Abstract
Pyrococcus furiosus laminarinase (LamA, PF0076) is an endo-glycosidase that hydrolyzes β-1,3-gluco-oligosaccharides, but not β-1,4-gluco-oligosaccharides. We studied the specificity of LamA towards small saccharides by using 4-methylumbelliferyl β-glucosides with different linkages. Besides endo-activity, wild-type LamA has some exo-activity, and catalyzes the hydrolysis of mixed-linked oligosaccharides (Glcβ4Glcβ3Glcβ-MU (Glc = glucosyl, MU = 4-methylumbelliferyl)) with both β-1,4 and β-1,3 specificities. The LamA mutant E170A had severely reduced hydrolytic activity, which is consistent with Glu170 being the catalytic nucleophile. The E170A mutant was active as a glycosynthase, catalyzing the condensation of α-laminaribiosyl fluoride to different acceptors. The best condensation yields were found at pH 6.5 and 50 °C, but did not exceed 30%. Depending on the acceptor, the synthase generated either a β-1,3 or a β-1,4 linkage.
Keywords: enzymatic synthesis, glycosyl fluoride, glycosylation, glycosynthase mutant, hydrolysis, laminarinase, nucleophile
Introduction
For the biocatalytic production of oligosaccharides, glycoside hydrolases are of interest because of their generally simple heterologous production and the availability of low-cost substrates. Synthesis of oligosaccharides is achieved by thermodynamically controlled reverse hydrolysis and kinetically controlled transglycosylation, which both require high substrate concentrations and low water activity, often achieved by elevated temperatures and the addition of organic solvents. Final yields, however, do not exceed 50% because the product is a substrate of the enzyme’s hydrolase activity. Increased yields have been obtained at higher temperatures with thermophilic enzymes (Boon et al. 1998, Kaper et al. 2001). Currently, a maximal yield can be accomplished only by using glycosynthases, glycoside hydrolases in which the catalytic nucleophile has been replaced by a small non-catalytic amino acid residue (Mackenzie et al. 1998). Glycosynthases couple activated glycosyl donors, which have the opposite anomeric configuration to that of the normal substrate, to a range of acceptors. Because these enzymes cannot hydrolyze the product, they carry out transglycosylation efficiently (Figure 1). Typical product yields are 65–80%, but some studies have reported yields of up to 100% (for reviews, see Moracci et al. 2001, Planas and Faijes 2002, Williams and Withers 2002). The glycosynthase strategy has now been applied to five exo-β-glycosidases (Mackenzie et al. 1998, Trincone et al. 2000, Nashiru et al. 2001, Jakeman and Withers 2002, Perugino et al. 2003), four endo-β-glucanases (Malet and Planas 1998, Fort et al. 2000, Hrmova et al. 2002, Jahn et al. 2003) and an exo-α-glucosidase (Okuyama et al. 2002). The combination of two endo-glycosynthases resulted in the efficient synthesis of hexasaccharides through the sequential transglycosylation of different disaccharides (Faijes et al. 2001).
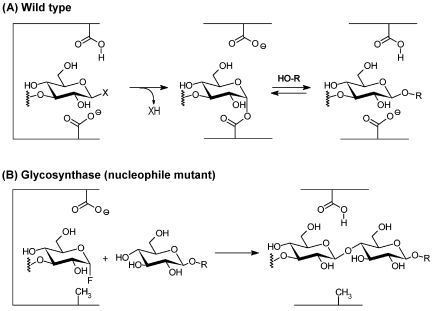
Figure 1.
Transglycosylation and glycosynthase reactions catalyzed by retaining glycosidases. (A) Transglycosylation or hydrolysis (R = hydrogen) catalyzed by a wild-type glycosidase. (B) Glycosynthase reaction catalyzed by a nucleophile mutant glycosidase.
The β-glucansynthase from Bacillus licheniformis is an efficient enzyme, mainly because of its strict regio- and stereospecificity (Malet and Planas 1998, Faijes et al 2003), traits that are also observed in the original wild-type hydrolase (Malet and Planas 1997, Planas 2000). The endo-β-1,3-1,4-glucanase hydrolyzes the β-1,4 glycosidic bond in a 3-O-substituted glucopyranosyl unit of mixed-linked glucans (Planas 2000). The enzyme is classified in Family 16 of glycoside hydrolases (Henrissat 1991), which also comprises endo-β-1,3-glucanases. Both types of Family 16 enzymes have an amino acid homology of 15–30%. The endo-β-1,3-glucanases, also called laminarinases, hydrolyze mixed-linked glucans such as lichenan, but prefer β-1,3-glucans such as laminarin, in contrast to the endo-β-1,3-1,4-glucanases, or lichenases. Therefore, we were interested in the behavior of a laminarinase, not only as a hydrolase, but also as a glycosynthase, particularly with respect to its specificity and efficiency.
The laminarinase (LamA, PF0076) from Pyrococcus furiosus is an extremely thermostable (t½ 100 °C = 16 h) and thermoactive (Topt = 100–105 °C) endo-β-1,3-glucanase (Gueguen et al. 1997, Chiaraluce et al. 2002) that has been reported to hydrolyze laminarin-oligosaccharides, but not cello-oligosaccharides and polysaccharides (Driskill et al. 1999). Hydrolysis of mixed-linked oligosaccharides has never been reported. However, polysaccharides containing both β-1,3 and β-1,4 linkages are hydrolyzed by LamA to a certain extent, but the glycosidic linkages of the products have not been identified (Gueguen et al. 1997). The same holds true for products generated in glycosylation reactions with laminarin-oligosaccharides (Driskill et al. 1999). Therefore, it is still unclear whether the enzyme hydrolyzes or synthesizes β-1,3 glycosidic linkages only or also β-1,4 bonds. Here we report on the hydrolytic specificity of LamA towards a series of small synthetic 4-methylumbelliferyl (MU) glucosides. In addition, the glycosynthase activity of a LamA nucleophile mutant was studied and compared with the activity of 1,3-1,4-β-glucansynthase from B. licheniformis.
