Abstract
A hyperthermophilic archaeal strain, KOD1, isolated from a solfatara on Kodakara Island, Japan, has previously been reported as Pyrococcus sp. KOD1. However, a detailed phylogenetic tree, made possible by the recent accumulation of 16S rRNA sequences of various species in the order Thermococcales, indicated that strain KOD1 is a member of the genus Thermococcus. We performed DNA–DNA hybridization tests against species that displayed high similarity in terms of 16S ribosomal DNA sequences, including Thermococcus peptonophilus and Thermococcus stetteri. Hybridization results and differences in growth characteristics and substrate utilization differentiated strain KOD1 from T. peptonophilus and T. stetteri at the species level. Our results indicate that strain KOD1 represents a new species of Thermococcus, which we designate as Thermococcus kodakaraensis KOD1 sp. nov.
Keywords: Archaea, classification, hyperthermophile, taxonomy, Thermococcales
Introduction
The archaeal order Thermococcales is currently composed of two major genera, Pyrococcus and Thermococcus (Huber and Stetter 2001, Itoh 2003), and a third genus, Palaeococcus (Takai et al. 2000). Members of the three genera are metabolically similar; they are obligate heterotrophs, growing on organic substrates, usually in the presence of elemental sulfur (S°), which is reduced to hydrogen sulfide (Amend and Shock 2001). Depending on the species, they utilize peptides, yeast extract, carbohydrates and some organic acids as carbon and energy sources.
Strain KOD1 is a hyperthermophilic archaeon isolated from a solfatara (102 °C, pH 5.8) on the shore of Kodakara Island, Kagoshima, Japan (Morikawa et al. 1994). Strain KOD1 is obligately heterotrophic and grows well on complex organic compounds in the presence of elemental sulfur. Cells are irregular cocci with several polar flagella. The strain contains tetra-ether and di-ether type lipids in its membrane (Morikawa et al. 1994), a typical characteristic of microorganisms from the domain Archaea(Konings et al. 2002). Strain KOD1 was, therefore, presumed to be a member of the thermophilic S°-reducing archaea and reported as Pyrococcus sp. KOD1 (Morikawa et al. 1994).
Since the isolation of strain KOD1, an abundant number of genes and their protein products from this archaeon have been examined (Imanaka and Atomi 2002). The first gene disruption system for a hyperthermophile has been established with this strain (Sato et al. 2003). We have further extended our studies and have recently sequenced the entire genome (unpublished data). Consequently, this strain has become one of the best characterized hyperthermophilic organisms. With the accumulation of 16S rRNA sequences available in the databases, we noticed that strain KOD1 clustered with various Thermococcus species in a branch distinct from that of the Pyrococcus species. Therefore, we undertook further studies to assess the classification of this strain.
Materials and methods
Media
The nutrient-rich complex medium (MA-YT medium) contained (per liter): 4.8 and 24.4 g Marine Art SF agents A and B, respectively (Senju Seiyaku, Osaka, Japan), 5 g yeast extract (Nacalai Tesque, Kyoto, Japan), 5 g tryptone (Nacalai Tesque) and 1.0 mg resazurin sodium salt. Strain KOD1 was also grown in an MA-YT medium in which the yeast extract and tryptone were replaced with 5 g yeast extract alone, 5 g tryptone alone, or 5 g peptone (BD Diagnostic Systems, Sparks, MD). After sterilizing the medium by autoclaving at 121 °C for 20 min, the bottles were transferred to an anaerobic chamber (CO2:H2:N2 (5:5:90)) (EAN-140, Espec, Osaka, Japan), and 5.0% Na2S·9H2O was added until the color of the resazurin sodium salt became clear. Afterwards, when necessary, 2.0 g l–1 elemental sulfur was added. For MA-YT medium (without elemental sulfur) supplemented with soluble starch (0.5%), maltooligosaccharides (0.5%), or sodium pyruvate (0.5%), the substrates were added prior to autoclaving. In an investigation of possible electron acceptors, the following compounds were added to the MA-YT medium at 0.15% (w/v): elemental sulfur, sodium thiosulfate pentahydrate, sodium sulfate, sodium sulfite, sodium nitrate and sodium nitrite. Ferric chloride hexahydrate was added at a concentration of 5 mg l–1.
