3B). Both the TATA and BRE sequences were identified upstream of Promoter 1 (P1) and Promoter 2 (P2). The relative levels of expression from both promoters were identical regardless of the carbon source (data not shown). Comparison of the promoter regions of tyrC in P. furiosus, P. abyssi and P. horikoshii revealed some conservation around the BRE and TATA sequences of P2, whereas little identity was found in the elements around P1.
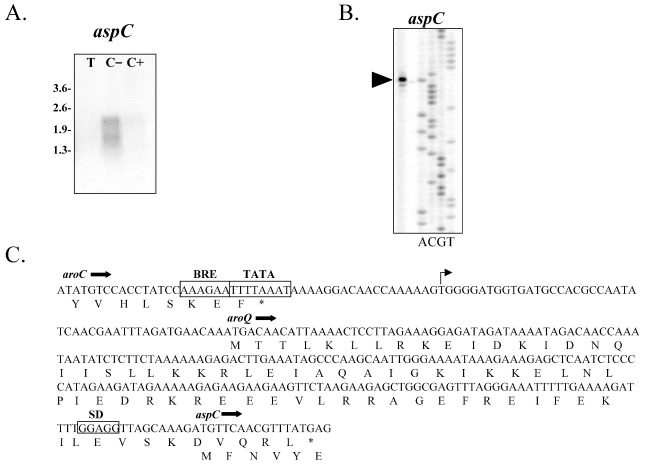
Expression of the aspC gene was detected only when the cells were grown in the absence of the aromatic amino acids. Two transcripts of about 2.5 and 1.8 kb were identified using an internal fragment of aspC as a probe (Figure 4A). Assuming transcription initiation just upstream of aroQ (see below), the transcript site suggests co-transcription of the aroQ– aspC–tyrA gene cluster, which is about 1.7 kb in length. The larger transcript is most likely the result of read-through. The addition of tryptone or the aromatic amino acids abolished expression of the aspC transcripts. A low basal level of AspAT activity was found in all extracts regardless of the carbon source (Table 4). This activity could be due to overlapping activities found in AroAT II and AlaAT. The transcriptional start site of the aspC transcript was mapped at a thymine base 46 bp upstream of the aroQ translational initiation site. Potential TATA and BRE sequences were identified in the promoter region. The leader sequence was longer than usually observed in P. furiosus, because the transcriptional start site is often found within a few base pairs of the ribosome binding site. A ribosome binding site located upstream of aroQ is absent in P. furiosus. The predicted translational start site is, however, in good agreement with the smaller monofunctional chorismate mutases, like that of M. jannaschii, and is the only possible start codon. The promoter region and the aroQ gene were completely sequenced in the primer extension experiments and the sequences are in excellent agreement with the genome sequencing project, thereby ruling out the possibility of a frame shift.
Figure 4.
(A) Northern analysis of the P. furiosus AspAT (aspC). Pyrococcus furiosus was grown in a chemically-defined medium with cellobiose as the carbon source in the presence (C+) or absence (C–) of the aromatic amino acids, or with tryptone (T). (B) Identification of the transcriptional start site for the aspC by primer extension analysis, indicated by the arrow. (C) The transcriptional start site as determined by primer extension analysis is designated by an arrow. The TATA and BRE sequences are boxed.
Discussion
The AspAT from P. furiosus is distinct from the other archaeal and thermophilic AspATs because of its relatively broad substrate specificity with both amino and α-keto acids, and its metabolic role. The enzymes from Methanococcus aeolicus, Thermus thermophilus and the halophilic archaeon Haloferax mediterranei are all specific for aspartate and appear to be involved in the catabolism of aspartate (Marino et al. 1988, Muriana et al. 1991, Xing and Whitman 1992, Okamoto et al. 1996). The AspAT from S. solfataricus is similar to the P. furiosus enzyme in that it shares similar substrate specificity regarding the amino acids and is capable of using pyruvate as an amino acceptor, but appears to be involved in the catabolism of aspartate (Marino et al. 1988). Except that the AspAT activity in crude extracts from S. solfataricus and M. aeolicus was identical regardless of the carbon or nitrogen source, little is known about the regulation of these enzymes.