Materials and methods
General
The NMR spectra were recorded on a Varian Gemini-300 spectrometer (Varian, Palo Alto, CA) in deuterium oxide (D2O). Chemical shifts (δ in ppm) were referenced to residual DMSO (δH 2.49) or d6DMSO (δC 39.7) as internal standards for 1H or 13C NMR, respectively. Mass spectra were recorded on a VG Autospec spectrometer (Waters-Micromass Technologies, Milford, MA) by the fast atomic bombardment (FAB) technique with an NaCl matrix.
Enzymes
The LamA mutant, E170A, was prepared by overlap extension polymerase chain reaction (PCR) (Ho et al. 1989) with the primers BG892, 5′-AAATTGTGGAGCGATCGACATAT GGAGT-3′ (sense) and BG893, 5′-ACTCCATTATGTCTAT CGCTCCACAATTT-3′ (antisense). Both recombinant wild-type and E170A mutant LamA proteins were expressed in Escherichia coli BL21 (DE3)(Stratagene, La Jolla, CA) and purified as previously reported (Gueguen et al. 1997, Kaper et al. 2001). The purity of the enzyme preparations was greater than 95% as judged by SDS-PAGE, according to Laemmli (1970). Enzyme concentrations were determined by UV spectrometry based on an ε280 = 83,070 M–1 cm–1 and a molecular mass of 30,085 g mol–1, as calculated by ProtParam (Gill and Von Hippel 1989).
Substrates
Substrates used in this study are shown in Figure 2. 4-Methylumbelliferyl β-laminaribioside (1), 4-methylumbelliferyl 3-O-β-cellobiosyl-β-D-glucopyranoside (2) and 4-methylumbelliferyl 3-O-β-cellotriosyl-β-D-glucopyranoside (3) were synthesized as reported previously (Malet et al. 1995). 4-Methylumbelliferyl β-cellobioside (4) was obtained as described by van Tilbeurgh et al. (1982), and 4-methylumbelliferyl β-D-glucoside (5) was obtained from Fluka (Buchs, Switzerland). α-Laminaribiosyl fluoride (6) was prepared as described by Malet and Planas (1998) by reacting laminaribiose peracetate with hydrogen fluoride in pyridine, followed by de-O-acetylation with sodium methoxide in methanol.
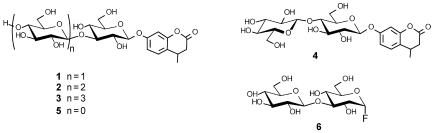
Figure 2.
Substrates for wild type and E170A LamA used in this study: 1 = 4-methylumbelliferyl β-laminaribioside (Glcβ3Glcβ3-MU); 2 = 3-O-β-cellobiosyl- β-D-glucopyranoside (Glcβ4Glcβ3Glcβ- MU); 3 = 3-O-β-cellotriosyl-β-D-glucopyranoside (Glcβ4Glcβ4Glcβ3Glcβ-MU); 4 = 4-methylumbelliferyl β-cellobioside (Glcβ4Glcβ-MU); 5 = 4-methylumbelliferyl β-D-glucoside (Glcβ-MU); and 6 = α-laminaribiosyl fluoride (Glcβ3GlcαF).
Hydrolytic activity of wild-type and E170A enzymes
Kinetics with laminarin as substrate
Reactions were performed with 0.5% laminarin in 100 mM sodium phosphate buffer, pH 6.5, and 0.02 µM wild-type enzyme or 1 µM E170A mutant, at 80 °C. Aliquots of the reaction were withdrawn at 10-min intervals, and the increase in reducing power was determined by the dinitrosalicylic acid (DNS) method (Miller 1959), with glucose as the standard.
Kinetics with 4-methylumbelliferyl glycoside substrates Kinetics with Substrates1 and 2 were performed by measuring changes in UV absorbance at 365 nm due to the release of 4-methylumbelliferone with a Varian Cary 4 spectrophotometer with a Peltier temperature control system. Rates of enzyme-catalyzed hydrolysis were determined by incubating the substrate (0–8 mM) in phosphate buffer (100 mM, pH 6.5) for 5 min in a thermostated cell holder at 80 °C, followed by addition of the corresponding enzyme (0.02 µM for wild type or 1 µM for E170A) and monitoring the absorbance change at λ = 365 nm (molar extinction coefficient, Δε = 4452 M–1cm–1).
High performance liquid chromatography (HPLC)
Reaction mixtures containing the substrate (4 mM) and wild-type LamA (20–200 nM) in phosphate buffer (100 mM), pH 6.5, were incubated at 80 °C. Aliquots were taken at different time intervals and analyzed by HPLC in a Nova-Pak C18 column (4 µm, 3.9 × 150 mm) (Waters, Milford, MA). The flow rate was 1 ml min–1, with 16% MeOH in 50 mM phosphate buffer (pH 6.5) as the mobile phase, and detection at 316 nm. Only products containing the 4-methylumbelliferyl chromophore were detected at this wavelength. Yields were calculated from the peak areas based on the corresponding response factor determined for free 4-methylumbelliferone (8770 area mM–1) and for 4-methylumbelliferyl glycosides (7603 area mM–1). Chromatographic peaks were identified by co-injection with independent standards.
