Abstract
The recombinant amidase from the hyperthermophylic archaeon Sulfolobus solfataricus (SSAM) a signature amidase, was cloned, purified and characterized. The enzyme is active on a large number of aliphatic and aromatic amides over the temperature range 60–95 °C and at pH values between 4.0 and 9.5, with an optimum at pH 5.0. The recombinant enzyme is in the form of a dimer of about 110 kD that reversibly associates into an octamer in a pH-dependent reaction. The pH dependence of the state of association was studied using gel permeation chromatography, analytical ultracentrifugation and dynamic light scattering techniques. At pH 7.0 all three techniques show the presence of two species, in about equal amounts, which is compatible with the existence of a dimeric and an octameric form. In decreasing pH, the dimers formed the octameric species and in increasing pH, the octameric species was converted to dimers. Above pH 8.0, only dimers were present, below pH 3.0 only octamers were present. The association of dimers into octamers decreased in non-polar solvents and increased with temperature. A mutant (Y41C) was obtained that did not show this behavior.
Keywords: amidase signature, amide metabolism, hyperthermophile
Introduction
Amidases, a large class of enzymes broadly spread over all three domains of living organisms, act on C–N bonds other than peptide bonds and catalyze the hydrolysis of various endogenous and foreign aliphatic and aromatic amides by transferring an acyl group to water producing free acids and ammonia. The discovery of an amidase signature gene family (Mayaux et al. 1991) allowed grouping of the primary structure of the codified enzymes into two classes, the first sharing a characteristic signature and the second lacking this signature. Amidase signature family enzymes include at least 200 proteins (most of these are putative, being derived from DNA data) from 90 different organisms distributed among Bacteria, Archaea and Eukarya (Altschul et al. 1997). These enzymes are characterized by the presence of the sequence GGSS(S/G)GS in a conserved stretch of about 130 amino acids (the “amidase signature sequence”) in the center of the protein (Mayaux et al. 1991, Chebrou et al. 1996). The biological functions of the amidase signature family enzymes include the formation of Gln-tRNAGln through the transamidation of misacylated Glu-tRNAGln by amidolysis of glutamine in many bacteria (Curnow et al. 1997, Schon et al. 1998), the formation of indole-3-acetic acid in pathogenic plant bacteria (Gaffney et al. 1990), the metabolic turnover of foreign and endogenous amides in prokaryotes and eukaryotes (Mayaux et al. 1991, Gomi et al. 1991) and the catabolism of neuromodulatory fatty acid amides in mammals (Koutek et al. 1994, Cravatt et al. 1996, Patricelli and Cravatt 2000).
In a previous study, we cloned and expressed the signature amidase from the archaeon Sulfolobus solfataricus MT4 as a fusion protein with a His-tag at the N-terminus (Scotto d’Abusco et al. 2001). Preliminary experiments on samples enzymatically deprived of the His-tag showed that the protein may undergo association-dissociation processes induced by pH. To further investigate this finding, we expressed the recombinant protein without the tag. The association-dissociation processes of the protein were investigated by gel permeation chromatography (GPC), analytical ultracentrifugation (AUC) and dynamic light scattering (DLS), and the results are presented in this paper. Random mutagenesis was performed followed by selection for mutants that did not undergo pH-induced association. Among the mutants obtained, the mutant Y41C, which has a fifth cysteine in addition to the four that form the two disulfide bridges in the wild type (wt) protein, attracted our attention. Unlike the wt protein, the Y41C mutant did not exhibit the reversible pH-dependent association.
The 3D structure of S. solfataricus amidase (SSAM) is not available; therefore, we computed a low-resolution model of the protein based on the peptide amidase (PAM) from Stenotrophomonas maltophila (Labahn et al. 2002).
Materials and methods
Protein expression and purification
Amidase expression was obtained by amplifying the amidase gene through PCR and inserting the PCR product into pET3d expression vector (Invitrogen, Carlsbad, CA). The clone λC5, previously described (Scotto d’Abusco et al. 2001), was used as the template.
The PCR reaction was performed with the primers AMS (5′GTCCATGGGAATTAAGTTACCCA3′) and AMS (5′ GAGAGGATCCTTATTTTTTGATTCT′) with the following conditions: 94 °C for 1 min; 56 °C for 1 min; and 72 °C for 2 min for 30 cycles. The PCR product was digested with Nco I and Bam HI and inserted into pET3d, digested with the same pair of enzymes. The construct obtained, pASC/2E, confirmed by DNA sequencing, was transformed into E. coli BL21 (DE3)pLysS strain (Invitrogen, Carlsbad, CA) for protein expression. Cells were grown at 37 °C in Luria-Bertani (LB) medium with ampicillin (100 µg ml–1) and 0.4 mM of isopropyl-β-D-thiogalactopyranoside (IPTG) was added when the culture reached 1.6 OD600. Cells were harvested by centrifugation and disrupted by sonication. The recombinant enzyme was found in the insoluble fraction, which was extracted three times with 20 mM sodium phosphate buffer, pH 7.8, containing 500 mM NaCl and 0.25% Tween 20. Samples containing 5 mg of protein were applied on a Hiload 16/60 Superdex 200 column (Amersham Biosciences Europe) and eluted with 20 mM sodium phosphate buffer, pH 7.0, containing 150 mM NaCl at 1 ml min–1. The purified protein was concentrated with an Amicon pressurized cell equipped with a PM 30 membrane (Amicon, Beverly, MA). The purity of the protein was 98% as checked through gel electrophoresis performed either under native or denaturing conditions.
Random mutagenesis
The mutagenesis reaction was performed with the DiversifyTM Random Mutagenesis Kit (Clontech, Palo Alto, CA) according to the instruction manual. The conditions for PCR mutagenesis were optimized to produce an average of two base pair substitutions per molecule within the amidase gene (wt) from S. solfataricus. The template was pASC/2E. The forward primer was -T7 (5′-TAATACGACTCACTATAGGG-3′), and the reverse primer was -T7R (5′-ATCAATAACGAGTCGCCACC-3′). The PCR reaction was carried out at 94 °C for 30 s; 46 °C for 30 s; and 72 °C for 1.5 min for 25 cycles. The PCR reaction was checked by agarose gel and the amplified DNA fragments were purified with a Nucleospin extract Kit (Macherey-Nagel, Düren, Germany). The amplified DNA fragments and pET3d were digested separately by Nco I and Bam HI restriction enzymes and ligated. The resulting plasmid was used to transform E. coli BL21 (DE3)pLysS strain. Five clones were analysed by sequencing. The expression and purification of the mutant protein were performed as reported above for the wt recombinant protein.
Protein determination
Protein concentration was determined at 280 nm based on an extinction coefficient of 54,290 M–1 cm–1, calculated according to Gill and von Hippel (1989) as reported by Scotto d’Abusco et al. (2001).
Enzyme assay
The standard enzyme assay was performed at 70 °C for 10 min with 7.5 mM benzamide in 50 mM citrate buffer at pH 5.0. The amount of benzoic acid produced was determined by high performance liquid chromatography (Scotto d’Abusco et al. 2001). The pH of buffers was checked at the assay temperatures.