The biosynthesis of the aromatic amino acids in P. furiosus appears to occur by way of the classical shikimate pathway based on the genes present on the chromosome. The argument that AspAT is involved solely in the biosynthesis of the aromatic amino acids is supported by several observations. The gene encoding AspAT is located between the aro and trp operons, which collectively encode the genes necessary for the biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan. In addition, aspC is expressed when P. furiosus is grown in the absence of the aromatic amino acids. In contrast, expression of aspC is not observed in the presence of proteinaceous substrates such as yeast extract and tryptone, thereby ruling out a catabolic role during proteolytic fermentation. Recently a similar mode of regulation has been shown with the trp operon in Thermococcus kodakaraensis KOD1 in which expression was tryptophan-dependent and was induced when the cells were grown in the absence of the amino acid (Tang et al. 1999). The broad substrate specificity of the pyrococcal enzyme is also ideally suited for amino acid biosynthesis because it is capable of utilizing a number of amino acids as amino donors for the biosynthesis of aromatic amino acids. This broad specificity of AspAT hasalso been observed in one other organism, Trypanosoma cruzi (Montemartini et al. 1993). It has been proposed that these broad-specificity enzymes may be the closest relative to the last common ancestor for the aminotransferase superfamily, also referred to as a contemporary “dinosaur” (Jensen and Gu 1996).
In T. litoralis and M. aeolicus there are two AroATs, and a similar situation is observed in P. furiosus (Xing and Whitman 1992, Andreotti et al. 1994). In all three organisms, both of the AroATs are specific for the aromatic amino acids; in addition, the AroAT I and AroAT II enzymes of the three organisms have similar kinetic features. A significant difference between these AroATs is found in the regulation of their transcription. The AroAT I was expressed under all conditions tested and the highest levels of expression were found when cells were grown with tryptone as the primary carbon and energy source. It is likely that the AroAT I is the dominant aminotransferase involved in the catabolism of the aromatic amino acids. However, an alternative metabolic role cannot be excluded because significant amounts of the tyrB transcript were present in all growth conditions tested, and it is not uncommon to see aminotransferases with overlapping activities and metabolic functions (Whitaker et al. 1982). Likewise, AroAT II has a low basal level of expression, which appears to be slightly induced in the presence of tryptone. Another alternative is that the AroAT II is active, and possibly induced by a yet to be identified substrate. It has been shown that the P. horikoshii AroAT II homolog was also active with aliphatic substrates, with maximal activity with the 8-carbon substrate 2-amino octanoic acid (Matsui et al. 2000). It is possible that the true substrate for the AroAT II has not yet been identified.
There are now four distinct aminotransferases that have been identified and characterized in P. furiosus, all of which belong to Subfamily I: AlaAT, AroAT I, AroAT II and AspAT. Aminotransferases play crucial roles in the fermentative metabolism of P. furiosus and are involved in energy production and biosynthesis (catabolism and anabolism; this work), as well as in the maintenance of the redox balance in the cell (Ward et al. 2000). Each aminotransferase appears to play distinct metabolic roles and is regulated differently. Details on the regulatory mechanisms of the modulation of aminotransferase expression as reported in this study, however, remain to be elucidated.
Acknowledgments
The research was partly supported by the EU BIOTECH (Contract BIO2-CT93-0274).
References
- R1.Andreotti G., Cubellis M.V., Nitti G., Sannia G., Mai X., Marino G., Adams M.W.W. Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis . Eur. J. Biochem. 1994;220:543–549. doi: 10.1111/j.1432-1033.1994.tb18654.x. [DOI] [PubMed] [Google Scholar]
- R2.Andreotti G., Cubellis M.V., Nitti G., Sannia G., Mai X., Adams M.W.W., Marino G. An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus . Biochim. Biophys. Acta. 1995;1247:90–96. doi: 10.1016/0167-4838(94)00211-x. [DOI] [PubMed] [Google Scholar]
- R3.Bell S.D., Kosa P.L., Sigler P.B., Jackson S.P. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Christen P., Metzler D. New York: John Wiley; 1985. Transaminases. [Google Scholar]
- R5.Fiala G., Stetter K.O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch. Microbiol. 1986;161:168–175. [Google Scholar]
- R6.Gonzalez J.M., Masuchi Y., Robb F.T., Ammerman J.W., Maeder D.L., Yanagibayashi M., Tamaoka J., Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- R7.Hayashi H., Inoue K., Nagata T., Kuramitsu S., Kagamiyama H. Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry. 1993;32:12229–12239. doi: 10.1021/bi00096a036. [DOI] [PubMed] [Google Scholar]