Glycosynthase activity of E170A LamA
α-Laminaribiosyl fluoride (6) (1 mM) and different glycosyl acceptors (5 mM) were dissolved in phosphate buffer (100 mM, pH 6.5) and pre-incubated for 5 min. The E170A mutant (2–7.5 µM) was added and the reactions were incubated at 35, 50 or 76 °C. Aliquots were taken at different time intervals, diluted with H2O and analyzed by HPLC as described previously (mobile phase 16–18% MeOH). New chromatographic peaks were first tentatively identified by co-injection with independent standards. Control reactions without enzyme were performed to assess the absence of any uncatalyzed condensation. The thermal stability of α-laminaribiosyl fluoride was analyzed in phosphate buffer (100 mM, pH 6.5) by monitoring fluoride anion release with a fluoride selective electrode (Sentek, Essex, U.K.), interfaced with a CiberScan Bench pH/ion meter (Eutech Instruments, Nijkerk, The Netherlands) as previously reported (Faijes et al. 2003).
Preparative glycosynthase reactions
α-Laminaribiosyl fluoride donor (10.6 mg, 0.031 mmol, 1 equivalent), acceptor 1 or 2 (5 equivalents) and E170A LamA mutant (0.6 mg) were dissolved in phosphate buffer (100 mM, pH 6.5, 5.5 ml) and incubated at 50 °C for 2 days. Samples (10 µl) were withdrawn, diluted 1:10 with H2O and analyzed by HPLC (Nova-Pak C18, MeOH 18%). During the reaction, more donor was added (final conditions: 6.9 ml, 40 mg donor). After centrifugation to remove traces of precipitate, the reaction mixture was loaded directly onto a reverse-phase Lichoprep RP-18 Lobar-A column (Merck, Whitehouse Station, NJ) and eluted with an H2O:MeOH gradient from 0 to 25%. Fractions were freeze-dried.
(a) Glcβ3GlcαF (6) + Glcβ-MU (5) glycosynthase reaction
Unreacted acceptor was eluted first. Then, the trisaccharide Glcβ3Glcβ4Glcβ-MU was obtained: m/z (FAB) 685 [M+Na]+. 1H NMR (D2O, 30 °C): δ2.42 (s, 3H, CH3), 3.34–4.08 (m, 18H, H-2I–III, H-3I–III, H-4I–III, H-5I–III, H-6aI–III, H-6bI–III), 4.60 (d, J1,2 = 8.1 Hz ,1H), H-1II, 4.80 (d, 1H, H-1III), 5.23 (d, J1,2 = 7.5 Hz, 1H, H-1I), 6.22 (s, 1H, H-3′), 7.04 (s, 1H, H-8′), 7.10 (d, J5′,6′ = 9.6 Hz, 1H, H-6′), 7.69 (d, J5′,6′ = 9.0 Hz, 1H, H-5′). 13C NMR (D2O, 30 °C) δ18.6 (CH3), 60.4–61.4 (C-6I–III), 68.7, 70.2 (C-4II,III), 73.2–76.6 (C-2I–III, C-3I,III, C-5I–III), 78.8 (C-4I), 84.7 (C-3II), 100.2–104.2 (C-1I–III, C-8′), 111.9 (C-3′), 114.5 (C-6′), 116.0 (C-4a′), 127.3 (C-5′), 154.5 (C-4′), 156.8 (C-1a′), 160.1 (C-7′), 165.2 (C-2′).
The third fraction was a minor compound tentatively assigned as the trisaccharide Glc3Glc3Glcβ-MU. It was treated with wild-type LamA and the disaccharide Glcβ3Glcβ-MU was detected by HPLC.
(b) Glcβ3GlcαF (6) + Glcβ3Glcβ-MU (1) glycosynthase reaction
First, unreacted acceptor was eluted, followed by the tetrasaccharide Glcβ3Glcβ3Glcβ3Glcβ-MU: m/z (FAB): 847.251 [M+Na]+ (calculated 847.248). 1H NMR (D2O, 30 °C): δ 2.42 (s, 3H, CH3), 3.34–4.01 (m, 24H, H-2I–IV, H-3I–IV, H-4I–IV, H-5I–IV, H-6aI–IV, H-6bI–IV), 4.76–4.87 (3H, H-1I–III), 5.23 (d, J1,2 = 7.5 Hz, 1H, H-1I), 6.23 (s, 1H, H-3′), 7.06 (s, 1H, H-8′), 7.10 (d, J5′,6′ = 9.6 Hz, 1H, H-6′), 7.69 (d, J5′,6′ = 9.0 Hz, 1H, H-5′). 13C NMR (D2O, 30 °C): δ18.7 (CH3), 61.0, 61.3 (C-6I–IV), 68.4–70.5, (C-4I–IV), 73.2–76.6 (C-2I–IV, C-3IV, C-5I–IV), 84.4–84.8 (C-3I–III), 100.0–104.2 (C-1I–III, C-8′), 111.8 (C-3′), 114.4 (C-6′), 115.8 (C-4a′), 127.2 (C-5′), 154.3 (C-4′), 156.6 (C-1a′), 159.9 (C-7′), 164.9 (C-2′).
A second condensation product corresponding to the tetrasaccharide Glcβ3Glcβ4Glcβ3Glcβ-MU was isolated: m/z: calculated for C34H48O23Na: 847.250; found: 847.248. 1H and 13C NMR are identical to those reported in Faijes et al. (2003).
Results and discussion
Hydrolase activity and specificity of wild-type LamA
The glucoside (5) and the cellobioside (4) were not hydrolyzed by wild-type LamA, whereas the 4-methylumbelliferyl laminaribioside (1) and the mixed-linked oligosaccharides Glcβ4Glcβ3Glcβ-MU (2) and Glcβ4Glcβ4Glcβ3Glcβ-MU (3) were substrates. Cleavage specificity was determined by HPLC (Figures 3A–C). Only chromophoric products (free MU and MU-glycosides) were analyzed (UV detection at 316 nm) and identified by co-injection with independent standards. Reactions were performed at low enzyme concentrations to extend the time course and allow the detection of transient intermediates.
