Abstract
Complete archaeal genomes were probed for the presence of long (≥ 25 bp) oligonucleotide repeats (words). We detected the presence of many words distributed in tandem with narrow ranges of periodicity (i.e., spacer length between repeats). Similar words were not identified in genomes of non-archaeal species, namely Escherichia coli, Bacillus subtilis, Haemophilus influenzae, Mycoplasma genitalium and Mycoplasma pneumoniae. BLAST similarity searches against the GenBank nucleotide sequence database revealed that these words were archaeal species-specific, indicating that they are of a signature character. Sequence analysis and genome viewing tools showed these repeats to be restricted to non-coding regions. Thus, archaea appear to possess a non-coding genomic signature that is absent in bacterial species. The identification of a species-specific genomic signature would be of great value to archaeal genome mapping, evolutionary studies and analyses of genome complexity.
Keywords: Archaea, bioinformatics, comparative genomics, genome signature, oligonucleotide frequencies
Introduction
Long (≥ 25 bp) oligonucleotide repeats (words) have been identified in prokaryotic genomes; however, investigations into the distribution patterns of these repeats have only recently been possible with the increasing availability of complete prokaryotic genomes. This type of analysis is important because repeat regions may function as part of regulatory elements within the genome (Pesole et al. 1992, Van Helden et al. 1998). The frequency and periodicity of short repeat elements (up to 10 bp) have previously been studied (Karlin and Burge 1995, 1996, Cole et al. 2001). Similar characterization of oligonucleotide words will likely clarify the functional significance of genomic sequence repeats (Heringa 1998).
Detection of repeats requires the implementation of specific statistical methods to evaluate the significance of repeat frequencies and periodic distributions. It is known that the sensitivity of repeat detection is positively correlated with sequence length. Several statistical techniques, based on the Markov model of sequence pattern prediction, have been developed to detect repeat sequence motifs as small as six to ten nucleotides in length (Pesole et al. 1992). However, use of Markov chain models for the prediction of long repeat sequences has drawbacks. Although the assumption that (n–1)-mers (n represents the size of the repeat) and (n–2)-mers are randomly distributed is valid for short-length repeats, it is not always true for high- order repetitive sequences. For example, if n = 30, it would have to be assumed that 29- and 28-mers were randomly distributed throughout the genome, which is unlikely. Investigations of the nature of the distribution of smaller derivatives of high-order repeats within complete genome sequences requires significant computational resources.
Repeats with highly significant frequencies and periodic distributions may have an important structural role, affecting the overall biological characteristics of the sequence. Furthermore, nonrandom nucleotide sequence patterns have a higher probability of being biologically active. Statistical search tools have been developed based on this model of repeat sequence frequency (Cox and Mirkin 1997).
Prokaryotic genomes tend to be optimized toward compactness, suggesting that the presence of long oligonucleotide repeats would be evolutionarily unfavorable. Nevertheless, repeat sequences have been identified in genomes of bacteria and organelles at a relatively high frequency, although analysis of the genomic distribution of all abundant repeats has indicated that they are virtually excluded from coding sequences. Therefore, these repeats might participate in a variety of events relevant to prokaryotic genome plasticity, namely amplification, deletion, inversion, translocation or transposition (Romero et al. 1999). Most investigations have focused on short repeats (up to 10 bp), which are present in genomes at high frequencies, and many tools have been developed to provide a graphical representation of word frequency within the analyzed sequences (Levy et al. 1998, Deschavanne et al. 1999). In this study, we investigated the presence of periodically distributed oligonucleotide repeats ~30 bp long in complete genomes of archaeal and bacterial species. Such repeat sequences may play a functionally significant role in the maintenance of DNA structure.
Materials and methods
Sequence collection
The complete genomes of seven archaeal species (Aeropyrum pernix (NC_000854), Archaeoglobus fulgidus (NC_000917), Methanococcus jannaschii (NC_000909), Methanothermobacter thermoautotrophicus (NC_000916), Pyrococcus abyssi (NC_000868), Pyrococcus horikoshii (NC_000961) and Thermoplasma volcanium (NC_002689, Kawashima et al. 1999, 2000)) and six bacterial species (Escherichia coli K-12 (NC_000913), Bacillus subtilis (NC_000964), Haemophilus influenzae (NC_000907), Mycoplasma genitalium (NC_ 000908), Synechocystis trididemni PCC6803 (NC_000911) and Mycoplasma pneumoniae (NC_000912)) (Table 1) were downloaded from GenBank (www.ncbi.nlm.nih.gov/Entrez/, February 2001 release) or The Institute of Genome Research (www.tigr.org). Taxonomic positions were determined for each species using the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html).