- R8.Jensen R.A., Gu W. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 1996;178:2161–2171. doi: 10.1128/jb.178.8.2161-2171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Kengen S.W., Luesink E.J., Stams A.J.M., Zehnder A.J. Purification and characterization of an extremely thermostable α-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus . Eur. J. Biochem. 1993;213:305–312. doi: 10.1111/j.1432-1033.1993.tb17763.x. [DOI] [PubMed] [Google Scholar]
- R10.Kengen S.W.M., Stams A.J.M., De Vos W.M. Sugar metabolism of hyperthermophiles. FEMS Microbiol. Rev. 1996;18:119–137. [Google Scholar]
- R11.MacBeath G., Kast P., Hilvert D. A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii . Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- R12.Mai X., Adams M.W. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 1994;269:16726–16732. [PubMed] [Google Scholar]
- R13.Mai X., Adams M.W. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus . J. Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Marino G., Nitti G., Arnone M.I., Sannia G., Gambacorta A., De Rosa M. Purification and characterization of aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus . J. Biol. Chem. 1988;263:12305–12309. [PubMed] [Google Scholar]
- R15.Matsui I., Matsui E., Sakai Y., Kikuchi H., Kawarabayasi Y., Ura H., Kawaguchi S., Kuramitsu S., Harata K. The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii . J. Biol. Chem. 2000;275:4871–4879. doi: 10.1074/jbc.275.7.4871. [DOI] [PubMed] [Google Scholar]
- R16.McPhalen C.A., Vincent M.G., Jansonius J.N. X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase. J. Mol. Biol. 1992;225:495–517. doi: 10.1016/0022-2836(92)90935-d. [DOI] [PubMed] [Google Scholar]
- R17.Mehta P.K., Hale T.I., Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- R18.Montemartini M., Santome J.A., Cazzulo J.J., Nowicki C. Purification and partial structural and kinetic characterization of tyrosine aminotransferase from epimastigotes of Trypansoma cruzi . Biochem. J. 1993;292:901–906. doi: 10.1042/bj2920901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Muriana F.J., Alvarez Ossorio M.C., Relimpio A.M. Purification and characterization of aspartate aminotransferase from the halophile archaebacterium Haloferax mediterranei . Biochem J. 1991;278:149–154. doi: 10.1042/bj2780149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Nobe Y., Kawaguchi S., Ura H., Nakai T., Hirotsu K., Kato R., Kuramitsu S. The novel substrate recognition mechanism utilized by aspartate aminotransferase of the extreme thermophile Thermus thermophilus HB8. J. Biol. Chem. 1998;273:29554–29564. doi: 10.1074/jbc.273.45.29554. [DOI] [PubMed] [Google Scholar]
- R21.Okamoto A., Higuchi T., Hirotsu K., Kuramitsu S., Kagamiyama H. X-ray crystallographic study of pyridoxal 5′-phosphate-type aspartate aminotransferases from Escherichia coli in open and closed form. J. Biochem. (Tokyo) 1994;116:95–107. doi: 10.1093/oxfordjournals.jbchem.a124509. [DOI] [PubMed] [Google Scholar]
- R22.Okamoto A., Kato R., Masui R., Yamagishi A., Oshima T., Kuramitsu S. An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. (Tokyo) 1996;119:135–144. doi: 10.1093/oxfordjournals.jbchem.a021198. [DOI] [PubMed] [Google Scholar]
- R23.Oue S., Okamoto A., Nakai Y., Nakahira M., Shibatani T., Hayashi H., Kagamiyama H. Paracoccus denitrificans aromatic amino acid aminotransferase: a model enzyme for the study of dual substrate recognition mechanism. J. Biochem. (Tokyo) 1997;121:161–171. doi: 10.1093/oxfordjournals.jbchem.a021561. [DOI] [PubMed] [Google Scholar]
- R24.Paris C.G., Magasanik B. Purification and properties of aromatic amino acid aminotransferase from Klebsiella aerogenes . J. Bacteriol. 1981;145:266–271. doi: 10.1128/jb.145.1.266-271.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Tang X., Ezaki S., Fujiwara S., Takagi M., Atomi H., Imanaka T. The tryptophan biosynthesis gene cluster trpCDEGFBA from Pyrococcus kodakaraensis KOD1 is regulated at the transcriptional level and expressed as a single mRNA. Mol. Gen. Genet. 1999;262:815–821. doi: 10.1007/s004380051145. [DOI] [PubMed] [Google Scholar]
- R26.Ward D.E., Kengen S., Van Der Oost J., De Vos W.M. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 2000;182:2559–2566. doi: 10.1128/jb.182.9.2559-2566.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Watrin L., Martin Jezequel V., Prieur D. Minimal amino acid requirements of the hyperthermophilic archaeon Pyrococcus abyssi, isolated from deep-sea hydrothermal vents. Appl. Environ. Microbiol. 1995;61:1138–1140. doi: 10.1128/aem.61.3.1138-1140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Whitaker R.J., Gaines C.G., Jensen R.A. A multispecific quintet of aromatic aminotransferases that overlap different biochemical pathways in Pseudomonas aeruginosa . J. Biol. Chem. 1982;257:13550–13556. [PubMed] [Google Scholar]
- R29.Xing R.Y., Whitman W.B. Characterization of amino acid aminotransferases of Methanococcus aeolicus . J. Bacteriol. 1992;174:541–548. doi: 10.1128/jb.174.2.541-548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]