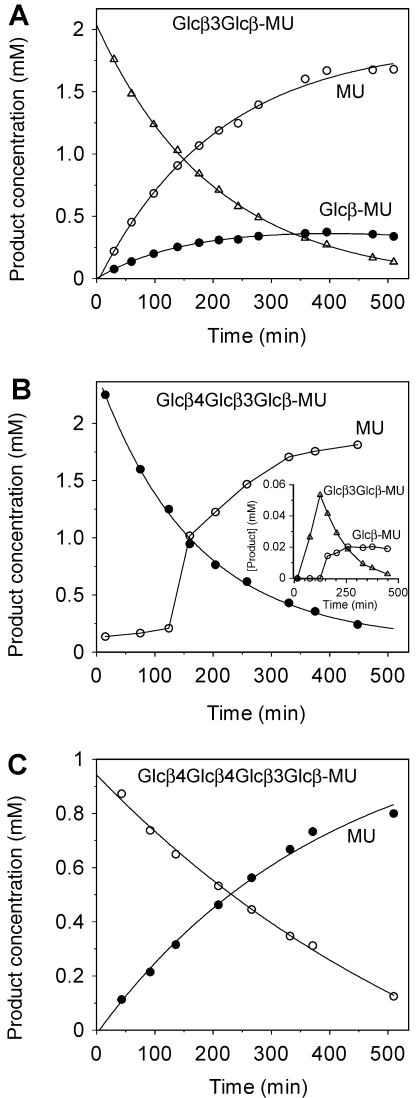
Figure 3.
Time course of the hydrolysis of 4-methylumbelliferyl β-glycosides catalyzed by 0.02 µM wild-type LamA from Pyrococcus furiosus in 100 mM phosphate buffer, pH 6.5, at 80 °C. (A) Substrate Glcβ3Glcβ-MU (1); (B) substrate Glcβ4Glcβ3Glcβ-MU (2); and (C) substrate Glcβ4Glcβ4Glcβ3Glcβ-MU (3).
Hydrolysis of 4-methylumbelliferyl laminaribioside (Glcβ3Glcβ-MU) (1) by LamA produced an MU concentration of up to 70% of the initial substrate concentration, and 15% Glcβ-MU (5) was formed after 400 min (10% Glcβ3Glcβ-MU substrate was not hydrolyzed). Therefore, we concluded that the laminaribioside (1) was hydrolyzed at the glycosidic bond with the aglycon and at the β-1,3 bond with a cleavage ratio of 4.5:1. The disaccharide isomer, Glcβ4Glcβ-MU (4), was not hydrolyzed by LamA, indicating that the enzyme does not accept β-1,4 linkages between the glucosyl units in subsites –1 and –2, but accepts β-1,3 linkages in this position. (Enzyme subsites are defined as the set of amino acid residues in the binding site (cleft) that interact with a monosaccharyl unit of the oligomeric substrate, and are numbered as –1, –2, ..., –n, for subsites on the non-reducing end, and +1, +2, ..., +n on the reducing end of the saccharide ligand from the site of cleavage (scissile glycosidic bond).)
With the Glcβ4Glcβ3Glcβ-MU (2) substrate (Figure 3B), Glcβ3Glcβ-MU (1) was initially formed (up to 3%), but was then hydrolyzed, and Glc-MU (1%) and MU (85%) were the final hydrolysis products after 400 min of reaction (10% of the substrate was not hydrolyzed). Because Glcβ4Glcβ-MU (4) was not hydrolyzed, the Glcβ-MU formed as the final product was unlikely to have been formed by cleavage of the β-1,3 bond of the trisaccharide, but was likely formed by hydrolysis of the initial disaccharide product 1. Formation of this transient disaccharide indicates that the enzyme can also hydrolyze the β-1,4 glycosidic bond of the β1,4-β-1,3-trisaccharide substrate. Finally, the tetrasaccharide Glcβ4- Glcβ4Glcβ3Glcβ-MU (3) was hydrolyzed with the release of MU (Figure 3C), and no other shorter chromophoric oligosaccharides were detected.
Even though LamA is an endo-glycosidase, as previously established for the enzyme-catalyzed hydrolysis of β-1,3-glucans (Gueguen et al. 1997), the cleavage pattern for Glcβ3Glcβ-MU (1) and Glcβ4Glcβ3Glcβ-MU (2) shows that it has some detectableexo-activity on simple oligosaccharides, with both β-1,4 and β-1,3 specificity. Comparison with other laminarinases indicates that this behavior is unique. However, it should be noted that studies on substrate specificity of laminarinases are not as extensive as those on lichenases from Family 16. LamR from Rhodothermus marinus has endo-activity on laminarin, but shows some exo-activity on oligosaccharides (Petersen et al. 2000, Borriss et al. 2003). Additionally, both β-1,3 and β-1,4 specificities have been reported for LamR; the enzyme hydrolyzes mixed-linked β-1,3-1,4 glucans with β-1,4 glycosidic bonds with higher specific activity than β-1,3 glucans. Moreover, activity on cello-oligosaccharides, but not on polysaccharides, has also been reported (Petersen et al. 2000). Hydrolysis of β-1,4 bonds of carboxymethyl cellulose has been observed with the putative laminarinases GluA and GluC from Lysobacter enzymogenes (Palumbo et al. 2003). However, the lichenase Lic16A from Clostridium thermocellum hydrolyzes lichenan and barley β-glucan, but with β-1,3 cleavage specificity (Fuchs et al. 2003). These examples illustrate that the specificities of laminarinases and lichenases in Family 16 of glycoside hydrolases cover a broad spectrum and require reclassification based on sequence similarity and substrate specificity criteria.
