Abstract
A gene encoding for a putative Family inorganic pyrophosphatase (PPase, EC 3.6.1.1) from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 was cloned and the biochemical characteristics of the resulting recombinant protein were examined. The gene (Accession No. 1907) from P. horikoshii showed some identity with other Family I inorganic pyrophosphatases from archaea. The recombinant PPase from P. horikoshii (PhPPase) has a molecular mass of 24.5 kDa, determined by SDS-PAGE. This enzyme specifically catalyzed the hydrolysis of pyrophosphate and was sensitive to NaF. The optimum temperature and pH for PPase activity were 70 °C and 7.5, respectively. The half-life of heat inactivation was about 50 min at 105 °C. The heat stability of PhPPase was enhanced in the presence of Mg2+. A divalent cation was absolutely required for enzyme activity, Mg2+ being most effective; Zn2+, Co2+ and Mn2+ efficiently supported hydrolytic activity in a narrow range of concentrations (0.05– 0.5 mM). The Km for pyrophosphate and Mg2+ were 113 and 303 µM, respectively; and maximum velocity, Vmax, was estimated at 930 U mg–1.
Keywords: archaea, hyperthermophile
Introduction
Inorganic pyrophosphatases (PPases, EC 3.6.1.1) are widely distributed in all living cells and catalyze the hydrolysis of pyrophosphate (PPi) into two orthophosphates (Pi) (Chen et al. 1990, Lundin et al. 1991). In many organisms, the bulk of PPase activity is soluble, localized in the cytosol and serves mainly to remove the PPi formed in biosynthetic reactions driven by nucleotide triphosphates (Kornberg 1962). Pyrophosphatases from the Bacteria, Archaea and Eukarya have been characterized. Bacterial and archaeal PPases exist as either homotetramers or homohexamers, with a subunit molecular mass of about 20 kDa. In contrast, eukaryotic PPases form mostly homodimers, with a subunit molecular mass of about 28–35 kDa. To date, the soluble PPases have been classified into Family I and Family II PPases (Shintani et al. 1998, Young et al. 1998). Family I PPases have a highly conserved active site structure formed by 14–16 amino acid residues and three to four Mg2+ ions and have similar catalytic properties (Harutyunyan et al. 1996, Heikinheimo et al. 1996, Baykov et al. 1999). Family II PPases are activated by Mn2+ and display 10–20 times greater activity than Family I PPases (Shintani et al. 1998, Parfenyev et al. 2001). Enzymes derived from microorganisms growing at extreme temperatures are of biotechnological use as highly thermostable biocatalysts. Thermostable PPases are commonly used to suppress pyrophosphorolysis in DNA sequencing reactions with DNA polymerases (Tabor and Richardson 1990, Vander Horn et al. 1997). Some extremely thermostable PPases have been isolated and characterized from archaea (Sulfolobus (Wakagi et al. 1992), Methanobacterium (van Alebeek et al. 1994), Methanococcus (Kuhn et al. 2000), Thermus (Satoh et al. 1998) and Thermoplasma (Richter and Schafer 1992)). However, no information is available on the biochemical properties of thermostable PPase from hyperthermophilic archaeon Pyrococcus spp. In the genome database of the hyperthermophilic archaeon Pyrococcus horikoshii OT3, we found the homologous PPase gene. The putative PhPPase gene has been classified as a member of the Family I PPases (Young et al. 1998, Shintani et al. 1998), and the structure of P. horikoshii PPase has already been reported (Liu et al. 2004). In this study, we report on the cloning of P. horikoshii PPase and compare the properties of the recombinant PhPPase with those of other microbial PPases.
Materials and methods
Materials
The DNA primers and substrates were prepared by FASMA (Kanagawa, Japan). The plasmid pET-21a was purchased from Novagen (Madison, WI, USA). The KOD DNA polymerase was purchased from Toyobo (Osaka, Japan). We obtained ATP, ADP and AMP from Sigma (St. Louis, MO). Sodium tripolyphosphate (PPPi), sodium pyrophosphate (PPi), sodium phosphate (Pi), phosphoenolpyruvate (PEP) and p-nitrophenyl phosphate (pNP) were obtained from Wako Pure Chemical Industries (Tokyo, Japan).