The pH optimum
The pH optimum for the amidase reaction was determined at 70 °C for 10 min, using the following buffers: citrate buffer for pH 3.0–5.8, sodium phosphate buffer for pH 6.0–7.9 and BIS-TRIS propane buffer for pH 6.5–9.5. The concentration of all buffers was 50 mM and their pH was checked at the assay temperature. The substrate was 7.5 mM benzamide.
Biochemical characterizations
The kinetic constants with substrates listed in Table 1 were determined at 70 °C in 200 µl of 50 mM citrate buffer at pH 5.0, by measuring the free acid produced by HPLC, as described by Scotto d’Abusco et al. (2001). Kinetic data were calculated by nonlinear regression analysis using ENZFITTER (Leatherbarrow 1990). For each calculation, there were at least seven velocity-substrate data pairs.
Table 1.
Kinetic properties of Sulfolobus solfataricus amidase and its Y41C mutant (in bold). The turnover number (kcat) was obtained dividing by the molecular mass of the active monomer (55,784 Da); temperature was 70 °C; there were 1–10 µg of enzyme in a total volume of 200 µl incubated for 2–10 min in 50 mM citrate buffer at pH 5.0. Abbreviation: Km = Michaelis constant.
| Substrate | Km (mM) | kcat (sec–1) | kcat/Km (M–1 sec–1) × 102 |
| Acetamide | 22.0 | 28.9 | 13.1 |
| 37.0 | 30.1 | 8.1 | |
| Propionamide | 10.0 | 82.9 | 82.9 |
| 10.0 | 99.4 | 99.4 | |
| Butyramide | 14.0 | 34.3 | 24.5 |
| 3.2 | 17.5 | 54.7 | |
| Isobutyramide | 7.0 | 48.9 | 69.9 |
| 6.5 | 58.4 | 89.8 | |
| Pentanamide | 2.5 | 26.8 | 107.2 |
| 1.5 | 28.6 | 190.6 | |
| Hexanamide | 4.5 | 19.5 | 43.3 |
| 4.5 | 21.6 | 48.0 | |
| Metacrylamide | 0.3 | 4.8 | 160.0 |
| 0.5 | 3.0 | 60.0 | |
| Benzamide | 0.9 | 6.7 | 74.4 |
| 0.6 | 7.2 | 120.0 | |
| p-OH benzamide | 0.1 | 1.9 | 190.0 |
| 0.1 | 2.1 | 210.0 | |
| p-NH2benzamide | 0.2 | 0.8 | 40.0 |
| 0.3 | 2.5 | 83.3 | |
| o-toluamide | 0.1 | 0.4 | 40.0 |
| 0.2 | 0.8 | 40.0 | |
| p-toluamide | 0.2 | 12.2 | 610.0 |
| 0.3 | 14.2 | 473.3 | |
| Nicotinamide | 1.5 | 6.4 | 42.6 |
| 0.4 | 2.2 | 55.0 | |
| 3-phenylpropionamide | 3.0 | 15.3 | 51.0 |
| 2.5 | 18.0 | 72.0 | |
| Indol-3-acetamide | 0.8 | 2.2 | 27.5 |
| 0.8 | 1.2 | 15.0 | |
Chemical modifications
Arginine residues were reacted with phenylglyossal as reported elsewhere (Riordan and Scandurra 1972) and arginine modification was deduced with an amino acid analyzer after hydrolysis of the protein in 6 N HCl at 110 °C in vacuo for 22 h.
Gel permeation chromatography
Protein samples were incubated overnight at room temperature in 50 mM citrate buffer for pH 3.0–5.0, in sodium phosphate for pH 6.0–7.0 and BIS-TRIS propane buffer for pH 8.0–9.0 and then injected on a Superdex 200 column (Amersham Biosciences Europe) equilibrated with 20 mM sodium phosphate buffer, pH 7.0, containing 0.15 M NaCl. The protein was eluted at room temperature in the same buffer at 0.4 ml min–1. The eluate was monitored at 226 and 280 nm. The column was calibrated with the following standards (Amersham Biosciences Europe): thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovoalbumin (43 kDa) and ribonuclease (13.7 kDa). The molecular mass of amidase was calculated by plotting the log of molecular mass versus elution volume (Ve) using a linear regression curve of the Unicorn 4.10 package for control and evaluation data of the Akta Chromatograph from Amersham Biosciences.
Analytical ultracentrifugation
Sedimentation velocity experiments were conducted at 1.17 × 105g and 20 °C in a Beckman Optima XL-A analytical ultracentrifuge equipped with absorbance optics and an An60- Ti rotor at a protein concentration of 0.5 mg m1–1. The buffer used was 50 mM sodium phosphate at pH 6.0 and 8.0. Equilibration with the desired buffer was achieved through dialysis or by passage through a Sephadex G25 column. Data were collected at 278 nm at a spacing of 0.005 cm with three averages in a continuous scan mode every 4 min and analyzed with the program Sedfit (Schuck and Rossmanith 2000), provided by Dr. Peter Schuck (NIH, Bethesda, MD).
Dynamic light scattering
Dynamic light scattering measurements were performed with a DynaPro-801 instrument with temperature control (Protein Solution, Charlottsville, VA), where the scattered light was collected at an angle of 90° through a fiber optic and converted to an electrical signal by an avalanche photodiode. The time-dependent autocorrelation function of the photon current was monitored with a 20-channel software correlator (based on a Digital Signal Processor unit) provided by the manufacturer. The first sampling time was 3.84 µs. The length of the subsequent channels increases in a quasi-logarithmic fashion. The samples were gently injected into the cell through a series of Whatman filters with decreasing porosity, from 0.22 to 0.02 µm. Protein concentrations for both the wt amidase and the Y41C mutant ranged from 3.0 to 0.2 mg ml–1. After an overnight incubation in the appropriate buffer, measurements were performed first at 3.0 mg ml–1 and then, after dilution with the same buffer, repeated at 2.6, 1.3, 0.5, 0.3 and 0.2 mg ml–1. Buffers used for measurements were 20 mM sodium phosphate at pH 8.0 and 20 mM sodium citrate at pH 4.0. Autocorrelation functions were measured every 60 s, based on 0.18 × 105 to 3.3 × 105 counts each. Data analyses were performed with Protein Solutions’ Dynamics analysis software. Measurements were made at 20.1 ± 0.2 °C.
Spectroscopic analyses
Intrinsic fluorescence emission measurements were made with a Perkin-Elmer LS50B spectrofluorimeter with a 1 cm pathlength quarz cuvette. Fluorescence emission spectra were recorded between 300 and 450 nm (1 nm sampling interval) at 20 °C with the excitation wavelength set at 295 nm. All the spectra were recorded at 0.1 mg ml–1 protein concentration after an overnight incubation in 20 mM sodium phosphate buffer at pH 8.0 or 20 mM sodium citrate buffer at pH 5.0. Experiments with the fluorescent dye 8-anilinonaphthalene-1-sulfonic acid ammonium salt (ANS) were performed at 20 °C by incubating the protein and ANS at a 1:10 molar ratio. After 5 min, fluorescence emission spectra were recorded at 400– 600 nm with the excitation wavelength set at 390 nm. The CD spectra were recorded on a Jasco J-720 spectropolarimeter. The results are expressed as mean residue ellipticity assuming a mean residue of 110 per amino acid residue.
