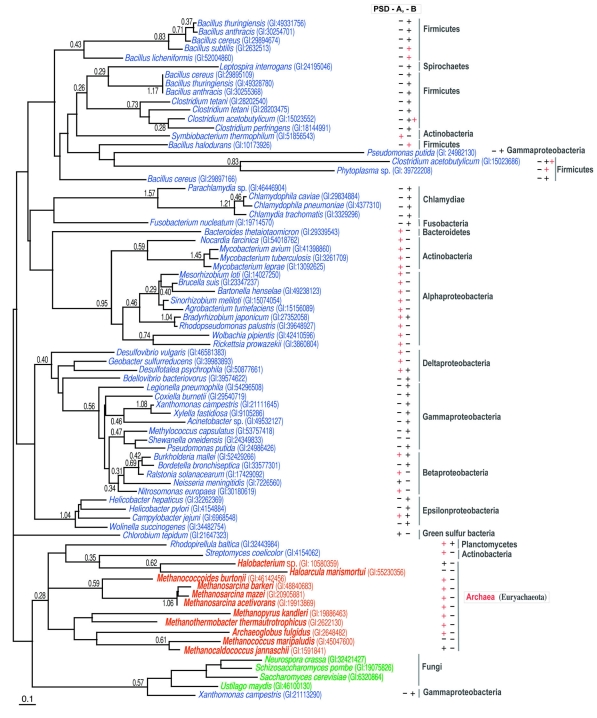
Figure 3.
An unrooted maximum likelihood (ML) tree of phosphatidylserine synthase/archaetidylserine synthase (PSS/ASS). Each sequence is indicated by the source name and the GI number from the National Center for Biotechnology Information. The archaeal, bacterial and eukaryotic proteins are colored red, blue and green, respectively. The reliability index of a cluster is only shown at the root node of the cluster when the value is equal to or greater than 0.25. The log likelihood (lnL) of the ML tree was –12209.94 and the optimal shape parameter alpha for gamma distribution was 0.84 (129 sites were compared). The bar under the tree corresponds to 0.1 amino acid substitutions/site. The presence or absence of phosphatidylserine decarboxylase-A or -B (PSD-A or PSD-B) in each species is indicated by a symbol, ‘+’ or ‘–’, after the source name. The symbol, ‘+’, colored red, shows that the genes or open reading frames for PSS/ASS and PSD/archaetidylserine decarboxylase (ASD) are tandemly encoded in the genome.