Enzyme construction and expression
Chromosomal DNA of P. horikoshii OT3 was prepared as reported previously (Gonzalez et al. 1998). The gene (PH1907) was amplified by PCR using the chromosomal DNA as the template and two primers, PP1 (forward): 5′-AGGGGGTACATATGAACCCGTTCCACGACCT-3′; and PP2 (reverse): 5′-AATAAAGTCGACTCACTCCTTCTTGCCGA-3′, which was designed based on an open reading frame (ORF) sequence for a protein of 178 amino acids. Amplification by PCR was carried out with KOD DNA polymerase at 94 °C for 30 s, 55 °C for 2 s, 74 °C for 30 s for 30 cycles. An amplified 0.53-kbp DNA fragment was digested with restriction enzymes NdeI and SalI (the NdeI and SalI sites are underlined in primers PP1 and PP2, respectively) and inserted into the pET-21a vector. The nucleotide sequences of the inserted genes were confirmed with an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer, Wellesley, MA). Escherichia coli BL21(DE3) cells containing the expression plasmid were grown in 2YT medium (1% yeast extract, 1.6% tryptone, 0.5% NaCl) containing ampicillin (100 µg ml–1) to an OD600 of approximately 0.6. Gene expression was then induced with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 4 h at 37 °C.
Purification of the recombinant proteins
Escherichia coli cells containing the expressed recombinant enzyme were centrifuged and frozen at −70 °C. The thawed cells were then disrupted by sonication in buffer A (50 mM Tris-HCl, pH 8.0, 15 mM MgCl2, 0.1 mM EDTA). The suspension of disrupted cells was centrifuged at 27,000 g for 30 min and the supernatant fraction was heat-treated at 85 °C for 30 min, followed by recentrifugation. The supernatant was loaded on a HiTrap Q column (Amersham Biosciences, Piscataway, NJ) equilibrated in buffer A, and the bound protein was eluted with a linear gradient of NaCl (0 to 1.0 M in the same buffer). The protein solution was concentrated using a centricon 10 filter from Amicon (Millipore, Bedford, MA) and dialyzed against buffer B (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 15 mM MgCl2). The dialyzed solution was loaded on a HiPrep Sephacryl S-200 HR 26/60 column (Amersham Biosciences) and eluted with buffer B. The purity of the recombinant protein was assessed by SDS-PAGE. Protein concentration was determined using a Bio-Rad protein assay system (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
Enzyme assay
In the standard assay, PPase activity was determined by the formation of inorganic phosphate at 60 °C in the reaction mixture containing 50 mM Tris-HCl (pH 7.5 at 60 °C), 5 mM MgCl2 and 1 mM sodium phosphate. The reaction was started by the addition of enzyme, and 100-µl samples were taken at 30-s time intervals and put on ice. The phosphate present was determined by adding 1 ml of phosphate reagent (1% ammonium heptamolybdate (w/v), 0.83 M sulfuric acid, 8% iron (II) sulfate (w/v)) and the absorbance of the phosphomolybdate complex was measured at 660 nm after 30 min (van Alebeek et al. 1994). Activities were developed with 0.1 µmol of standard sodium phosphate. The activity towards other substrates such as sodium tripolyphosphate (PPPi), phosphoenolpyruvate (PEP), p-nitrophenyl phosphate (pNP), ATP, ADP and AMP was determined by the same method as above except that PPi was replaced by each substrate. One unit of activity is defined as the amount of enzyme required to hydrolyze 1 µmol PPi per min at 60 °C.