Table 1.
Archaeal and bacterial species analyzed. The six bacterial species listed below served as negative (non-archaeal) controls for the study.
| Domain | Kingdom | Phylum | Order | Family | Species |
| Archaea | Crenarchaeota | Desulfurococcaceae | Aeropyrum pernix | ||
| Euryarchaeota | Archaeoglobaceae | Archaeoglobus fulgidus | |||
| Methanococcaceae | Methanococcus jannaschii | ||||
| Methanobacteriaceae | Methanothermobacter thermoautotrophicus | ||||
| Thermoplasmataceae | Thermoplasma volcanium | ||||
| Thermococcaceae | Pyrococcus abyssi | ||||
| Pyrococcus horikoshii | |||||
| Bacteria | Proteobacteria | Enterobacteriaceae | Escherichia coli | ||
| Pasteurellaceae | Haemophilus influenzae | ||||
| Cyanobacteria | Chroococcales | Synechocystis trididemni | |||
| Firmicutes | Bacillaceae | Bacillus subtilis | |||
| Mycoplasmataceae | Mycoplasma genitalium | ||||
| Mycoplasma pneumoniae | |||||
Data analysis
Long sequence repeats (words ≥ 25 bp) were analyzed with two computer programs developed in-house: GenCount and OligoCount (available from the authors on request). GenCount is a C-based bioinformatics tool that identifies repeat sequences of a user-defined unit length and determines their periodic distribution. OligoCount is a Perl-based program that counts n-mer oligonucleotides in a sequence, generates the expected occurrences based on an n–2 Markov chain, calculates percent composition, chi-squared and z-scores, and tracks the positions of the oligonucleotides. OligoCount calculates the expected number of occurrences of a given oligonucleotide, assuming a weighted random distribution. A chi-squared value is calculated to facilitate comparison of the observed and expected occurrences. The program outputs information for each oligonucleotide that has a chi-squared value greater than the significance threshold, and z-scores are calculated based on a formula from Rocha et al. (1998). Only repeats with statistically significant frequencies were evaluated further during this analysis.
Results
We identified high-order oligonucleotide repeats of 30 bp in completely sequenced archaeal genomes (Table 1). Many of these repeats were statistically significant with respect to repeat number, and were periodically distributed; i.e., they occurred with a statistically significant copy number, in tandem on the sense strand, separated by spacers of more or less fixed length (Figures 1 and 2). Furthermore, such repetitive elements were not identified in the non-archaeal control genomes listed in Table 1, except in S. trididemni, which contained a 30 bp repeat with a low copy number that was not statistically significant.
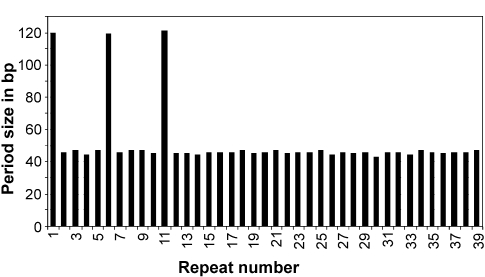
Figure 1.
Periodic distribution of the GTAAGAAAGGGAGGCTCC TGAAAATGGAGA repeat in A. fulgidus (giô2689296 section 134 of 172 of the complete genome; only specific repeats on the plus strand were considered). This repeat was found to be specific to A. fulgidus by BLAST searches against the entire GenBank nucleotide database. The x-axis represents the number of unit length repeats, i.e., each consecutive occurrence of the repeat sequence was assigned a number from 1 through n (repeat number) where n is the total copy number of the repeat within the genome. The y-axis represents the periodicity, i.e., the spacer length between consecutive repeats.
Figure 2.
Distribution of the ATTTCAATCCCATTTTGGTCTGAT TTTAAC (30 bp) repeat within the complete genome sequence of M. thermoautotrophicus. The x-axis represents the number of unit length repeats, i.e., each consecutive occurrence of the repeat sequence was assigned a number from 1 through n (repeat number) where n is the total copy number of the repeat within the genome. The y-axis represents the period distance or periodicity, i.e., the spacer length between consecutive repeats.