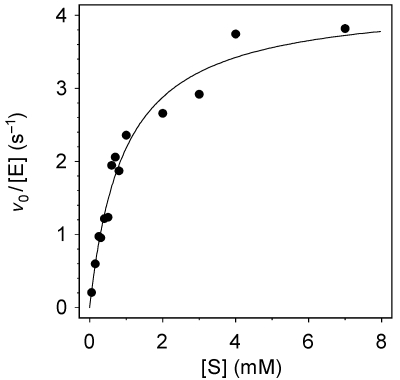
The substrate Glcβ3Glcβ-MU (1) was selected as the reference substrate for wild-type LamA glycosidase activity because it is preferentially hydrolyzed with release of the chromophoric aglycon, MU. The reaction follows Michaelis-Menten kinetics (Figure 4) with kcat = 4.22 ± 1.69 s–1 and KM = 0.93 ± 0.79 mM.
Figure 4.
Kinetics of hydrolysis of Glcβ3Glcβ-MU (1) catalyzed by 0.02 µM wild-type LamA from Pyrococcus furiosus in 100 mM phosphate buffer, pH 6.5, at 80 °C.Abbreviations: v0= initial velocity; [E] = enzyme concentration; and [S] = substrate concentration.
Hydrolase and glycosynthase activity of E170A
Residue Glu170 of Pyrococcus furiosus laminarinase is proposed to act as the catalytic nucleophile (Gueguen et al. 1997), based on sequence similarity with other Family 16 glycosyl hydrolases for which the catalytic residues have been experimentally identified (Juncosa et al. 1994, Viladot et al. 1998, Planas 2000). The Glu170 residue was replaced by alanine to produce the mutant LamA, E170A, to confirm the role of Glu170 as an essential catalytic residue in the hydrolytic mechanism, and to evaluate its potential as a glycosynthase.
The hydrolase activity of the E170A mutant was measured with laminarin and several 4-methylumbelliferyl β-glycosides as substrates, and compared with the hydrolase activity of the wild-type enzyme (Table 1). With laminarin, the E170A mutant retained only 0.05% activity relative to the wild-type enzyme, whereas a 104-fold reduction was obtained with the laminaribioside substrate (1). This effect is consistent with Glu170 being the catalytic nucleophile.
Table 1.
Hydrolase activity of wild-type and E170A LamA on laminarin and 4-methylumbelliferyl β-glycosides. Reactions took place at 80 °C in 100 mM phosphate buffer, pH 6.5, containing 0.5% laminarin or 4 mM MU-glycoside. Activity was determined as release of the chromophoric aglycon (MU = 4-methylumbelliferone).
| Substrate | Wild type | E170A |
| Laminarin | 832 U mg–1 | 0.46 U mg–1 |
| Glcβ3Glcβ-MU (1) | 3.75 s–1 | 4 × 10–4 s–1 |
| Glcβ4Glcβ3Glcβ-MU (2) | 0.41 s–1 | 1.5 × 10–4 s–1 |
| Glcβ4Glcβ-MU (4) | < 10–4 s–1 | < 10–4 s–1 |
| Glcβ-MU (5) | < 10–4 s–1 | < 10–4 s–1 |
Because a laminaribiosyl unit binds productively in subsite –2/–1 (as deduced from hydrolysis of Glcβ3Glcβ-MU (1) by wild-type LamA), α-laminaribiosyl fluoride (6) was used as the glycosyl donor to determine whether the E170A mutant has glycosynthase activity with different acceptors. Reactions were monitored by HPLC to detect MU-glycoside products. With Glcβ-MU (5) as the acceptor and a reaction temperature of 35 °C, a trisaccharide product (yield ≈ 2% after 3 h reaction) was obtained. When the reaction was performed at 50 °C, the same trisaccharide was produced with a maximum yield of 20% at 24 h (4.5% after 3 h reaction). The yield did not increase with longer incubation or with an increase in temperature to 76 °C, at which the enzyme is probably more active because of its thermophilic nature. The initial rate of condensation was higher at 76 °C (17.7 × 10–4 s–1) than at 50 °C (5.7 × 10–4 s–1), but the yield of condensation product did not exceed 10% at the higher temperature, most likely because of spontaneous hydrolysis of the α-glycosyl fluoride donor (Faijes et al. 2003). The reaction at 50 °C was performed on a semi-preparative scale (10 mg of donor, 0.6 mg enzyme, two equivalents of acceptor) (Table 2). Two main products were isolated after chromatographic separation of the reaction mixture. The first (20% yield, as determined by HPLC) was identified as Glcβ3Glcβ4Glcβ-MU (7) by NMR spectroscopy: a new signal at δ 4.60 ppm (J = 8.1 Hz), characteristic of H-1 for a β-1,4 glycosidic bond, was observed in the 1H NMR spectrum, whereas the 13C NMR spectrum showed the 4I and 3II carbons involved in glycosidic linkages at δ 78.8 and 84.7 ppm, respectively. This product was identical to that reported for the glycosynthase reaction of the E134A mutant of Bacillus licheniformis endo-1,3-1,4-β-glucanase with the same donor and acceptor molecules (Malet and Planas 1998). A second product (3% yield as determined by HPLC) was isolated; it was also a trisaccharide, tentatively assigned as Glcβ3Glcβ3Glcβ-MU (8), because addition of wild-type LamA to the glycosynthase reaction mixture yielded Glcβ3Glcβ-MU (1) as well as Glcβ4Glcβ-MU (4), resulting from hydrolysis of 8 and 7, respectively.
Table 2.