Monomer detection
The dissociation of the dimer was attempted by adding either urea (up to 2 M) or guanidinium chloride (up to 0.8 M), with thiocyanate (up to 0.5 M) to a solution of wt protein or Y41C mutant or by increasing the pH to 11.0. Protein samples in 50 mM BIS-TRIS propane buffer pH 8.0 (where only dimers are present) were incubated for 2 h at 20 °C with the mentioned compounds and injected in a GPC column.
Thermoactivity
The temperature dependence of the amidase reaction was studied under standard conditions over the range 40–100 °C.
Thermal stability
The measurements were conducted at 70, 80 and 90 °C in Eppendorf tubes (0.2 ml) using 0.2 mg ml–1 protein in 70 µl 50 mM citrate buffer at pH 5.0. The tubes were sealed and incubated for the reported times in a Thermomixer compact (Eppendorf, Hamburg, Germany). After incubation a 50 µl sample was assayed for standard activity.
Differential scanning calorimetry
Heat capacity versus temperature profiles were obtained with a VP-DSC differential scanning calorimeter (MicroCal.Inc., Northampton, MA) at a heating rate of 60 °C h–1. Protein solutions (0.1–0.3 mg ml–1) in the appropriate buffer were degassed at 25 °C before the calorimetric experiments. The reference cell was filled with the same solvent mixture used for the sample, but lacking the protein. Both cells were kept under an excess pressure of 200 Pa to avoid bubbling during the scan. At the end of the run the solutions were subjected to a second heating cycle under the same conditions to determine the reversibility of the transitions. Thermograms were corrected by subtracting the instrumental baseline, obtained with both cells filled with the same solvent, and normalized for protein concentration. The Tm (temperature at which excess heat capacity reaches a maximum, °C) was determined with the ORIGIN software provided by MicroCal, after subtraction of a cubic baseline connecting the pre- and post-transition traces.
Construction of the model
Three signature amidases with different metabolic roles are presently known at the atomic resolution: malonamidase E2 (MAE2) from Bradyrhizobium japonicum (Shin et al. 2003), the fatty acid amide hydrolase from Rattus norvegicus (FAAH) (Bracey et al. 2002) and the peptide amidase (PAM) from Stenotrophomonas maltophila (Labahn et al. 2002). The SSAM 3D model was built with PAM as a template. This choice was suggested by a better sequence alignment of the target/template (complete coverage, identity level = 30%) compared with the other two sequences and also because SSAM and PAM use similar substrates. The 3D structure was computed after template/target alignment with Modeller v. 6.2 (Sali and Blundell 1993). After prediction of the interaction patches with ISPRED (Fariselli et al. 2002), the dimer form was built with the ZDOCK tool (Chen and Weng 2002).
Results
The expression of recombinant wt S. solfataricus amidase by the pET3d expression vector, allowed us to obtain the enzyme in its native form. Furthermore, random mutagenesis was applied in order to find mutants showing physico-chemical properties or substrate specificities or both that are different from those found in the recombinant S. solfataricus wt amidase. Mutant amidases from all the clones obtained were expressed and purified and their physico-chemical properties analyzed. Mutant amidase with the Y41C substitution was selected because it behaved differently from the wt protein as a function of pH.
pH optimum and kinetic parameters
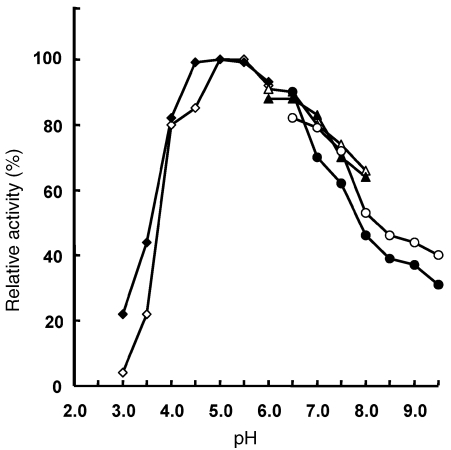
As reported in Figure 1, the activities of recombinant wt amidase and its mutant, measured at 70 °C, showed pH optima at pH 5.0. Therefore this pH was chosen for measurements of kinetic parameters on a number of aromatic and aliphatic amides (Table 1).
Figure 1.
Effect of pH on the activity of Solfolobus solfataricus recombinant wild type (wt) amidase (filled symbols) and its Y41C mutant (open symbols). Enzyme activity was measured at 70 °C for 10 min, with 7.5 mM benzamide in 50 mM citrate buffer at pH 4.0–5.8 (diamonds), sodium phosphate buffer at pH 6.0–7.9 (triangles) and BIS-TRIS propane buffer at pH 6.5–9.5 (circles). The pH of each buffer was checked at 70 °C. The 100% activity corresponds to 7.2 and 7.8 units mg–1 of wt and mutant proteins, respectively (1 enzyme unit = 1 µmol of free acid formed per minute).
Gel permeation chromatography
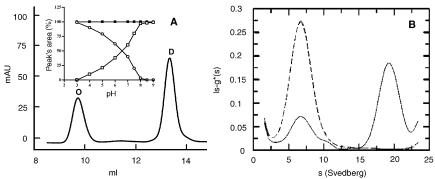
After overnight incubation at room temperature of the wt protein in various 50 mM buffers at various pHs, the samples were injected onto a Superdex 200 column eluted at 0.4 ml min–1 with 20 mM sodium phosphate buffer at pH 7.0 and 0.15 M NaCl. The elution pattern of a sample incubated in 50 mM BIS-TRIS propane buffer pH 7.2 is shown in Figure 2A. Two peaks with elution volumes corresponding to a molecular mass of about 83 kDa and 400 kDa, compatible with a dimeric (D) and octameric (O) species, respectively, were obtained. The ratio of the two species was dependent on pH. The distribution of the area of the two peaks as a function of pH is shown in the inset of Figure 2A: a decrease in pH resulted in increasing formation of the oligomeric species (open circles) and a parallel reduction of the dimeric species (open squares). Conversely, the latter species increased with increasing pH: from pH 8.0 upward, only the dimeric species was present, whereas below pH 3.0, only the octameric species was present. Conversion of one species of the wt protein to the other induced by pH was completely reversible within 20 minutes and was independent of the concentration of the buffer (20–100 mM).
Figure 2.
Effect of pH on the oligomeric state. (A) Gel permeation chromatography: 30 µg of protein was incubated overnight in 50 mM BIS-TRIS propane buffer at pH 7.2, then injected onto the Superdex 200 column, eluted at room temperature at 0.4 ml min–1 with 20 mM phosphate buffer containing 150 mM NaCl and monitored at 226 nm. Abbeviations: O = octamer; D = dimer. The inset shows the percentage of each peak’s area from the elution patterns of wild type (wt) protein samples obtained after overnight incubation with buffers at various pH values (open circles = octamer; open squares = dimer). The filled squares show the percentage of the dimer’s area obtained with Y41C mutant samples after overnight incubation with buffers at various pH values. (B) Analytical ultracentrifugation:the wt protein concentration was 0.5 mg ml–1 in 50 mM phosphate buffer at pH 6.0 (solid line) and 8.0 (dashed line). Experiments were conducted at 1.17 × 105 g and 20 °C, at a wavelength of 278 nm after reaching equilibrium (12 h). Peak (80%) with Sw,20 = 18 corresponds to the octamer (about 400 kDa). Peak (20%) with Sw,20 = 6 corresponds to the dimer (about 100 kDa).