The minimal ASW-AA medium, which supported growth of strain KOD1, was composed of: 1.25-fold dilution of artificial seawater (0.8 × ASW) (Robb and Place 1995) supplemented with 5.0 ml l–1 modified Wolfe’s trace minerals (0.5 g MnSO4·2H2O, 0.1 g CoCl2,0.1 g ZnSO4, 0.01 g CuSO4·5H2O, 0.01 g AlK(SO4)2, 0.01 g H3BO3 and 0.01 g NaMoO4·2H2O per liter), 5.0 ml l–1 vitamin mixture (Robb and Place 1995), 20 amino acids (250 mg cysteine-HCl·H2O; 200 mg each of glutamic acid and glycine; 125 mg each of arginine-HCl and proline; 100 mg each of asparagine·H2O, histidine-HCl·H2O, isoleucine, leucine, lysine-HCl, threonine and tyrosine; 75 mg each of alanine, methionine, phenylalanine, serine and tryptophan; and 50 mg each of aspartic acid, glutamine and valine per liter), 1.0 mg l–1 resazurin sodium salt and 2.0 g l–1 elemental sulfur (pH was adjusted to 6.9 with NaOH). The medium was sterilized by autoclaving before adding elemental sulfur and the vitamin mixture. After transferring the bottles to an anaerobic chamber, filter-sterilized vitamin mixture was added with the 5.0% Na2S·9H2O and elemental sulfur as described above.
Phylogenetic analysis of 16S ribosomal DNA sequences
Sequences of 16S ribosomal DNA of various Thermococcus species available from the EMBL/GenBank/DDBJ databases were collected and aligned. Regions spanning 1231 bp (between 83-GGCGGACGG and GCGTGTCAT-1313 in the Thermococcus kodakaraensis sequence) were used to calculate phylogeny. The multiple alignment of DNA sequences was performed using the ALIGN program contained within the ClustalW program (Thompson et al. 1994) provided by the DNA Data Bank of Japan (DDBJ). The phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei 1987) and by the maximum-likelihood method (Felsenstein 1981) with the fastDNAmL program (Olsen et al. 1994). Bootstrap resampling was performed 1000 times. The phylogenetic tree was visualized with the TreeView program.
DNA–DNA hybridization studies
Data from DNA–DNA hybridizations between strain KOD1 and Thermococcus peptonophilus JCM9653T, Thermococcus gorgonarius JCM10552T, Thermococcus fumicolans JCM10128T and Thermococcus guaymasensis JCM10136T were kindly provided by Dr. Takashi Itoh at the Japan Collection of Microorganisms (JCM), RIKEN, Saitama, Japan and Dr. Francesco Canganella at the Department of Agrobiology and Agrochemistry, University of Tuscia, Italy. For our own hybridization experiment, Thermococcus stetteri JCM8559T, Thermococcus profundus JCM9378T (Kobayashi et al. 1994) andThermococcus fumicolans JCM10128T (Godfroy et al. 1996) were obtained from JCM. Thermococcus celer ATCC35543T (Zillig et al. 1983) was obtained from the American Type Culture Collection (Manassas, VA). All culturing manipulations were performed in an anaerobic chamber. Stock cultures were inoculated in MA-YT medium containing 0.5% elemental sulfur. Culture bottles containing the medium were anaerobically sealed and incubated at 85 or 95 °C for 24 to 40 h. Cells were harvested and lysed with detergent (1% N-lauroylsarcosine, Nacalai Tesque), and chromosomal DNA was purified by CsCl density gradient ultra-centrifugation (Optima XL-90, Beckman, Fullerton, CA). The DNA–DNA hybridization experiments were carried out at Higeta Shoyu (Chiba, Japan) using methods described elsewhere (Ezaki et al. 1989).