Results and discussion
Identification of inorganic pyrophosphatase gene
In the Pyrococcus horikoshii OT3 genome database (http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID= ot3_ G1) (Kawarabayasi et al. 1998), we identified an ORF (Accession No. PH1907) encoding a putative inorganic pyrophosphatase (PhPPase). The putative PhPPase gene encodes a protein of 178 amino acids, with a predicted molecular mass of 20,833 Da and an isoelectric point of 4.81. From the data of sequence homology (Shintani et al. 1998, Young et al. 1998), PhPPase is classified as a member of Family I PPase. The deduced amino acid sequence showed a high identity with other archaeal PPases from Pyrococcus furiosus (94%) (Robb et al. 2001), Methanopyrus kandleri (69%) (Slesarev et al. 2002), Methanobacterium thermoautotrophicum (61%) (Smith et al. 1997) and Thermoplasma acidophilum (54%) (Richter and Schafer 1992). Structural and multiple sequence aligments reveal a group of 14 conserved, mostly polar, amino acid residues (Kankare et al. 1994). The 14 amino acids have been found to be identical in all other PhPPase amino acid sequences. The ORF (PH1907) was amplified by PCR from a P. horikoshii genomic DNA, cloned and sequenced to confirm the sequences in the database. The gene was expressed in E. coli cells, and the recombinant protein was purified as described in the Materials and methods. Expression and subsequent purification yielded 3.2 mg of PhPPase from a 1-l culture.
Molecular properties
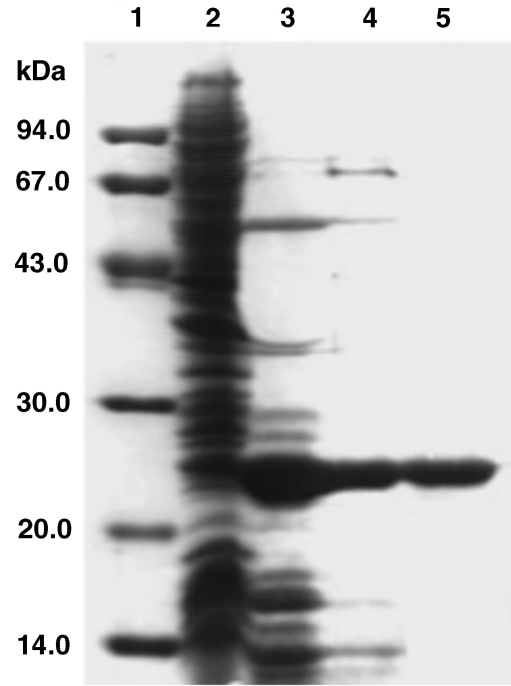
The molecular mass of the protein from PH1907 was estimated to be about 24.5 kDa by SDS-PAGE (Figure 1). This value is inconsistent with the size (20,833 Da) calculated from the amino acid sequence. This discrepance may be explained by the many positively charged amino acid residues of PhPPase. Recently, a three-dimensional crystal structure of PhPPase showed that the enzyme is an oligomeric hexamer (Liu et al. 2004).
Figure 1.
Purification of recombinant Pyrococcus horikoshii pyrophosphatase (PhPPase). Lane 1, molecular mass markers; Lane 2, crude extract of induced cells; Lane 3, supernatant of crude extract after heat treatment at 85 °C for 30 min; Lane 4, HiTrap Q column peak fractions; and Lane 5, Sephacryl S-200 HR 26/60 column peak fractions. The gel was stained with Coomassie brilliant blue.
Catalytic properties
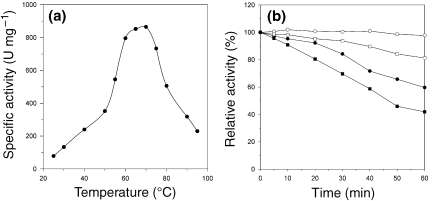
We measured the activity of the recombinant PhPPase in the presence of 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2 and 1 mM sodium pyrophosphate. The recombinant PhPPase showed maximal activity in the temperature range from 60 to 75 °C, with a temperature optimum of 70 °C (Figure 2a). The optimal pH for the PhPPase was 7.5 at 60 °C. Heat inactivation of the enzyme was estimated by measuring the residual PPase activity after heat treatment at two different temperatures in the presence or absence of 5 mM Mg2+ (Figure 2b). The recombinant PhPPase showed high thermostability, and the half-life of heat inactivation was about 50 min at 105 °C. The heat stability of PhPPase was much higher than that reported previously for other thermostable PPases from Aquifex aeolicus (90 min at 95 °C) (Hoe et al. 2001) and Sulfolobus acidocaldarius (150 min at 95 °C) (Hansen et al. 1999). Furthermore, the heat stability of PhPPase was enhanced by the addition of Mg2+ to the incubation mixture, as reported for the other PPases (Hachimori et al. 1979, Ichiba et al. 1988). The difference between the optimum temperature of PhPPase and the optimum growth temperature of P. horikoshii suggests that PhPPase requires co-factors for its thermophilicity in cells.