In A. fulgidus, the repeat sequence CTTTCAATCCCATTTTGGTCTGATTTTAAC was found in two locations within the genome. The repeat was present from 976801 to 992232 and from 1471880 to 1482686, with a narrow range of periodicity in each case. In addition, a reverse complement of this sequence, GTTGAAATCAGACCAAAATGGGATTGAAAG, was distributed 60 times in the A. fulgidus genome (Table 2) with a periodicity of 39 ± 3 bp (with a few exceptions). Parallel analysis of the other archaeal genomes revealed similar periodicity except in A. pernix, which possessed no high-order repeat sequences (Table 2).
Table 2.
The most common 30 bp repeats with a narrow range of periodicity in different archaeal genomes. Repeats that recur more than 20 times in tandem are considered high-order repeats. The proportion of the sense strand of the genome represented by these repeats (percent of genome), the absolute number (copy number) and the mean ± SD spacer length between consecutive repeats, excluding outliers (periodicity; bp), are given. Where repeat sequences occur in two locations within the genome, mean periodicity of both locations are given. Underlined fragments depict common repetitive elements within each organism.
| Organism | Repeat sequence | Percent of genome | Copy number | Periodicity |
| Archaeoglobus fulgidus | GTAAGAAAGGGAGGCTCCTGAAAATGGAGA | 0.0018 | 41 | 46 ± 2 |
| TAAGAAAGGGAGGCTCCTGAAAATGGAGAT | 0.0019 | 42 | 46 ± 2 | |
| AAGAAAGGGAGGCTCCTGAAAATGGAGATT | 0.0019 | 42 | 46 ± 2 | |
| AGAAAGGGAGGCTCCTGAAAATGGAGATTG | 0.0019 | 42 | 46 ± 2 | |
| GAAAGGGAGGCTCCTGAAAATGGAGATTGA | 0.0019 | 42 | 46 ± 2 | |
| AAAGGGAGGCTCCTGAAAATGGAGATTGAA | 0.0019 | 42 | 46 ± 2 | |
| AAGGGAGGCTCCTGAAAATGGAGATTGAAA | 0.0019 | 42 | 46 ± 2 | |
| GGGAGGCTCCTGAAAATGGAGATTGAAAG | 0.0019 | 42 | 47 ± 3 | |
| AGTTGAAATCAGACCAAAATGGGATTGAAA | 0.0011 | 23 | 39 ± 2 | |
| GTTGAAATCAGACCAAAATGGGATTGAAAG | 0.0028 | 60 | 39 ± 3 | |
| CTTTCAATCCCATTTTGGTCTGATTTCAAC | 0.0022 | 47 | 39 ± 3 | |
| Aeropyrum pernix | GTCCCGGGTTCAAATCCCGGCGGGCCCGCC | 0.0004 | 7 | NA |
| TCCCGGGTTCAAATCCCGGCGGGCCCGCCA | 0.0004 | 7 | NA | |
| Methanococcus jannaschii | AATTAAAATCAGACCGTTTCGGAATGGAAA | 0.0017 | 29 | 40 ± 3 |
| ATTAAAATCAGACCGTTTCGGAATGGAAAT | 0.0027 | 45 | 39 ± 3 | |
| GTTAAAATCAGACCTCTTGGAGGATGGAAA | 0.0020 | 33 | 42 ± 3 | |
| Methanothermobacter | ATTTCAATCCCATTTTGGTCTGATTTTAAC | 0.0071 | 124 | 37 ± 3 |
| thermoautotrophicus | GTTAAAATCAGACCAAAATGGGATTGAAAT | 0.0024 | 60 | 37 ± 3 |
| AATTTCAATCCCATTTTGGTCTGATTTTAA | 0.0022 | 39 | 38/105 | |
| TATTTCAATCCCATTTTGGTCTGATTTTAA | 0.0021 | 37 | 38/103 | |
| TTTCAATCCCATTTTGGTCTGATTTTAACT | 0.0021 | 36 | 36/104 | |
| TTTCAATCCCATTTTGGTCTGATTTTAACC | 0.0020 | 35 | 36/104 | |
| TTTCAATCCCATTTTGGTCTGATTTTAACA | 0.0018 | 31 | 36/105 | |
| GATTTCAATCCCATTTTGGTCTGATTTTAA | 0.0015 | 27 | 36/105 | |
| Pyrococcus abyssi | CTTTCAATTCTATTTTAGTCTTATTGGAAC | 0.0012 | 21 | 38 ± 3 |
| GTTCCAATAAGACTAAAATAGAATTGAAAG | 0.0015 | 26 | 38 ± 3 | |
| TGTTCCAATAAGACTAAAATAGAATTGAAA | 0.0007 | 12 | 39 ± 3 | |
| Pyrococcus horikoshii | TCTTTCCACACTATTTAGTTCTACGGAAAC | 0.0018 | 32 | 42 ± 2 |
| CTTTCCACACTATTTAGTTCTACGGAAACA | 0.0015 | 26 | 43 ± 3 | |