Results and discussion
Growth characteristics of strain KOD1
Strain KOD1 has been reported to grow on a variety of complex nutrients in the presence of elemental sulfur (Morikawa et al. 1994). Here we further examined the growth of the strain in various media at 85 °C. In the presence of elemental sulfur, strain KOD1 grew on 0.5% yeast extract/0.5% tryptone (MA-YT), 0.5% yeast extract, 0.5% tryptone, or 0.5% peptone. When we replaced the elemental sulfur in MA-YT medium with sodium thiosulfate pentahydrate, sodium sulfate, sodium sulfite, sodium nitrate, sodium nitrite or ferric chloride hexahydrate, no growth was observed during a 5-day period. When soluble starch (0.5%) or sodium pyruvate (0.5%) was added to MA-YT medium, the requirement for elemental sulfur was relieved. Consumption of these substrates during growth was confirmed, and substrate depletion correlated well with the growth of the strain. Under these conditions, significant amounts of acetate, succinate, isobutyrate and isovalerate and/or 2-methylbutyrate accumulated in the medium. Low concentrations of formate and lactate were also detected. Molecular hydrogen and carbon dioxide were the main gases accumulating in the gas phase. Strain KOD1 utilized maltooligosaccharides and cyclodextrins in a similar manner in the absence of elemental sulfur. We found that strain KOD1 could also grow in a minimal ASW-AA medium (see Materials and methods) with free amino acids as the sole carbon source.
Phylogenetic analysis of 16S ribosomal DNA sequences
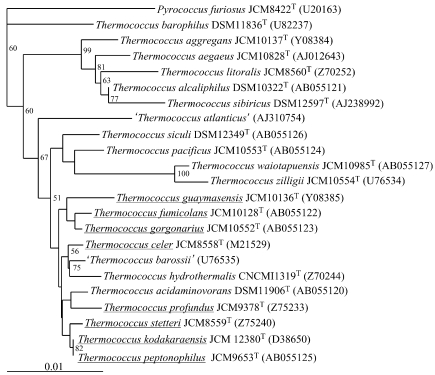
We first constructed a phylogenetic tree with various Thermococcus species using Pyrococcus furiosus (Fiala and Stetter 1986) as an outgroup. The 16S rRNA gene sequences of the strains shown in Figure 1 were selected for multiple alignment and construction of a phylogenetic tree using the neighbor-joining method. The phylogenetic position of strain KOD1 was located within the genus Thermococcus. With the sequences we applied, strain KOD1 clustered with T. peptonophilus (González et al. 1995) and T. stetteri (99%) (Miroshnichenko et al. 1989). When the maximum-likelihood method was applied, the phylogenetic position of several strains changed, but the clustering among T. peptonophilus, T. stetteri and strain KOD1 was unaltered. The results indicate that strain KOD1 is a member of Thermococcus and suggest that it is most closely related to T. peptonophilus and T. stetteri.
Figure 1.
Phylogenetic tree showing the position of strain KOD1. The tree was constructed using the neighbor-joining method as described in the Materials and methods. The scale bar represents the expected number of changes per sequence position. Bootstrap values greater than 50% are shown in the tree. Strains used in DNA–DNA hybridization tests are underlined. Strains and accession numbers for all sequences are indicated.