Figure 2.
Thermophilicity and thermostability of Pyrococcus horikoshii pyrophosphatase (PhPPase). (a) The PPase activity was determined at the indicated temperatures, as described in the Materials and methods. Negative control reactions in the absence of the enzyme were performed in parallel. (b) Enzyme (1 µM PhPPase in 50mM Tris-HCl, pH 7.5) was incubated at 100 °C in the presence (open circles) or absence (filled circles) of Mg2+ and at 105 °C in the presence (open squares) or absence (filled squares) of Mg2+, and the residual PPase activity of samples was measured at 60 °C.
Divalent cation requirement
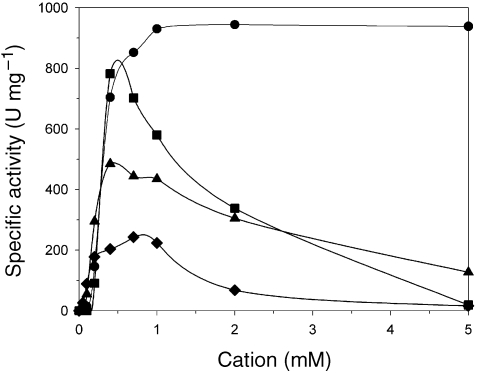
Family I PPase has shown strong metal cation-dependency, with Mg2+ conferring the highest efficiency (Cooperman 1982) and sensitivity to inhibition by Ca2+ (Yang and Wensel 1992). In contrast, Family II PPase prefers Mn2+ overMg2+ (Parfenyev et al. 2001). Pyrocococcus horikoshii pyrophosphatase is dependent on the presence of divalent cations for catalytic activity, with the highest activity in the presence of Mg2+. Other cations (Zn2+ Co2+ Mn2+) could efficiently replace Mg2+, but the effectiveness of the latter cations is limited to a narrow range of concentrations (0.05–0.5 mM) (Figure 3). Neither Ni2+ nor Ca2+ could activate the enzyme for catalysis. These results support the conclusion that PhPPase belongs to the Family I PPases. The enzyme responded sensitively to changes in the cation concentration of the reaction mixture. In particular, a drastic decrease in enzyme activity was observed at Zn2+, Co2+ and Mn2+ concentrations above 0.5 mM (Figure 3). The inhibition at high cation concentrations is probably due to the presence of free cations in the assay (Celis and Romero 1987).
Figure 3.
Effect of divalent cation concentration on pyrophosphate (PPi) hydrolysis. Assay conditions were as follows: 50mM Tris-HCl, pH 7.5, 1 mM sodium pyrophosphate (NaPPi), and concentrations of divalent cations were varied as indicated. Symbols: circles = Mg2+; squares = Zn2+; triangles = Co2+; and diamonds = Mn2+.
Substrate specificity
The substrate specificity of PhPPase was investigated (Table 1) and found to be high with respect to PPi. The release activities of inorganic phosphate (Pi) from PPPi, ATP and ADP were determined to be 2.7, 5.9 and 2.9% of that from PPi, respectively. Hydrolysis of AMP, PEP and pNP by PhPPase was not observed. The reaction rate of PhPPase at different PPi and Mg2+ concentrations did not follow normal Michaelis-Menten kinetics. Therefore, kinetic data were analyzed according to the procedure of Hill by the EZ-Fit program (Perrella 1988). At pH 7.5 and 60 °C, the values of Km and maximum velocity (Vmax) for PPi were determined to be 113 ± 12 µM and 930 ± 40 U mg–1, respectively, which corresponds to a kcat of 744 s–1. An apparent Km for Mg2+ was determined to be 303 ± 9 µM.
Table 1.