Partial reassociation may also be induced by elevated temperature: a wt protein solution in 50 mM phopshate buffer or in 50 mM BIS-TRIS propane at pH 8.0 was heated for 1 h at 80 °C and immediately injected on a Superdex column. The elution pattern indicated that 60% of the dimeric species had been converted to the octameric species.
Different behavior was observed when the same experiments were conducted with the Y41C mutant. In the GPC experiments, this mutant displayed only one peak, corresponding to the dimeric species, over the whole pH range examined either at 20 or at 80 °C (filled squares in the inset of Figure 2A).
To test whether the association of wt dimers into octamers was caused by hydrophobic interactions among dimers, the polarity of the protein environment was decreased by addition of hydrophobic agents. Experiments were conducted on a protein solution brought to pH 8.0 with 50 mM BIS-TRIS propane buffer, to induce dissociation of octamers into dimers, followed by the addition of 5, 10 and 15% methanol, ethanol and n-propanol and 5 and 10% of n-butanol; after 10 min the pH was brought to 4.0 with 100 mM citrate buffer to induce oligomerization; after 20 min at 20 °C the solution was injected onto the Superdex column. Neither methanol nor ethanol inhibited formation of the oligomer at pH 4.0: only 15% n-propanol or 10% n-butanol was able to block formation of the oligomer.
To test whether insertion of a bulky residue at many points in the protein alters the association process induced by pH, the protein was reacted with phenylglyossal. The reagent modified at least fourteen arginine residues per dimer out of a total of 42: the chemically modified protein was fully active. The solution of the chemically modified protein was brought to pH 4.0 with citrate buffer to induce oligomerization and injected on the Superdex column: no octameric species were detected.
Analytical ultracentrifugation and dynamic light scattering
Analytical ultracentrifugation experiments performed on the wt protein yielded two peaks, with sedimentation coefficients of 6S and 18S, in a ratio that varied as a function of pH (Figure 2B). At 20 °C and pH 6.0, the 18S species was predominant (solid line), whereas at pH 8.0 only, the 6S species was present (dashed line). The value of 18S observed for the heavier species corresponds to a molecular mass of about 400 kDa, in agreement with the GPC results. The molecular mass of the lighter species is around 100 kDa, consistent with a dimeric form of the protein and also in agreement with the GPC data.The Y41C mutant exhibited only the 6S peak at both pH 6.0 and pH 8.0 (data not shown).
The size of the recombinant amidase and of the Y41C mutant was assessed by DLS. The DLS apparatus was used to determine the hydrodynamic radius (RH) of the photon scattering amidase molecules in buffer, by analyzing the time scale of the fluctuations in the amount of scattered light. Determination of the RH provides an estimate of the molecular mass of the protein species under study. This estimate is useful for discriminating, for example, between a dimer and a monomer. The RH of amidase was calculated to be 4.23 nm, according to the classical Stokes-Einstein relation for a sphere (Table 2). Software-based conversion of RH measurements estimated a molecular mass of 95.4 kDa, which accounted for an amidase dimer. This value did not change significantly, even at the lowest possible concentration attainable with the DLS instrument, ranging from 4.23 to 4.05 nm (molecular mass of 95.4 kDa versus 86.3 kDa). A decrease in the polydispersity of the protein preparation (from 40 to 20% polydispersity) was observed by decreasing the protein concentration, but no conversion from dimer to monomer was detected, indicating that the dimeric form was the most stable one in the concentration range tested (3.0–0.2 mg ml–1). Even with a polydispersity value of 40%, the measured radius perfectly fitted a monomodal gaussian distribution, not a bimodal size distribution. Therefore we can exclude the presence of species other than the dimer. At pH 4.0, RH increased to 7.52 nm (with a polydispersity of less than 20%), indicating an oligomeric state of amidase which contained more than one dimer. This value corresponded to an estimated molecular mass of 385.7 kDa, compatible with a tetramer of dimers (Table 2). In contrast, light scattering experiments with the Y41C mutant, conducted at a protein concentration ranging from 3.0 to 0.2 mg ml–1 either at pH 4.0 or 8.0, always yielded the presence of a species compatible with a dimer (Table 2). Also in the DLS experiments, the measured radii fitted a monomodal gaussian distribution, not a bimodal size distribution.
Table 2.
Estimation of molecular mass by dynamic light scattering measurements with the wild type (wt) amidase and the Y41C mutant. Abbreviation: RH = hydrodynamic radius.
| pH 8.0 | pH 4.0 | pH 8.0 | pH 4.0 | |
| RH (nm)1 | RH (nm)1 | Estimated molecular mass (kDa)2 | Estimated molecular mass (kDa)2 | |
| Wt amidase | 4.2 | 7.5 | 95.4 | 385.7 |
| Y41C mutant | 4.4 | 4.5 | 106.0 | 111.0 |
1 Mean hydrodynamic radius derived from the measured translational diffusion coefficient using the Stokes-Einstein equation. 2 Molecular mass estimated from RH assuming that the particles are spherical and of standard density.
Monomer detection
To determine if the dimers could be chemically dissociated, solutions of the wt and mutant proteins in 50 mM BIS-TRIS propane buffer pH 8.0 (having only dimers) were treated with urea (up to 2 M), guanidinium chloride (up to 0.8 M) or thiocyanate (up to 0.5 M) (Ptitsyn et al. 1990). At all concentrations the protein maintained full activity and when injected on the Superdex column, no dissociation of the dimer occurred. Likewise, incubation of the two proteins in 50 mM Tris adjusted to pH 11.0 (Almatawah et al. 1999) did not cause dissociation of the dimers.
Spectroscopic analyses
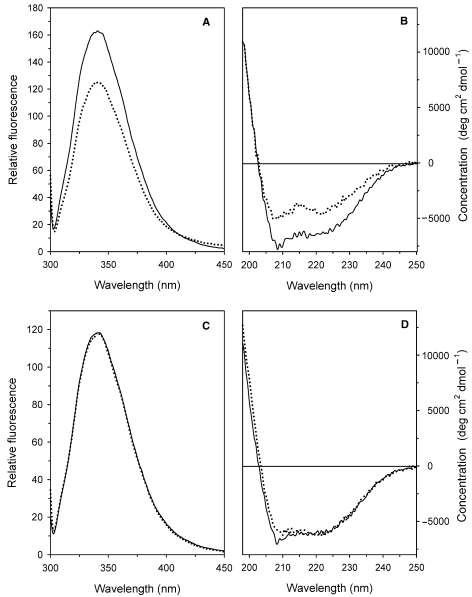
To assess whether the behavior of the wt and Y41C proteins as a function of pH was accompanied by pH-dependent conformational changes, circular dichroism (CD) and intrinsic fluorescence emission spectra of both proteins were recorded at pH 5.0 and 8.0 (Figure 3). A significant modification of the tertiary and secondary structure of the wt protein occurred on transition between these pH values (Figures 3A and B) as indicated by the decrease in fluorescence (Figure 3A) and ellipticity (Figure 3B). No structural modifications in the Y41C mutant were detected (Figures 3C and D).
Figure 3.