DNA–DNA hybridization analyses
The genomic DNA sequence similarities between strain KOD1 and the phylogenetically related Thermococcus species were assessed on the basis of DNA–DNA hybridization studies. Results of hybridizations between strain KOD1 and four Thermococcus species were kindly provided by Drs. Takashi Itoh and Francesco Canganella. With the DNA of strain KOD1 fixed, they found %hybridization was 56% with T. peptonophilus, 16% with T. gorgonarius, 22% with T. fumicolans and 16% with T. guaymasensis (%hybridization with strain KOD1 set at 100%). In further experiments, with the DNA of strain KOD1 fixed, we found %hybridization with DNA was 13% with T. stetteri, 32% with T. profundus, 31% with T. celer and 15% with T. fumicolans (%hybridization with strain KOD1 set at 100%). The hybridization scores between strain KOD1 and the Thermococcus strains thus suggest that strain KOD1 is a novel species of Thermococcus.
Comparison with T. peptonophilus and T. stetteri
Phylogenetic analysis of 16S rRNA sequences indicated that, among the previously reported Thermococcus species, strain KOD1 was most closely related to T. peptonophilus and T. stetteri. However, the %hybridization between strain KOD1 and T. stetteri was low (13%). Further, T. stetteri JCM8559T (=DSM 5262T) grows only in the presence of elemental sulfur and not with free amino acids (Miroshnichenko et al. 1989). Strain KOD1 can grow without elemental sulfur in the presence of starch, maltooligosaccharides and pyruvate. The strain also grows well with free amino acids in the presence of elemental sulfur. The optimal growth temperature of T. stetteri K-3T (JCM8559T) is also much lower (73–77 °C) than that of strain KOD1 (85 °C). These differences indicate that strain KOD1 can be distinguished from T. stetteri at the species level.
Thermococcus peptonophilus OG-1T (JCM9653T) is a fast-growing, piezophilic strain isolated from deep-sea hydrothermal areas (González et al. 1995) with growth rates as high as 1.65 h–1. At 85 °C, we have observed a maximum growth rate of 0.7 h–1 when KOD1 cells were grown in a batch culture in MA-YT medium with 1% sodium pyruvate and 0.2% elemental sulfur. In continuous culture, we have not observed growth rates greater than 0.8 h–1.
Among substrates, T. peptonophilus grows well on peptides and does not utilize substrates such as pyruvate, starch or a mixture of amino acids (González et al. 1995). In contrast, we show here that strain KOD1 grows well on all of these substrates. This may reflect the different environments from which these strains were isolated; T. peptonophilus was isolated from deep-sea hydrothermal areas, whereas strain KOD1 was isolated from a hot water environment on the coast of Kodakara Island (Morikawa et al. 1994).
Consistent with the observed growth on starch and maltooligosaccharides, we found that strain KOD1 harbors a variety of genes involved in polysaccharide degradation, including 4-α-glucanotransferase, α-amylase and cyclodextrin glucanotransferase. The enzyme activities of the respective protein products have been biochemically characterized (Tachibana et al. 1996, 1997, Rashid et al. 2002a). Strain KOD1 grows well under gluconeogenic conditions with pyruvate and amino acids, and the key enzyme of gluconeogenesis, fructose 1,6-bisphosphatase (FBPase), has been examined. The enzyme represents a novel archaeal-type FBPase and is strictly regulated at the transcriptional level responding to the carbon source (Rashid et al. 2002b). The major enzymes involved in chitin assimilation by strain KOD1 have been characterized (Tanaka et al. 2001, 2003), and it has been found that the expression of these enzymes is induced only in the presence of chitin and its degradation products (Tanaka et al. 2003). These features indicate that strain KOD1 can assimilate various carbon sources, and has evolved strict regulatory systems, enabling it to use these substrates efficiently. These characteristics distinguish strain KOD1 from T. peptonophilus at the species level. Representative differences among the three strains are summarized in Table 1. These results, in addition to those of the DNA–DNA hybridizations, indicate that strain KOD1 is a novel species of Thermococcus, designated as Thermococcus kodakaraensis.
Table 1.