Substrate specificity of the Pyroccocus horikoshii pyrophosphatase (PhPPase). Activities were determined as described in Materials and methods. Each substrate was tested at a concentration of 1 mM in the presence of 5 mM Mg2+.
| Substrate | Relative activity (%) |
| PPi | 100 |
| PPPi | 2.7 |
| ATP | 5.9 |
| ADP | 2.9 |
| AMP | 0 |
| PEP | 0 |
| pNP | 0 |
Effect of fluoride
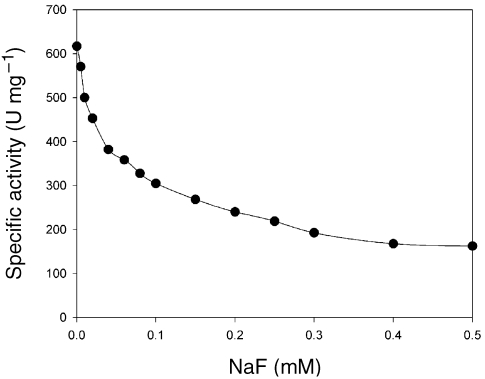
The influence of fluoride, a potent inhibitor of PPases, was examined. A plot of activity for NaF concentration (Figure 4) indicated that 0.5 mM NaF inhibits PPase activity by 75%. The I50 (concentration for 50% inhibition) was 0.1 mM. The fluoride sensitivity of PhPPase is higher than that of bacterial and archaeal PPases: E. coli (91%, 1 mM) (Josse 1966), Thiobacillus thiooxidans (81%, 1.25 mM) (Tominaga and Mori 1977), Rhodopseudomonas palustris (50%, 5 mM) (Schwarm et al. 1986) and Methanobacterium thermoautotrophicum (50%, 0.9 mM) (van Alebeek et al. 1994). In conclusion, we have identified a gene that codes for PPase protein from the P. horikoshii genome and also compared the properties of recombinant PhPPase with those of other PPases. The PhPPase is more similar to bacterial and archaeal enzymes than to eukaryotic enzymes, based on the molecular mass of a single subunit. Cytoplasmic PPase has no function in energy conservation. Its function seems to keep the cellular PPi concentration low, through the hydrolysis of PPi. Whether PhPPase is located in the cytoplasm has not been reported. This work increased our understanding of the metabolism of a hyperthermophilic archaeon. The hyperthermostability of PhPPase suggests that it can be used to suppress pyrophosphorolysis in sequencing reactions using thermostable DNA polymerases (Tabor and Richardson 1990, Vander Horn et al. 1997).
Figure 4.
Inhibition of PhPPase activity by NaF. The specific activity was determined at the indicated NaF concentrations, as described in Materials and methods.
Acknowledgments
This work was performed as part of the fellowship program supported by the New Energy Industrial Technology Development Organization (NEDO).
References
- R1.Baykov A.A., Cooperman B.S., Goldman A., Lahti R. Cytoplasmic inorganic pyrophosphatase. In: Schroder H.C., editor. Inorganic Polyphosphates. Berlin: Vol. 23. Springer-Verlag; 1999. pp. 127–150. [DOI] [PubMed] [Google Scholar]
- R2.Celis H., Romero I. The phosphate-pyrophosphate exchange and hydrolytic reactions of the membrane-bound pyrophosphatase of Rhodospirillum rubrum: effects of pH and divalent cations. J. Bioenerg. Biomembr. 1987;19:255–272. doi: 10.1007/BF00762416. [DOI] [PubMed] [Google Scholar]
- R3.Chen J., Brevet A., Fromant M., Lévêque F., Schmitter J.M., Blanquet S., Plateau P. Pyrophosphatase is essential for growth of Escherichia coli . J. Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Cooperman B.S. The mechanism of action of yeast inorganic pyrophosphatase. Methods Enzymol. 1982;87:526–548. doi: 10.1016/s0076-6879(82)87030-4. [DOI] [PubMed] [Google Scholar]
- R5.Gonzalez J.M., Masuchi Y., Robb F.T., Ammerman J.W., Maeder D.J., Yanagibayashi M., Tamaoka J., Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- R6.Hachimori A., Shiroya Y., Hirato A., Miyahara T., Samejima T. Effects of divalent cations on thermophilic inorganic pyrophosphatase. J. Biochem. 1979;86:121–130. [PubMed] [Google Scholar]