Glycosynthase-catalyzed condensations of α-laminaribiosyl fluoride donor (6) with acceptors 5 and 1. Conditions: 100 mM phosphate buffer, pH 6.5, 50 °C, [donor] = 1 mM, [acceptor] = 5 mM, [enzyme] = 7.5 µM. Yields were determined by HPLC after a 24-h reaction. Boldface numbers indicate the linkage formed in the condensation reaction.
| Acceptor | Product | Yield (%) |
| Glcβ-MU (5) | Glcβ3Glcβ4Glcβ-MU (7) | 20 |
| Glcβ3Glcβ3Glcβ-MU (8) | 3 | |
| Glcβ3Glcβ-MU (1) | Glcβ3Glcβ4Glcβ3Glcβ-MU (9) | 2 |
| Glcβ3Glcβ3Glcβ3Glcβ-MU (10) | 25 | |
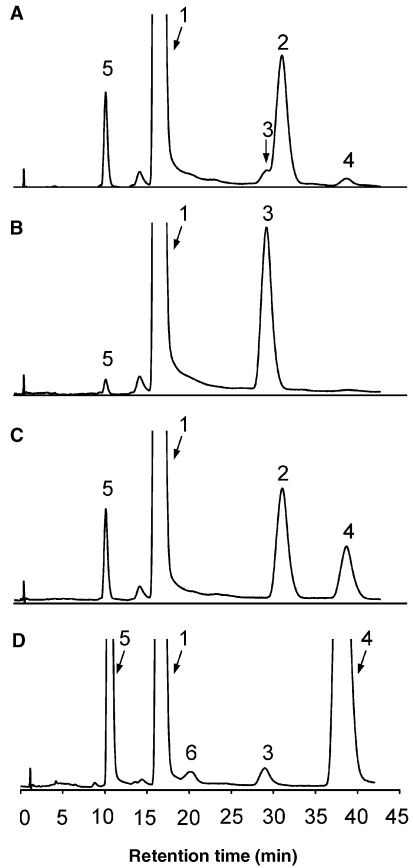
The reaction with E170A and Glcβ3Glcβ-MU (1) acceptor was also analyzed at 50 °C (Table 2). Two tetrasaccharide products were detected by HPLC with an overall initial rate (v0/[E]) of 16.1 × 10–2 s–1 and 27% yield after 10 h (Figure 5A). To elucidate the regioselectivity of the glycosynthase reaction, the mixture was digested with two glycosidases: LamA (1,3-β-glucanase) and LicA (endo-1,3-1,4-β-glucanase from B. licheniformis), which only hydrolyzes β-1,4 glycosidic bonds on 3-O-substituted glucosyl units (Planas 2000). Analysis by HPLC revealed that LicA degraded only the minor tetrasaccharide product of the glycosynthase reaction (Figure 5C), indicating that the newly formed glycosidic bond was β-1,4 (Glcβ3Glcβ4Glcβ3Glcβ-MU (9)). In contrast, LamA degraded the major tetrasaccharide product (Figure 5D), which was then assigned to Glcβ3Glcβ3Glcβ3Glcβ-MU (10). The structure of both tetrasacharides was confirmed by 1H and 13C NMR spectroscopy. Tetrasaccharide 9 was identical to the product reported for the glycosynthase reaction of the E134A mutant 1,3-1,4-β-glucanase (Faijes et al. 2003). The major tetrasaccharide 10 (25% yield) was unambiguously identified: the three signals corresponding to H-1 in inter-glucosidic bonds were in the δ 4.76–4.87 ppm region in the 1H NMR spectrum (characteristic of β-1,3 bonds), whereas the 13C NMR spectrum showed that three C-3 carbons were involved in glycosidic bonds (δ 84.4–84.8 ppm).
Figure 5.
The HPLC analysis of reaction products. (A) Glycosynthase reaction of E170A LamA with α-laminaribiosyl fluoride (6) and Glcβ3Glcβ-MU (1); (B) glycosynthase reaction of E134A (endo-β-1,3-1,4-glucanase from B. licheniformis) with α-laminaribiosyl fluoride (6) and Glcβ3βGlcβ-MU (1); (C) digestion of reaction mixture A with wild-type endo-β-1,3-1,4-glucanase from B. licheniformis; (D) digestion of reaction mixture A with wild-type LamA. Peaks in the chromatogram were identified by coinjection with independed standards. Peak labels: 1. Glcβ3Glc-MU (1) with retention time, tR = 17.2 min; 2. Glcβ3Glcβ3Glcβ3Glc-MU (10), tR = 31.1 min; 3. Glcβ3Glcβ4Glcβ3Glc-MU (9), tR = 29.5 min; 4. MU, tR = 38.8 min; 5. Glcβ-MU (5), tR = 10.5 min; and 6. unidentified hydrolysis product, tR = 20.1 min.