Effects of pH on the intrinsic fluorescence and on far UV CD spectra of the Solfolobus solfataricus recombinant wild type (wt) amidase and the Y41C mutant. Fluorescence emission spectra (A, C) were recorded with the excitation wavelength at 295 nm. Far UV CD spectra were recorded in a 0.1 cm path-length quartz cuvette. All spectra were recorded with 0.1 mg ml–1 protein concentrations at 20 °C after overnight incubation of the wt (A, B) and the Y41C mutant (C, D) at pH 8.0 (20 mM sodium phosphate, solid line) and pH 5.0 (20 mM sodium citrate, dotted line).
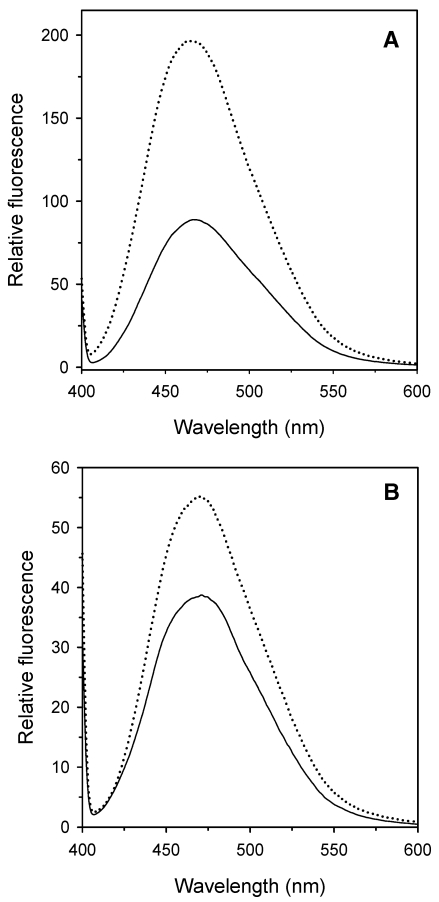
The experiments with ANS reported in Figure 4, show that the extrinsic fluorescence emission spectrum of wt protein with ANS at pH 5.0 was almost twice that measured at pH 8.0, whereas the spectrum of the Y41C mutant was only slightly increased (Figures 4A and B). These results demonstrate that the dye bound more tightly to the wt protein at pH 5.0 than at pH 8.0, whereas pH had no effect on the binding of dye to the Y41C mutant protein.
Figure 4.
Relative accessibility of hydrophobic residues in Solfolobus solfataricus wild type (wt) amidase (A) and its Y41C mutant (B) monitored by the extrinsic fluorescence of 8-anilinonaphtalene-1-sulfonic acid ammonium salt (ANS). The ANS (25 µM) was added to the protein (2.5 µM), which had previously been incubated overnight at pH 8.0 (20 mM sodium phosphate, solid line) and at pH 5.0 (20 mM sodium citrate, dotted line). Fluorescence emission spectra (390 nm) were recorded at 20 °C, 10 min after the addition of ANS.
Thermoactivity, thermostability and thermal unfolding
Thermoactivity and thermostability of the wt and Y41C proteins were studied at pH 5.0 and 8.0. Thermoactivity of the two proteins at both pH 5.0 and pH 8.0 was similar, with a maximum at 95 °C (data not shown). Thermal stability of the wt enzyme and its mutant Y41C at pH 5.0 and 70 °C, were similar, with half-life values over 70 h (Table 3). At 80 °C no differences were detected when both proteins were tested at pH 5.0, but at pH 8.0, the half-life of the wt protein was reduced by more than half, whereas that of the mutant was reduced by more than 99%.
Table 3.
Thermal stability (half-lives, h) of the wild type (wt) Solfolobus solfataricus amidase and its Y41C mutant measured at pH 5.0 and 8.0.
| pH | 70 °C | 80 °C | 90 °C | |
| 5.0 | WT | > 70 | 65 | 60 |
| Y41C | > 70 | 64 | 20 | |
| 8.0 | WT | > 70 | 25 | 0.5 |
| Y41C | > 70 | 0.5 | 0.25 | |
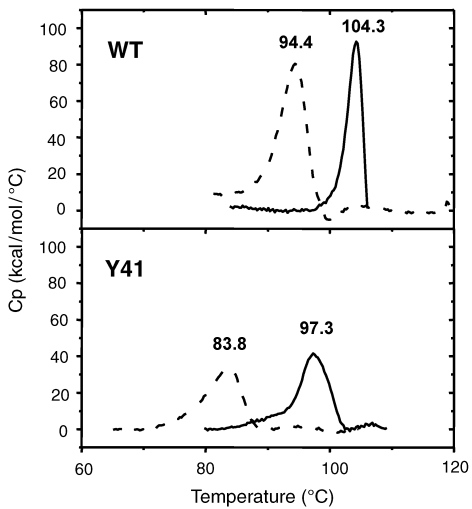
The DSC profile of both proteins is shown in Figure 5. Thermal unfolding of the wt protein at pH 8.0 gave a Tm of 94.4 °C. At pH 5.0 the wt protein showed a Tm of 104.3 °C. The Y41C mutant showed lower Tm values at both pH 8.0 (83.5 °C) and pH 5.0 (97.3 °C). No reversibility was found under the measured conditions.
Figure 5.
Differential scanning calorimetry thermograms of wild type (wt) amidase and its Y41C mutant at different pH values. Solid lines correspond to traces obtained in 20 mM citrate buffer at pH 4.0, dotted lines to those obtained in 20 mM sodium phosphate at pH 8.0. The concentration was always expressed as moles of dimer. Denaturation temperatures (Tm) are indicated above the corresponding peak.
Modeling of three dimensional structure
The 3D structure of SSAM was obtained by homology modeling. The selected template that best aligns with the target sequence is PAM from Stenotrophomonas maltophila (Labahn et al. 2002). Figure 6 shows the sequence alignment of SSAM, MAE2, FAAH and PAM, focusing on the highly conserved signature sequence and the active site (shown in color). The Ser-Ser-Lys catalytic triad is highlighted in red. After modeling the target on the template, the 3D structure of SSAM results in an alpha/beta protein with a Root Mean Square Deviation to PAM equal to 0.08 nm. The content of secondary structure is characterized by 25.7% of alpha, 12.2% of beta and 62.1% of coil structural types, respectively, in good agreement with the percentage calculated from the far UV CD spectra.
Figure 6.
Sequence alignment of the conserved stretch. The 130 amino acids, including the “amidase signature sequence” of Sulfolobus solfataricus amidase (SSAM) (GB AF290611), malonyl amidase E2 (MAE2) from Bradyrhizobium japonicum (PDB 1GR8), fatty acid amide hydrolase (FAAH) from Rattus norvegicus (PDB 1MT5) and peptidyl amidase (PAM) from Stenotrophomonas maltophila (PDB 1M22). Invariant residues are reported in green whereas conservative residues are in blue. The catalytic triad (K, S, S) is indicated in red.