Comparison of some properties among Thermococcus kodakaraensis, T. peptonophilus and T. stetteri.
| Properties | T. kodakaraensis | T. peptonophilus | T. stetteri |
| KOD1T = JCM12380T | OG-1T = JCM9653T | K-3T = JCM8559T | |
| Source of isolation | Solfataraon Kodakara Island | Deep-sea hydrothermal vent | Solfatarain Kraternaya |
| in the western Pacific Ocean | Cove | ||
| Temperature range of growth (optimum) | 60–100 °C (85 °C) | 60–100 °C (85 °C) | 55–94 °C (76 °C) |
| Carbon source | |||
| Free amino acids | Yes | No | No |
| Yeast extract | Yes | Yes | Not reported |
| Tryptone | Yes | Yes | Not reported |
| Peptone | Yes | Yes | Yes |
| Starch | Yes | No | Yes |
| Maltooligosaccharides | Yes | No | Not reported |
| Pyruvate | Yes | No | Not reported |
| Requirement for S°with | |||
| Free amino acids | Yes | No growth2 | No growth2 |
| Yeast extract | Yes | No | Not reported |
| Tryptone | Yes | No | Not reported |
| Peptone | Yes | No | Yes |
| Starch | No | No growth2 | Yes |
| Maltooligosaccharides | No | No growth2 | Not reported |
| Pyruvate | No | No growth2 | Not reported |
| G+C content | 52.0 mol%1 | 52 mol% | 50.2 mol% |
| Reference | This study | González et al. 1995 | Miroshnichenko et al. 1989 |
1 Calculated from the complete genome sequence. 2 No growth regardless of the presence or absence of elemental sulfur.
Description of Thermococcus kodakaraensis sp. nov.
Thermococcus kodakaraensis sp. nov.
ko.da.ka.ra.en′.sis L. gen. n. kodakaraensis pertaining to Kodakara Island, the source of the hot water sample from which the strain was isolated.
Cells are irregular cocci of 1–2 µm in diameter. Multiplication is by constriction. Highly motile with a polar tuft of multiple flagella. Cell envelope consists of two layers. Strictly anaerobic. Temperature range of growth is 60–100 °C, with an optimum of approximately 85 °C. The pH range of growth is between 5 and 9 with an optimum of approximately 6.5. Range of NaCl concentration allowing growth is between 1 and 5%, with an optimum of 3%. Elemental sulfur, as a terminal electron acceptor, is required for heterotrophic growth on yeast extract, tryptone, peptone and free amino acids (producing hydrogen sulfide). Grows without elemental sulfur through fermentation with the addition of starch, maltooligosaccharides, cyclodextrins or pyruvate (producing molecular hydrogen). The G+C content is 52.0 mol% (genome sequence). The organism was isolated from a solfatara (102 °C, pH 5.8) on the shore of Kodakara Island, Kagoshima, Japan.
Type strain: Thermococcus kodakaraensis KOD1T, JCM 12380T, ATCC BAA-918T.
Acknowledgments
The authors are deeply grateful for the constructive comments and advice from Dr. Takashi Itoh at JCM during manuscript preparation.