Within the M. thermoautotrophicus genome, 124 copies of the repeat sequence ATTTCAATCCCATTTTGGTCTGATT TTAAC were identified; the spacer length between these repeats was 37 ± 3 bp, with the exception of seven outliers (Figure 2). This repeat sequence contains the 25-nucleotide sub-sequence TTTCAATCCCATTTTGGTCTGATTT, which is common to most of the repeats found in M. thermoautotrophicus and is also found in a repeat sequence in A. fulgidus (CTTTCAATCCCATTTTGGTCTGATTTCAAC). Different repeats with significant copy numbers were found in P. abysii and M. jannaschii (Table 2). Comparison of all the repeats in P. abyssi and P. horikoshii, which are members of the same family, reveals the presence of a similar core sub-sequence (TTCCA). Within each studied archaeal organism, several repetitive elements with common sub-patterns were observed (see underlined fragments in Table 2). However, repetitive elements lacking the common core structure were also found in A. fulgidus, M. thermoautotrophicus and P. abysii (see non-underlined sequences in Table 2).
Based on a BLASTN analysis (www.ncbi.nlm.nih.gov/entrez/blast), these repeat sequences were unique to each individual archaeal genome. A BLAST search against the complete GenBank database using a 30 bp word returned significant hits only in the source archaeal genomes, suggesting that these long repeats are exclusive to archaea. Using the TIGR genome browser (www.tigr.org), the majority of long repeats were found in areas of low gene density and localized mainly in non-coding regions. For example, in M. thermoautotrophicus, long repeats were present between coding sequences.
Discussion
The complete sequencing of many genomes has made it possible to search for functionally significant sequence structures on a genome-wide scale in a large variety of organisms. Repetitive elements make up a large proportion of the non-coding portion of the genome and have traditionally hindered automated assembly of raw sequence data; hence, identification and characterization of such elements is significant technically as well as biologically. In this report, characteristic oligonucleotide repeat elements with regular, narrow periodicities were identified in archaeal genomes. Furthermore, similar repeats were not identified in the thermally labile bacterium E. coli or other non-archaeal control species (Table 1). The only exception to this rule was S. trididemni, in which a 30 bp repeat was identified; however, the repeat existed in low copy number and was not statistically significant.
BLASTN searches of the GenBank nucleotide database for each repeat element yielded hits mostly within the source archaeal organism, indicating signature character. However, certain repeats could be found in different species of the same family, such as in P. abysii and P. hirokoshii,or (with a few nucleotide mismatches) in unrelated species, as in M. thermoautotrophicus, A. fulgidus and P. abysii. The occurrence of these sequences among different species may have been facilitated by lateral DNA transfer. Different repetitive elements within the same organism were also observed, e.g., in A. fulgidus, M. thermoautotrophicus and P. abysii. These results are consistent with the recently reported identification of repeat loci in archaebacteria (Jansen et al. 2002a, 2002b).
Most repeats were located within non-coding, intergenic regions. However, in A. fulgidus, some repeats are reportedly transcribed into snmRNA and presumably play regulatory roles (Tang et al. 2002). The wide dispersion of these repeats in genomes suggests that they are mobile, which is in agreement with previous findings (Jansen et al. 2002a, 2002b). It is likely that these repetitive elements are propagated by forces similar to those acting on other mobile elements such as insertion sequences and transposons.
Resilience to DNA damage
The presence of such uniformly distributed words or patterns with high copy number suggests tolerance to DNA damaging agents such as ionizing radiation or chemicals. Because of inherent DNA sequence similarity, any damage could be effectively repaired by strand insertion, homologous recombination or non-homologous end joining.