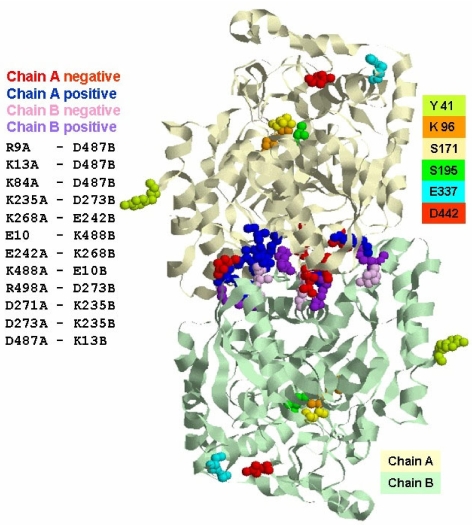
The putative interaction patches localized on the accessible surface of the protein were predicted with ISPRED, a neural-network based predictor for protein interaction sites that has been described previously (Fariselli et al. 2002). This allowed us to compute the protein putative dimer with ZDOCK (Chen and Weng 2002), a program based on complementarity shape and electrostatic interactions. Figure 7 shows the putative model of the SSAM dimer: charged ion-pair residues putatively involved in the electrostatic interactions between the two monomers are highlighted as red and pink spheres (negatively charged) and blue and magenta spheres (positively charged). The model also shows the catalytic triad (K96 (orange), S171 (yellow) and S195 (green)) at the active site, E337 (light blue) and D442 (red) at the entrance of the active site and Y41 in brilliant green. It is evident that, according to the model, Y41 is not involved in the interaction surface between monomers in the dimer. This is in agreement with the finding that the Y41C mutation does not affect protein dimerization but only the oligomerization of dimers.
Figure 7.
Model of the Sulfolobus solfataricus amidase (SSAM) dimer. Putative model of the SSAM dimer: charged ion-pair residues putatively involved in the electrostatic interactions (positively charged groups are blue and magenta, negatively charged groups are red and pink) between the two monomers are highlighted. Also shown are the catalytic triad (K96 (orange), S171 (yellow) and S195 (green)) at the active site, E337 (light blue) and D442 (red) at the entrance of the active site and Y41 in brilliant green.
Discussion
The most interesting feature of the recombinant wt amidase was its response to pH. The octameric species was predominant at acidic pH values, whereas the dimeric species was predominant at pH values higher than 7.0. This behavior is infrequent, with most proteins behaving in the opposite way, i.e., the association of protomers in multimers is impaired at acidic pH values (Ralston 1991, Zeng et al. 1997, Calvete et al. 1999, Hochgrebe et al. 2000, Madern et al. 2000, Madern et al. 2001, Wah et al. 2001, Lew and London 2003).
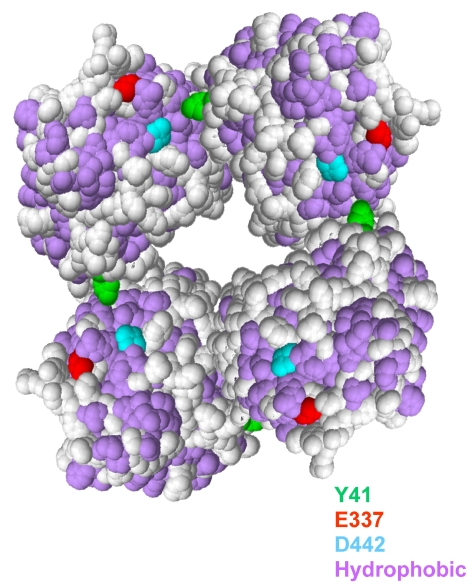
Our results suggest that the association of dimers into octamers is the consequence of conformational changes induced by the protonation of some ionizable group allowing the interaction of the segment containing Y41 (LLKLQLESYE-RLDSLP) with a hydrophobic patch exposed to solvent in the contiguous dimer as proposed in the scheme reported in Figure 8. Reducing the polarity of the environment by organic solvents such as n-butanol or n-propanol decreased the hydrophobic interactions and increased the inhibition of the dimer association in accordance with the effects of organic solvents on most enzymes (Williams et al. 1995).
Figure 8.
Proposed scheme of the Sulfolobus solfataricus amidase (SSAM) octamer. Each sphere represents a dimer. Hydrophobic residues are in magenta. Also shown are E337 (red) and D442 (light blue) at the entrance to the active site and Y41 in green.
The experimental results lead to the conclusion that acidic pH may induce changes in the tertiary structure of the wt protein (as revealed by fluorescence and CD spectra) which acquires a new spatial arrangement that permits new hydro- phobic interactions. The conformational changes induced in the dimers may lead to the exposure on their surface of sticky hydrophobic segments that could result in hydrophobic zipper-like structures, as reported for proteins such as ERK2 (Khokhlatchev et al. 1997), PAK-1 (Hoffman and Cerione 2000) and sulfotransferases (Petrotchenko et al. 2001), and in the pH-dependent transition of viral fusion proteins (Hsu et al. 2002, Yao et al. 2003). On this basis, the interactions among the dimers would then be mainly hydrophobic and Y41 could have the role of a trigger priming oligomerization. In turn, oligomerization would be impaired when Y41 is substituted with a cysteine residue, as in the Y41C mutant.
Such an interpretation is supported by the fluorescence experiments with ANS and by the chemical inhibition of oligomerization. The fluorescence experiments with ANS demonstrate a higher availability of hydrophobic residues in the wt protein at acidic pH values. Because ANS binds to hydrophobic groups of a protein (Anderson and Weber 1966), in the wt protein, at pH 5.0, more hydrophobic groups should be available for interaction with the dye, i.e., those groups embedded in the core of the protein at pH 8.0 are more exposed at pH 5.0.
The association of wt amidase dimers into octamers, as induced by acidic pH was hampered by reduced environmental polarity and by the insertion of a bulky residue such as phenylglyossal in many points of the protein, which could modify the conformation of dimers, rendering them unable to associate into octamers without modifying the specific activity of the enzyme.
The molecular mass of the SSAM as determined by the amino acid sequence is 55,784 Da. Ultracentrifugation analyses yielded two peaks with Sw,20 = 6S and 18S corresponding to a molecular mass of about 100 kDa and 400 kDa, respectively, which are compatible with a dimeric and an octameric species. These values were confirmed by the DLS and GPC experiments.
Attempts failed to dissociate the dimer of the wt protein and the Y41C mutant with urea or guanidinium chloride (generic chaothropes that affect the hydrogen bond network of a protein), a lipophilic salt such as thiocyanate (which can interfere with ion pairs located either on the surface or in the hydrophobic core of folded proteins (Ptitsyn et al. 1990)) or with treatment at pH 11.0 (Almatawah et al. 1999), suggesting that the two subunits are strongly bound, likely through the electrostatic interactions depicted in the dimer model of Figure 7.
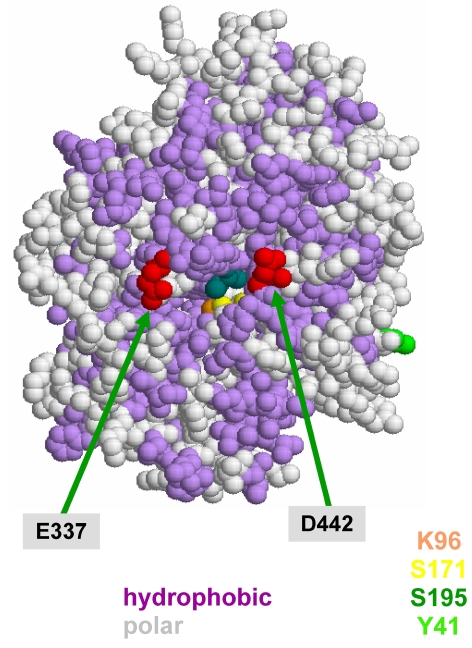
Maximal activity of the recombinant wt amidase and its mutant Y41C was found at pH 5.0. Aromatic substrates are better than aliphatic substrates for both enzymes, as reflected by the Michaelis constant (Km) values, for which aliphatic substrates were one order of magnitude higher than aromatic substrates. The aliphatic substrates entered the active site at pH 5.0 with greater difficulty, but the products left it more easily, as reflected by the higher turnover rate (kcat) relative to aromatic substrates. However, kcat values for aliphatic substrates are higher than those for aromatic substrates. As a consequence, enzymatic efficiency (expressed as kcat/Km) of both proteins was high both with aliphatic and aromatic substrates, but was higher with the latter. These results, taken together with the modeling data reported in Figure 7 and 9, suggest that at the active site of the enzyme, hydrophobic residues achieve a better interaction with aromatic substrates and some carboxylic groups which, with their negative charges, may drive away the negatively charged product of the reaction.