References
- R1.Amend J.P., Shock E.L. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 2001;25:175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x. [DOI] [PubMed] [Google Scholar]
- R2.Ezaki T., Hashimoto Y., Yabuuchi E. Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 1989;39:224–229. [Google Scholar]
- R3.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- R4.Fiala G., Stetter K.O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch. Microbiol. 1986;145:56–61. [Google Scholar]
- R5.Godfroy A., Meunier J.-R., Guezennec J., Lesongeur F., Raguénès G., Rimbault A., Barbier G. Thermococcus fumicolans sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent in the north Fiji Basin. Int. J. Syst. Bacteriol. 1996;46:1113–1119. doi: 10.1099/00207713-46-4-1113. [DOI] [PubMed] [Google Scholar]
- R6.González J.M., Kato C., Horikoshi K. Thermococcus peptonophilus sp. nov., a fast-growing, extremely thermophilic archaebacterium isolated from deep-sea hydrothermal vents. Arch. Microbiol. 1995;164:159–164. [PubMed] [Google Scholar]
- R7.Huber R., Stetter K.O. Discovery of hyperthermophilic microorganisms. Methods Enzymol. 2001;330:11–24. doi: 10.1016/s0076-6879(01)30367-1. [DOI] [PubMed] [Google Scholar]
- R8.Imanaka T., Atomi H. Catalyzing “hot” reactions: enzymes from hyperthermophilic archaea. Chem. Rec. 2002;2:149–163. doi: 10.1002/tcr.10023. [DOI] [PubMed] [Google Scholar]
- R9.Itoh T. Taxonomy of nonmethanogenic hyperthermophilic and related thermophilic archaea. J. Biosci. Bioeng. 2003;96:203–212. [PubMed] [Google Scholar]
- R10.Kobayashi T., Kwak Y.S., Akiba T., Kubo T., Horikoshi K. Thermococcus profundus sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Syst. Appl. Microbiol. 1994;17:232–236. [Google Scholar]
- R11.Konings W.N., Albers S.-V., Koning S., Driessen A.J.M. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek. 2002;81:61–72. doi: 10.1023/a:1020573408652. [DOI] [PubMed] [Google Scholar]
- R12.Miroshnichenko M.L., Bonch-Osmolovskaya E.A., Neuner A., Kostrikina N.A., Chernych N.A., Alekseev V.A. Thermococcus stetteri, sp. nov., a new extremely thermophilic marine sulfur-metabolizing archaebacterium. Syst. Appl. Microbiol. 1989;12:257–262. [Google Scholar]
- R13.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Olsen G.J., Matsuda H., Hagstrom R., Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- R15.Rashid N., Cornista J., Ezaki S., Fukui T., Atomi H., Imanaka T. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 2002;184:777–784. doi: 10.1128/JB.184.3.777-784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Rashid N., Imanaka H., Kanai T., Fukui T., Atomi H., Imanaka T. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 2002;277:30649–30655. doi: 10.1074/jbc.M202868200. [DOI] [PubMed] [Google Scholar]
- R17.Robb F.T., Place A.R. Archaea: A Laboratory Manual—Thermophiles. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. Media for thermophiles; pp. 167–168. [Google Scholar]
- R18.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- R19.Sato T., Fukui T., Atomi H., Imanaka T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2003;185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Tachibana Y., Leclere M.M., Fujiwara S., Takagi M., Imanaka T. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J. Ferment. Bioeng. 1996;82:224–232. [Google Scholar]
- R21.Tachibana Y., Fujiwara S., Takagi M., Imanaka T. Cloning and expression of the 4-α-glucanotransferase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J. Ferment. Bioeng. 1997;83:540–548. [Google Scholar]
- R22.Takai K., Sugai A., Itoh T., Horikoshi K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 2000;50:489–500. doi: 10.1099/00207713-50-2-489. [DOI] [PubMed] [Google Scholar]
- R23.Tanaka T., Fukui T., Imanaka T. Different cleavage specificities of the dual catalytic domains in chitinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biol. Chem. 2001;276:35629–35635. doi: 10.1074/jbc.M105919200. [DOI] [PubMed] [Google Scholar]
- R24.Tanaka T., Fukui T., Atomi H., Imanaka T. Characterization of an exo-β-D-glucosaminidase involved in a novel chitinolytic pathway from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2003;185:5175–5181. doi: 10.1128/JB.185.17.5175-5181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Zillig W., Holz I., Janekovic D., Schäfer W., Reiter W.D. The archaebacterium Thermococcus celer represents a novel genus within the thermophilic branch of the archaebacteria. Syst. Appl. Microbiol. 1983;4:88–94. doi: 10.1016/S0723-2020(83)80036-8. [DOI] [PubMed] [Google Scholar]