- R7.Hansen T., Urbanke C., Leppanen V.M., Goldman A., Brandenburg K., Schafer G. The extreme thermostable pyrophosphatase from Sulfolobus acidocaldarius: enzymatic and comparative biophysical characterization. Arch. Biochem. Biophys. 1999;363:135–147. doi: 10.1006/abbi.1998.1072. [DOI] [PubMed] [Google Scholar]
- R8.Harutyunyan E.H., Kuranova I.P., Vainshtein B.K., Hohne W.E., Lamzin V.S., Dauter Z., Teplyakov A.V., Wilson K.S. X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate. Eur. J. Biochem. 1996;239:220–228. doi: 10.1111/j.1432-1033.1996.0220u.x. [DOI] [PubMed] [Google Scholar]
- R9.Heikinheimo P., Lehtonen J., Baykov A., Lahti R., Cooperman B.S., Goldman A. The structural basis for pyrophosphatase catalysis. Structure. 1996;4:1491–1508. doi: 10.1016/s0969-2126(96)00155-4. [DOI] [PubMed] [Google Scholar]
- R10.Hoe H.S., Kim H.K., Kwon S.T. Expression in Escherichia coli of the thermostable inorganic pyrophosphatase from the Aquifex aeolicus and purification and characterization of the recombinant enzyme. Protein Expr. Purif. 2001;23:242–248. doi: 10.1006/prep.2001.1498. [DOI] [PubMed] [Google Scholar]
- R11.Ichiba T., Shibasaki T., Iizuka E., Hachimori A., Samejima T. Cation-induced thermostability of yeast and Escherichia coli pyrophosphatases. Biochem. Cell Biol. 1988;66:25–31. doi: 10.1139/o88-004. [DOI] [PubMed] [Google Scholar]
- R12.Josse J. Constitutive inorganic pyrophosphatase of Escherichia coli.I. Purification and catalytic properties. J. Biol. Chem. 1966;241:1938–1947. [PubMed] [Google Scholar]
- R13.Kankare J., Neal G.S., Salminen T., Glumoff T., Cooperman B.S., Lahti R., Goldman A. The structure of E.coli soluble inorganic pyrophosphatase at 2.7 Å resolution. Protein Eng. 1994;7:823–830. doi: 10.1093/protein/7.7.823. [DOI] [PubMed] [Google Scholar]
- R14.Kawarabayasi Y., Sawada M., Horikawa H., et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- R15.Kornberg A. On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions. In: Kasha M., Pullman B., editors. Horizons in Biochemistry. New York: Academic Press; 1962. pp. 251–264. [Google Scholar]
- R16.Kuhn N.J., Wadeson A., Ward S., Young T.W. Methanococcus jannaschii ORF mj0608 codes for a class C inorganic pyrophosphatase protected by Co2+ or Mn2+ ions against fluoride inhibition. Arch. Biochem. Biophys. 2000;379:292–298. doi: 10.1006/abbi.2000.1860. [DOI] [PubMed] [Google Scholar]
- R17.Liu B., Bartlam M., Gao R., Zhou W., Pang H., Liu Y., Feng Y., Rao Z. Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii . Biophys. J. 2004;86:420–427. doi: 10.1016/S0006-3495(04)74118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Lundin M., Baltscheffsky H., Ronne H. Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J. Biol. Chem. 1991;266:12168–12172. [PubMed] [Google Scholar]
- R19.Parfenyev A.N., Salminen A., Halonen P., Hachimori A., Baykov A.A., Lahti R. Quaternary structure and metal ion requirement of family II pyrophosphatases from Bacillus subtilis, Streptococcus gordonii, and Streptococcusmutans . J. Biol. Chem. 2001;276:24511–24518. doi: 10.1074/jbc.M101829200. [DOI] [PubMed] [Google Scholar]
- R20.Perrella F.W. EZ-FIT: a practical curve-fitting microcomputer program for the analysis of enzyme kinetic data on IBM-PC compatible computers. Anal. Biochem. 1988;174:437–447. doi: 10.1016/0003-2697(88)90042-5. [DOI] [PubMed] [Google Scholar]