Figure 9.
Hydrophobic and charged residues around the entrance of the Sulfolobus solfataricus amidase active site.
As expected for proteins from a hyperthermophilic organism, thermal stability for both proteins, reported in Table 3 as half-lives and in Figure 5 as Tm values, was high (a decrease in activity was observed only in the mutant at 90 °C, as expected in a protein with an additional cysteine) but differed with pH. As shown by GPC, DLS and AUC, the octameric species is predominant in the wt protein at acidic pH values, whereas the dimeric species is predominant at pH values higher than 7.0. In Y41C, only the dimeric species was present at all pH values. Combining all the information acquired through spectroscopic analyses, GPC, AUC, DLS and DSC experiments, we draw the following conclusions.
The Y41C dimer is less compact than the wt dimer. The thermal stability of Y41C at 80 °C and pH 8.0 is reduced to a half-life of 0.5 h compared with 25 h for the wt at the same pH and temperature. However, when Y41C is brought to pH 5.0, it assumes a structure and demonstrates behavior comparable to that of the wt protein at the same pH and temperature with a thermal stability of 64 h, although no octamers are formed. Microcalorimetry indicated that, at pH 4.0, Y41C has a Tm of 97.3 °C, which is close to that of the dimeric wt species at pH 8.0 (94.4 °C). At pH 4.0 to 5.0, Y41C, but not the wt protein, becomes more compact, resulting in increased ANS binding, although this was not detected spectrophotometrically.
In contrast, in the wt protein, acidic pH (4.0 or 5.0), as shown by GPC, AUC and DLS, lead to oligomerization, accompanied by the highest Tm (104.4 °C), changes in tertiary structure, as revealed by fluorescence and CD spectra, and larger amounts of ANS binding the protein.
At pH 5.0 or pH 8.0 both proteins were active up to 95 °C. This means that the dimer of Y41C (present at all pH values), the dimer of the wt (present at pH 8.0) and the octamer of the wt predominant at pH 5.0, are all thermoactive species.
We speculate that oligomerization could have a physiological role in the cell which exists in a highly acidic environment (pH 2.5) and at an average temperature of about 85 °C. The inner pH of the cell is 6.8 (De Rosa et al. 1975, Moll and Shafer 1988) but this value may not be constant because most of the cell’s energy is employed in contending with the acidic environment. Temperature too, could fluctuate owing to the discontinuous emission of hot gases. Therefore, oligomerization through an increase in hydrophobic interactions induced by pH and temperature could be one of the mechanisms increasing protein thermostability. Other examples of increased protein thermostability through oligomerization were suggested by Opitz et al. (1987) on the basis of fragmentation studies on lactate dehydrogenase, from modeling studies by Kohlhoff et al. (1996) on triosephosphate isomerase, by Villeret et al. (1998) on ornithine carbamoyltransferase and more recently by Shima et al. (2000) on formyltransferase from the hyperthermophilic Methanopyrus kandleri.
Acknowledgments
We are grateful to Prof. E. Chiancone for her criticism and helpful discussion and to Prof. B. Maras for amino acid determinations. This research was funded by Italian MIUR (Ministery of University and Research), by CNR Target Project on Biotechnology and by ASI (Italian Space Agency).
References
- R1.Almatawah A.A., Cramp R., Cowan A. Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles. 1999;3:283–291. doi: 10.1007/s007920050129. [DOI] [PubMed] [Google Scholar]
- R2.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Anderson S., Weber G. The reversibile acid dissociation and hybridization of lactic dehydrogenase. Arch. Biochem. Biophys. . 1966;116:207–223. doi: 10.1016/0003-9861(66)90028-2. [DOI] [PubMed] [Google Scholar]
- R4.Bracey M.H., Hanson M.A., Masuda K.R., Stevens R.C., Cravatt B.F. Structural adaptation in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;296:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- R5.Calvete J.J., Thole H.H., Raida M., Urbane C., Romero A., Grangeiro T.B. Molecular characterization and crystallization of Diocleinae lectins. Biochim. Biophys. Acta. 1999;1430:367–375. doi: 10.1016/s0167-4838(99)00020-5. [DOI] [PubMed] [Google Scholar]
- R6.Chebrou H., Bigey F., Arnaud A., Galzy P. Study of the amidase signature group. Biochim. Biophys. Acta. 1996;1298:285–293. doi: 10.1016/s0167-4838(96)00145-8. [DOI] [PubMed] [Google Scholar]
- R7.Chen R., Weng Z. Docking unbound proteins using shape complementarity, desolvation and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- R8.Cravatt B.F., Giang D.K., Mayfield S.P., Boger D.L., Lerner R.A., Gilula N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- R9.Curnow A.W., Hong K., Yuan R., Kim S., Martins O., Winkler W., Henkin T.M., Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.De Rosa M., Gambacorta A., Bulock D. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus acidocaldarius . J. Gen. Microbiol. . 1975;86:154–164. doi: 10.1099/00221287-86-1-156. [DOI] [PubMed] [Google Scholar]
- R11.Fariselli P., Pazos F., Valencia A., Casadio R. Prediction of protein-protein interaction sites in heterocomplexes with neural networks. Eur. J. Biochem. 2002;269:1356–1361. doi: 10.1046/j.1432-1033.2002.02767.x. [DOI] [PubMed] [Google Scholar]
- R12.Gaffney T.D., Da Costa E Silva O., Yamada T., Kosuge T. Indolacetic acid operon of Pseudomonas syringae subsp. savastanoi: transcription analysis and promoter identification. J. Bacteriol. 1990;172:5593–5601. doi: 10.1128/jb.172.10.5593-5601.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Gill S., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- R14.Gomi K., Kitamoto K., Kumagai C. Cloning and molecular characterization of the acetamidase-encoding gene (amdS) from Aspergillus orizae . Gene. 1991;108:91–98. doi: 10.1016/0378-1119(91)90491-s. [DOI] [PubMed] [Google Scholar]
- R15.Hochgrebe T., Pankhurst G.J., Wilce J., Easterbrook-Smith S.B. pH-dependent changes in the in vitro ligand-binding properties and structure of human clusterin. Biochemistry . 2000;15:1411–1419. doi: 10.1021/bi991581b. [DOI] [PubMed] [Google Scholar]
- R16.Hoffman G.R., Cerione R.A. Flipping the switch: the structural basis for signaling through the CRIB motif. Cell. 2000;102:403–406. doi: 10.1016/s0092-8674(00)00045-3. [DOI] [PubMed] [Google Scholar]