Deinococcus radiodurans is known to be resistant to a range of DNA damaging agents such as ionizing radiation, oxidizing agents and mutagens, as a result of extremely efficient DNA repair processes that are poorly understood. One factor may be that the genome is enriched in repetitive elements such as autonomous insertion sequence (IS)-like transposons and small intergenic repeats (Makarova et al. 2001). We therefore compared the distribution of periodic tandem repeats in the genome of this organism with those of archaea. Our results indicated the presence of a 23-mer repeat with low copy number, lacking a distinct periodic pattern of distribution (data not shown). Hence, although the repeats found in archaea and D. radiodurans may be beneficial to the host in terms of tolerance to DNA damage, they may be under different selective and evolutionary pressures.
Nucleosome forming potential
Similar to eukaryotic nucleosomal positional elements, oligonucleotide (dA) tracts or 5′-(G/C)3NN(A/T)3NN-3′ motifs are well-characterized, high-affinity histone octamer binding sites that direct the localized assembly of archaeal nucleosomes (reviewed in Bailey and Reeve 1999). Most of our patterns (Table 2) contain such motifs and may be involved in chromatin remodeling, and thus, may regulate gene expression.
Cis gene regulation
We searched for the presence of putative transcription factor binding sites in the identified repeat sequences with the MatInspector program (Quandt et al. 1995); however, no such sequences were found. It is possible that the repeats may be 3′or 5′ untranslated regions that modulate gene expression.
Stem-loop potential
Cox and Mirkin (1997) have shown that normalized over-representation of repeats corresponds to the probability of DNA structure formation and therefore, most enriched repeats have the potential to form DNA secondary structures such as H-DNA, Z-DNA, cruciforms and slipped structures. All of the identified repeats in our study adopt a stem-loop conformation (data not shown) when folded using Zuker’s MFOLD (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/). This observation may shed some light on the evolution of such large repeats. Ogata and Miura (2000) found that long DNA sequences of more than 20 kb can be synthesized from a short DNA segment with palindromic or quasi-palindromic repetitive structure by hairpin elongation in the absence of a complementary DNA template in a few tested hyperthermophilic archaea, including the Pyrococcus spp. (considered in our analysis). Genomic expansion by such a method, along with homologous recombination and strand slippage mechanisms, may be a feature of archaebacteria, which are considered to be the primordial ancestors of higher life forms (Ogata and Miura 2000). Furthermore, the formation of such structures may lend greater resilience to the genome under denaturing conditions such as high temperature, salt, pH or pressure. In addition, secondary structure-forming characteristics have been implied in recognition by protein factors and thus may play a role in archaeal gene expression and regulation.
Evolutionary origin and significance
Most repeats arise by tandem duplication, hyperploidization, strand slippage, transposition or double-strand break repair by insertion. A recent study of long repeats in bacterial and archaeal genomes showed that direct repeats are more common than inverted repeats and concluded that interspersed repeats are mostly created as tandem repeats followed by successive rounds of opposing processes such as recombination (to maintain high identity) and deletion (for shorter length) (Achaz et al. 2002). The repetitive elements described in this report are interspersed throughout the respective genomes and may be under the same influences as mobile genetic elements. Recent reports have identified different autonomous IS-like and non-autonomous miniature inverted repeat element (MITE)-like mobile elements in newly sequenced archaeal genomes that are propagated by transposases and contribute to evolution by genomic rearrangements. Insertion sequence elements are commonly found in bacteria, whereas MITEs are more prominent in archaea (Brugger et al. 2002). The mutation rate in such repetitive elements is probably low.
Achaz et al. (2002) examined long repeats in bacterial and archaeal genomes and identified a negative correlation between spacer size and sequence identity, and a positive correlation between spacer size and repeat length, which is in agreement with our results (Table 2). The origin of spacers is unknown, although they may have arisen by random events because they are dissimilar within the same repetitive element of a given organism.
We believe that the nucleotide composition of a genome exerts a strong influence on the presence of periodic repeat patterns such as those seen here. Achaz et al. (2002) found that a strong negative correlation exists between nucleotide composition and repeat density in bacterial genomes. Low complexity genomes would be expected to produce more tandem repeats because of a higher compositional bias, and unbiased genomes may generate repeats at random that are then duplicated by different mechanisms, giving rise to larger repeats.
Some of the repetitive elements identified in this study have recently been identified by another group (Jansen et al. 2002a, 2002b). Their pattern search algorithm identified repeats in 40 prokaryotic genomes, but none were found among viral or eukaryotic species and the distribution was skewed toward archaea.