In conclusion, LamA hydrolyzed mixed-linked oligosaccharides, confirming that the laminaribiose moiety is the preferred substrate unit; however, the enzyme also showed exo-activity, hydrolyzing both β-1,3 and β-1,4 linkages. The E170A LamA mutant possessed glycosynthase activity, catalyzing the condensation of α-laminaribiosyl fluoride (6) donor with Glcβ-MU (5) and Glcβ3Glcβ-MU (1) as acceptors. The reaction was not regiospecific because both β-1,3 and β-1,4 glycosidic bonds were formed; with the monosaccharide acceptor 5, a 7:1 ratio of β-1,4 to β-1,3 was obtained, whereas a 1:17 ratio was observed with the disaccharide acceptor 1. These results contrast those reported for a Family 17 1,3-β-glucanase from barley (Hrmova et al. 2002), in which the corresponding glycosynthase mutant (E231A) is regiospecific for β-1,3 glycosidic linkages. Family 16 laminarinases seem to include a wider range of specificities. Transglycosylation catalyzed by wild-type laminarinases from Oerskovia sp. and Spisula sachalinensis exclusively produced β-1,3 linkages (Borriss et al. 2003), whereas LamR from R. marinus catalyzed the synthesis of both β-1,3 and β-1,4 bonds (Krah et al. 1998). The efficiency of E170A LamA as a glycosynthase is rather poor, at least with α-glycosyl fluoride as the activated donor, with preparative yields not exceeding 30% in condensation products (yields not optimized). This is probably a consequence of the hyperthermophilic nature of LamA from Pyrococcus furiosus. The maximum temperature for hydrolase activity of wild-type LamA is 100–105 °C, and activity drops to 10% at 50 °C (Gueguen et al. 1997). The α-glycosyl fluoride donor is labile at high temperatures and is readily hydrolyzed above 50 °C. Therefore, the temperature used in this study is a compromise between donor stability and enzymatic activity that renders conditions far from optimal.
Acknowledgments
This research was supported by the Technology Foundation (STW), applied science division of NWO and the technology program of the Ministry of Economic Affairs, The Netherlands, and grant BIO2001-2064-C02-02 from the Ministry of Science and Technology, Spain.
References
- R1.Boon M.A., van der Oost J., de Vos W.M., Jansen A.E.M. Synthesis of oligosaccharides catalyzed by thermostable β-glucosidase from Pyrococcus furiosus . Appl. Biochem. Biotechnol. 1998;75:269–278. [Google Scholar]
- R2.Borriss R., Krah M., Brumer H., et al. Enzymatic synthesis of 4-methylumbelliferyl (1→3)-β-D-glucooligosaccharides. New substrates for β-1,3-1,4-D-glucanase. Carbohydr. Res. 2003;338:1455–1467. doi: 10.1016/s0008-6215(03)00199-x. [DOI] [PubMed] [Google Scholar]
- R3.Chiaraluce R., van der Oost J., Lebbink J.H., Kaper T., Consalvi V. Persistence of tertiary structure in 7.9 M guanidinium chloride: the case of endo-β-1,3-glucanase from Pyrococcus furiosus. Biochemistry. 2002;41:14624–14632. doi: 10.1021/bi026498u. [DOI] [PubMed] [Google Scholar]
- R4.Driskill L.E., Bauer M.W., Kelly R.M. Synergistic interactions among β-laminarinase, β-1,4-glucanase, and β-glucosidase from the hyperthermophilic Archaeon Pyrococcus furiosus during hydrolysis of β-1,4-, β-1,3-, and mixed-linked polysaccharides. Biotechnol. Bioeng. 1999;66:51–60. [PubMed] [Google Scholar]
- R5.Faijes M., Fairweather J.K., Driguez H., Planas A. Oligosaccharide synthesis by coupled endo-glycosynthases of different specificity: a straightforward preparation of two mixed-linkage hexasaccharide substrates of 1,3/1,4-β-glucanases. Chemistry. 2001;7:4651–4655. doi: 10.1002/1521-3765(20011105)7:21<4651::aid-chem4651>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- R6.Faijes M., Perez X., Perez O., Planas A. Glycosynthase activity of Bacillus licheniformis 1,3-1,4-β-glucanase mutants: specificity, kinetics, and mechanism. Biochemistry. 2003;42:13304–13318. doi: 10.1021/bi030131n. [DOI] [PubMed] [Google Scholar]
- R7.Fort S., Boyer V., Greffe L., Davies G., Moroz O., Christiansen L., Schulein M., Cottaz S., Driguez H. Highly efficient synthesis of β(1→4)-oligo- and polysaccharides using a mutant cellulase. J. Am. Chem. Soc. 2000;122:5429–5437. [Google Scholar]
- R8.Fuchs K.P., Zverlov V.V., Velikodvorskaya G.A., Lottspeich F., Schwarz W.H. Lic16a of Clostridium thermocellum, a non-cellulosomal, highly complex endo-β-1,3-glucanase bound to the outer cell surface. Microbiology. 2003;149:1021–1031. doi: 10.1099/mic.0.26153-0. [DOI] [PubMed] [Google Scholar]
- R9.Gill S.C., Von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- R10.Gueguen Y., Voorhorst W.G., van der Oost J., de vos W.M. Molecular and biochemical characterization of an endo-β-1,3-glucanase of the hyperthermophilic Archaeon Pyrococcus furiosus . J. Biol. Chem. 1997;272:31258–31264. doi: 10.1074/jbc.272.50.31258. [DOI] [PubMed] [Google Scholar]
- R11.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- R13.Hrmova M., Imai T., Rutten S.J., Fairweather J.K., Pelosi L., Bulone V., Driguez H., Fincher G.B. Mutated barley (1,3)-β-D-glucan endohydrolases synthesize crystalline (1,3)-β-D-glucans. J. Biol. Chem. 2002;277:30102–30111. doi: 10.1074/jbc.M203971200. [DOI] [PubMed] [Google Scholar]