- R17.Hsu C.H., Wu S.H., Chang D.K., Chen C. Structural characterizations of fusion peptide analog of influenza virus hemagglutinin. Implication of the necessity of a helix-hinge-helix motif in fusion activity. J. Biol. Chem. 2002;277:22725–22733. doi: 10.1074/jbc.M200089200. [DOI] [PubMed] [Google Scholar]
- R18.Khokhlatchev A., Xu S., English J., Wu P., Schaefer E., Cobb M.H. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J. Biol. Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- R19.Kohlhoff M., Dahm A., Hensel R. Tetrameric triosephosphate isomerase from hyperthermophilic Archaea. FEBS Lett. 1996;383:245–250. doi: 10.1016/0014-5793(96)00249-9. [DOI] [PubMed] [Google Scholar]
- R20.Koutek B., Prestwich G.D., Howlett A.C., Chin S.A., Salehani D., Akhavan N., Deutsch D.G. Inhibitors of arachidonoyl ethanolamide hydrolysis. J. Biol. Chem. 1994;269:22937–22940. [PubMed] [Google Scholar]
- R21.Labahn J., Neumann S., Buldt G., Kula M.R., Granzin J. An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 2002;322:1053–1064. doi: 10.1016/s0022-2836(02)00886-0. [DOI] [PubMed] [Google Scholar]
- R22.Leatherbarrow R.J. Use of nonlinear regression to analyze enzyme kinetic data: application to situations of substrate contamination and background subtraction. Anal. Biochem. 1990;184:274–278. doi: 10.1016/0003-2697(90)90680-8. [DOI] [PubMed] [Google Scholar]
- R23.Lew S., London E. The effect of interactions involving ionizable residues flanking membrane-inserted hydrophobic helices upon helix-helix interactions. Biochemistry . 2003;42:10833–10842. doi: 10.1021/bi034929i. [DOI] [PubMed] [Google Scholar]
- R24.Madern D., Ebel C., Mevarech M., Richard S.B., Pfister C., Zaccai G. Insights into the molecular relationship between malate and lactate dehydrogenases:structural and biochemical properties of monomeric and dimeric intermediates of a mutant of tetrameric L-(LDH-like) malate dehydrogenase from the halophilic archaeon Haloarcula marismortui . Biochemistry. 2000;39:1001–1010. doi: 10.1021/bi9910023. [DOI] [PubMed] [Google Scholar]
- R25.Madern D., Ebel C., Dale H.A., Lien T., Steen I.H., Birkeland N.K., Zaccai G. Differences in the oligomeric states of the LDH-like MalDH from the hyperthermophilic archaea Methanococcus jannaschii and Archaeoglobus fulgidus . Biochemistry . 2001;40:10310–10316. doi: 10.1021/bi010168c. [DOI] [PubMed] [Google Scholar]
- R26.Mayaux J.F., Cerebelaud E., Soubrier F., Yeh P., Blanche F., Petre D. Purification, cloning, and primary structure of new enantiomer-selective amidase from Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile hydratase. J. Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Moll R., Shafer G. Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius . FEBS Lett. 1988;32:359–363. [Google Scholar]
- R28.Opitz U., Rudolph R., Jaenicke R., Ericsson L., Neurath H. Proteolytic dimers of porcine muscle lactate dehydrogenase characterization, folding and reconstitution of the truncated and nicked polypeptide chain. Biochemistry . 1987;26:1399–1406. doi: 10.1021/bi00379a028. [DOI] [PubMed] [Google Scholar]
- R29.Patricelli M.P., Cravatt B.F. Clarifying the catalytic roles of conserved residues in the amidase signature family. J. Biol. Chem. 2000;275:19177–19184. doi: 10.1074/jbc.M001607200. [DOI] [PubMed] [Google Scholar]
- R30.Petrotchenko E.V., Pedersen L.C., Borchers C.H., Tome K.B., Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Lett. 2001;490:39–43. doi: 10.1016/s0014-5793(01)02129-9. [DOI] [PubMed] [Google Scholar]
- R31.Ptitsyn O.B., Pain R.H, Semisotnov G.V., Zerovnik E., Razguliaiev O.I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- R32.Ralston G.B. Temperature and pH dependence of the self-association of human spectrin. Biochemistry. 1991;30:4179–4186. doi: 10.1021/bi00231a011. [DOI] [PubMed] [Google Scholar]
- R33.Riordan J., Scandurra R. Essential arginyl residues in aspartate aminotransferases. Biochem. Biophys. Res. Commun. 1972;66:417–424. doi: 10.1016/s0006-291x(75)80344-5. [DOI] [PubMed] [Google Scholar]
- R34.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- R35.Schon A., Kannangara C.G., Gough S., Soll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1998;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- R36.Schuck P., Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- R37.Scotto d’Abusco A., Ammendola S., Scandurra R., Politi L. Molecular and biochemical characterization of the recombinant amidase from hyperthermophilic archaeon Sulfolobus solfataricus . Extremophiles. 2001;5:183–192. doi: 10.1007/s007920100190. [DOI] [PubMed] [Google Scholar]
- R38.Shima S., Thauer R.K., Ermler U., Durchschlag H., Tziatzios C., Shubert D. A mutation affecting the association equilibrium of formyltransferase from the hyperthermophilic Methanopyrus kandleri and its influence on the enzyme’s activity and thermostability. Eur. J. Biochem. . 2000;267:6619–6623. doi: 10.1046/j.1432-1327.2000.01756.x. [DOI] [PubMed] [Google Scholar]
- R39.Shin S., Yun Y.S., Koo H.M., Kim Y.S., Choi K.Y., Oh B.H. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J. Biol. Chem. 2003;278:24937–24943. doi: 10.1074/jbc.M302156200. [DOI] [PubMed] [Google Scholar]
- R40.Villeret V., Clantin B., Tricot C., Legrain C., Roovers M., Stalon V., Glansdorff N., Van Beeumen J. The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures. Proc. Natl. Acad. Sci. USA. 1998;95:2801–2806. doi: 10.1073/pnas.95.6.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Wah D.A., Romero A., Gallego Del Sol F., Cavad B.S., Ramos M.V., Grangeirp T.B., Sampaio A.H., Calvete J.J. Crystal structure of native an Cd/Cd-substituted Dioclea guaianensis seed lectin.A novel manganese-binding site and structural basis of dimer-tetramer association. J. Mol. Biol. 2001;310:885–894. doi: 10.1006/jmbi.2001.4814. [DOI] [PubMed] [Google Scholar]
- R42.Williams D.L. Jr., Rapanovich I., Russel A. Proteins in essential nonaqueous environments. In: Gregory R.B., editor. Protein-Solvent Interactions. New York: Marcel Dekker; 1995. pp. 327–341. [Google Scholar]
- R43.Yao Y., Ghosh K., Epand R.F., Epand R.M., Ghosh H.P. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology . 2003;310:319–332. doi: 10.1016/s0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]
- R44.Zeng X., Zhu H., Lashuel H.A., Hu J.C. Oligomerization properties of GCN4 leucine zipper e and g position mutants . Protein Sci. . 1997;6:2218–2226. doi: 10.1002/pro.5560061016. [DOI] [PMC free article] [PubMed] [Google Scholar]