- R21.Richter O.M., Schafer G. Cloning and sequencing of the gene for the cytoplasmic inorganic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum . Eur. J. Biochem. 1992;209:351–355. doi: 10.1111/j.1432-1033.1992.tb17296.x. [DOI] [PubMed] [Google Scholar]
- R22.Robb F.T., Maeder D.L., Brown J.R., Diruggiero J., Stump M.D., Yeh R.K., Weiss R.B., Dunn D.M. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- R23.Satoh T., Samejima T., Watanabe M., Nogi S., Takahashi Y., Kaji H., Teplyakov A., Obmolova G., Kuranova I., Ishii K. Molecular cloning, expression, and site-directed mutagenesis of inorganic pyrophosphatase from Thermus thermophilus HB8. J. Biochem. 1998;124:79–88. doi: 10.1093/oxfordjournals.jbchem.a022100. [DOI] [PubMed] [Google Scholar]
- R24.Schwarm H.M., Vigenschow H., Knobloch K. Purification and properties of a soluble inorganic pyrophosphatase from Rhodopseudomonas palustris . Biol. Chem. Hoppe-Seyler. 1986;367:119–126. doi: 10.1515/bchm3.1986.367.1.119. [DOI] [PubMed] [Google Scholar]
- R25.Shintani T., Uchiumi T., Yonezawa T., Salminen A., Baykov A.A., Lahti R., Hachimori A. Cloning and expression of a unique inorganic pyrophosphatase from Bacillus subtilis: evidence for a new family of enzymes. FEBS Lett. 1998;439:263–266. doi: 10.1016/s0014-5793(98)01381-7. [DOI] [PubMed] [Google Scholar]
- R26.Slesarev A.I., Mezhevaya K.V., Makarova K.S., et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Smith D.R., Doucette-Stamm L.A., Deloughery C., et al. Complete genome sequence of Methanobacterium thermoautotrophicum delta H: functional analysis and comparative genomics. J. Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Tabor S., Richardson C.C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase: effect of pyrophosphorysis and metal ions. J. Biol. Chem. . 1990;265:8322–8328. [PubMed] [Google Scholar]
- R29.Tominaga N., Mori T. Purification and characterization of inorganic pyrophosphatase from Thiobacillus thiooxidans . J. Biochem. 1977;81:477–483. doi: 10.1093/oxfordjournals.jbchem.a131481. [DOI] [PubMed] [Google Scholar]
- R30.van Alebeek G.J., Keltjens J.T., van Der Drift C. Purification and characterization of inorganic pyrophosphatase from Methanobacterium thermoautotrophicum (strain delta H) Biochim. Biophys. Acta. 1994;1206:231–239. doi: 10.1016/0167-4838(94)90213-5. [DOI] [PubMed] [Google Scholar]
- R31.Vander Horn P.B., Davis M.C., Cunniff J.J., et al. ThermoSequenase DNA polymerase and T. acidophilum pyrophosphatase: new thermostable enzyme for DNA sequencing. BioTechniques. 1997;22:758–765. doi: 10.2144/97224pf02. [DOI] [PubMed] [Google Scholar]
- R32.Wakagi T., Lee C.H., Oshima T. An extremely stable inorganic pyrophosphatase purified from the cytosol of a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius strain 7. Biochim. Biophys. Acta. 1992;1120:289–296. doi: 10.1016/0167-4838(92)90250-h. [DOI] [PubMed] [Google Scholar]
- R33.Yang Z., Wensel T.G. Inorganic pyrophosphatase from bovine retinal rod outer segments. J. Biol. Chem. 1992;267:24634–24640. [PubMed] [Google Scholar]
- R34.Young T.W., Kuhn N.J., Wadeson A., Ward S., Burges D., Cooke G.D. Bacillus subtilis ORF yybQ encodes a manganese-dependent inorganic pyrophosphatase with distinctive properties: the first of a new class of soluble pyrophosphatase? Microbiology. 1998;144:2563–2571. doi: 10.1099/00221287-144-9-2563. [DOI] [PubMed] [Google Scholar]