Conclusions
In summary, signature-like oligonucleotide repeats with narrow periodic distribution patterns were identified in the non-coding portions of archaeal genomes. Because no similar structures were identified in the genome sequences of several bacterial species, it is possible that these repeat regions serve an important structural role in the maintenance of DNA fidelity under harsh environmental conditions. Although the biological role of these highly conserved, long, archaea-specific repeats is unknown, we speculate that they are involved in both DNA sequence structure and evolution. Some of the hypotheses presented in this report may thus serve as the basis for further experimental or comparative investigations.
Acknowledgments
We thank the staff of the Bioinformatics Supercomputing Centre at the Hospital for Sick Children for providing technical support during the preparation of this work.
References
- R1.Achaz G., Rocha E.P., Netter P., Coissac E. Origin and fate of repeats in bacteria. Nucleic Acids Res. 2002;30:2987–2994. doi: 10.1093/nar/gkf391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Bailey K.A., Reeve J.N. DNA repeats and archaeal nucleosome positioning. Res. Microbiol. 1999;150:701–709. doi: 10.1016/s0923-2508(99)00122-9. [DOI] [PubMed] [Google Scholar]
- R3.Brugger K., Redder P., She Q., Confalonieri F., Zivanovic Y., Garrett R.A. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 2002;206:131–141. doi: 10.1111/j.1574-6968.2002.tb10999.x. [DOI] [PubMed] [Google Scholar]
- R4.Cole S.T., Supply P., Honore N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 2001;72:449–461. [PubMed] [Google Scholar]
- R5.Cox R., Mirkin S.M. Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. USA. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Deschavanne P.J., Giron A., Vilain J., Fagot G., Fertil B. Genomic signature: characterization and classification of species assessed by chaos game representation of sequences. Mol. Biol. Evol. 1999;16:1391–1399. doi: 10.1093/oxfordjournals.molbev.a026048. [DOI] [PubMed] [Google Scholar]
- R7.Heringa J. Detection of internal repeats: how common are they? Curr. Opin. Struct. Biol. 1998;8:338–345. doi: 10.1016/s0959-440x(98)80068-7. [DOI] [PubMed] [Google Scholar]
- R8.Jansen R., Van Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- R9.Jansen R., Van Embden J.D., Gaastra W., Schouls L.M. Identification of a novel family of sequence repeats among prokaryotes. Omics. 2002;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- R10.Karlin S., Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- R11.Karlin S., Burge C. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc. Natl. Acad. Sci. USA. 1996;93:1560–1565. doi: 10.1073/pnas.93.4.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Kawashima T., Yamamoto Y., Aramaki H. , et al. Determination of the complete genomic DNA sequence of Thermoplasma volcanium GSS1. Proc. Jpn. Acad. Ser. B. 1999;75:213–218. [Google Scholar]
- R13.Levy S., Compagnoni L., Myers E.W., Stormo G.D. Xlandscape: the graphical display of word frequencies in sequences. Bioinformatics. 1998;14:74–80. doi: 10.1093/bioinformatics/14.1.74. [DOI] [PubMed] [Google Scholar]
- R14.Makarova K.S., Aravind L., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Ogata N., Miura T. Elongation of tandem repetitive DNA by the DNA polymerase of the hyperthermophilic archaeon Thermococcus litoralis at a hairpin–coil transitional state: a model of amplification of a primordial simple DNA sequence. Biochemistry. 2000;39:13993–14001. doi: 10.1021/bi0013243. [DOI] [PubMed] [Google Scholar]
- R16.Pesole G., Prunella N., Liuni S., Attimonelli M., Saccone C. WORDUP: an efficient algorithm for discovering statistically significant patterns in DNA sequences. Nucleic Acids Res. 1992;20:2871–2875. doi: 10.1093/nar/20.11.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Rocha E.P., Viari A., Danchin A. Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res. 1998;26:2971–2980. doi: 10.1093/nar/26.12.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Romero D., Martinez-Salazar J., Ortiz E., Rodriguez C., Valencia-Morales E. Repeated sequences in bacterial chromosomes and plasmids: a glimpse from sequenced genomes. Res. Microbiol. 1999;150:735–743. doi: 10.1016/s0923-2508(99)00119-9. [DOI] [PubMed] [Google Scholar]
- R20.Tang T.H., Bachellerie J.P., Rozhdestvensky T., Bortolin M.L., Huber H., Drungowski M., Elge T., Brosius J., Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus . Proc. Natl. Acad. Sci. USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Van Helden J., Andre B., Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]