- R14.Jahn M., Stoll D., Warren R.A.J., Szabo L., Singh P., Gilbert H.J., Ducros V.M., Davies G.J., Withers S.G. Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides. Chem. Commun. 2003;2003:1327–1329. doi: 10.1039/b302380j. [DOI] [PubMed] [Google Scholar]
- R15.Jakeman D.L., Withers S.G. On expanding the repertoire of glycosynthases: mutant β-galactosidases forming β-(1,6)-linkages. Can. J. Chem. 2002;80:866–870. [Google Scholar]
- R16.Juncosa M., Pons J., Dot T., Querol E., Planas A. Identification of active site carboxylic residues in Bacillus licheniformis endo-1,3-1,4-β-D-glucan 4-glucanohydrolase by site-directed mutagenesis. J. Biol. Chem. 1994;269:14530–14535. [PubMed] [Google Scholar]
- R17.Kaper T., Verhees C.H., Lebbink J.H., et al. Characterization of β-glycosyl hydrolases from Pyrococcus furiosus . Methods Enzymol. 2001;330:329–346. doi: 10.1016/s0076-6879(01)30386-5. [DOI] [PubMed] [Google Scholar]
- R18.Krah M., Misselwitz R., Politz O., Thomsen K.K., Welfle H., Borriss R. The laminarinase from thermophilic eubacterium Rhodothermus marinus. Conformation, stability, and identification of active site carboxylic residues by site-directed mutagenesis. Eur. J. Biochem. 1998;257:101–111. doi: 10.1046/j.1432-1327.1998.2570101.x. [DOI] [PubMed] [Google Scholar]
- R19.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- R20.Mackenzie L.F., Wang Q., Warren R.A.J., Withers S.G. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 1998;120:5583–5584. [Google Scholar]
- R21.Malet C., Planas A. Mechanism of Bacillus 1,3-1,4-β-D-glucan 4-glucanohydrolases: kinetics and pH studies with 4-methylumbelliferyl β-D-glucan oligosaccharides. Biochemistry. 1997;36:13838–13848. doi: 10.1021/bi9711341. [DOI] [PubMed] [Google Scholar]
- R22.Malet C., Planas A. From β-glucanase to β-glucansynthase: glycosyl transfer to α-glycosyl fluorides catalyzed by a mutant endoglucanase lacking its catalytic nucleophile. FEBS Lett. 1998;440:208–212. doi: 10.1016/s0014-5793(98)01448-3. [DOI] [PubMed] [Google Scholar]
- R23.Malet C., Viladot J.L., Ochoa A., Gallego B., Brosa C., Planas A. Synthesis of 4-methylumbelliferyl-β-D-glucan oligosaccharides as specific chromophoric substrates of (1→3), (1→4)-β-D-glucan 4-glucanohydrolases. Carbohydr. Res. 1995;274:285–301. doi: 10.1016/0008-6215(95)00102-y. [DOI] [PubMed] [Google Scholar]
- R24.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. [Google Scholar]
- R25.Moracci M., Trincone A., Rossi M. Glycosynthases: new enzymes for oligosaccharide synthesis. J. Mol. Catal. B Enzym. 2001;11:155–163. [Google Scholar]
- R26.Nashiru O., Zechel D.L., Stoll D., Mohammadzadeh T., Warren R.A., Withers S.G. β-Mannosynthase: synthesis of β-mannosides with a mutant β-mannosidase. Angew. Chem. Int. 2001;40:417–420. doi: 10.1002/1521-3773(20010119)40:2<417::AID-ANIE417>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- R27.Okuyama M., Mori H., Watanabe K., Kimura A., Chiba S. α-Glucosidase mutant catalyzes “α-glycosynthase”-type reaction. Biosci. Biotechnol. Biochem. 2002;66:928–933. doi: 10.1271/bbb.66.928. [DOI] [PubMed] [Google Scholar]
- R28.Palumbo J.D., Sullivan R.F., Kobayashi D.Y. Molecular characterization and expression in Escherichia coli of three β-1,3-glucanase genes from Lysobacter enzymogenes Strain N4-7. J. Bacteriol. 2003;185:4362–4370. doi: 10.1128/JB.185.15.4362-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Perugino G., Trincone A., Giordano A., van der Oost J., Kaper T., Rossi M., Moracci M. Activity of hyperthermophilic glycosynthases is significantly enhanced at acidic pH. Biochemistry. 2003;42:8484–8493. doi: 10.1021/bi0345384. [DOI] [PubMed] [Google Scholar]
- R30.Petersen B.O., Krah M., Duus J.O., Thomsen K.K. A transglycosylating 1,3(4)-β-glucanase from Rhodothermus marinus. NMR analysis of enzyme reactions. Eur. J. Biochem. 2000;267:361–369. doi: 10.1046/j.1432-1327.2000.01008.x. [DOI] [PubMed] [Google Scholar]
- R31.Planas A. Bacterial 1,3-1,4-β-glucanases: structure, function and protein engineering. Biochim. Biophys. Acta. 2000;1543:361–382. doi: 10.1016/s0167-4838(00)00231-4. [DOI] [PubMed] [Google Scholar]
- R32.Planas A., Faijes M. Glycosidases and glycosynthases in enzymatic synthesis of oligosaccharides. An overview. Afinidad. 2002;59:295–313. [Google Scholar]
- R33.Trincone A., Perugino G., Rossi M., Moracci M. A novel thermophilic glycosynthase that effects branching glycosylation. Bioorg. Med. Chem. Lett. 2000;10:365–368. doi: 10.1016/s0960-894x(99)00700-3. [DOI] [PubMed] [Google Scholar]
- R34.van Tilbeurgh H., Claeyssens M., De Bruyne C.K. The use of 4-methylumbelliferyl and other chromophoric glycosides in the study of cellulolytic enzymes. FEBS Lett. 1982;149:152–156. [Google Scholar]
- R35.Viladot J.L., De Ramon E., Durany O., Planas A. Probing the mechanism of Bacillus 1,3-1,4-β-D-glucan 4-glucanohydrolases by chemical rescue of inactive mutants at catalytically essential residues. Biochemistry. 1998;37:11332–11342. doi: 10.1021/bi980586q. [DOI] [PubMed] [Google Scholar]
- R36.Williams S.J., Withers S.G. Glycosynthases: mutant glycosidases for glycoside synthesis. Aust. J. Chem. 2002;55:3–12. [Google Scholar]