Abstract
In Methanococcus maripaludis strain JJ, deletion of the homolog to cbiJ, which encodes the corrin biosynthetic enzyme precorrin 6-X reductase, yielded an auxotroph that required either cobamide or acetate for good growth. This phenotype closely resembled that of JJ117, a mutant in which tandem repeats were introduced into the region immediately downstream of the homolog of cbiJ. Mutant JJ117 also produced low quantities of cobamides, about 15 nmol g–1 protein or 1–2% of the amount found in wild-type cells. These results confirm the role of the cbiJ homolog in cobamide biosynthesis in the Archaea and suggest the presence of low amounts of a bypass activity in these organisms.
Keywords: archaea, cbiJ, vitamin B12 biosynthesis
Introduction
Methanococcus maripaludis is a facultative, autotrophic methanogen that produces methane from H2 + CO2 (Jones et al. 1983a). In methanogens, cobamide is a cofactor of the N5-methyltetrahydromethanopterin: CoM methyltransferase, which catalyzes an essential step of methane production (Kengen et al. 1992). The coenzyme is also a component of the acetyl-CoA decarbonylase/synthase complex (ACDS) that is required for autotrophic acetyl-CoA biosynthesis by the modified Ljungdahl-Wood pathway (Abbanat and Ferry 1991). Although many methanogens contain cobamide factor III (Coα-[(5-hydroxybenzimidazoyl)]-cobamide), methanococci contain pseudo vitamin B12 (Coα-[α-(7-adenyl)]-cobamide) (Stupperich and Kräutler 1988). Labeling studies and comparative analyses of genomic sequences suggest many similarities of the cobamide biosynthetic pathway in archaea to that in bacteria (Eisenreich and Bacher 1991, Selkov et al. 1997). However, there is direct evidence for only a few of the steps (Scherer et al. 1984, Blanche et al. 1991, Thomas and Escalante-Semerena 2000, Woodson et al. 2003, Woodson and Escalante-Semerena 2004).
In previous research, a mutant JJ117 of M. maripaludis was isolated by random mutagenesis and enrichment for an acetate auxotroph (Kim and Whitman 1999). Growth of this mutant depended on either acetate or cobalamin, and best growth was obtained with both compounds. The mutation was due to tandem insertion of a plasmid into a homolog of phosphopentose mutase (ppm), a gene that was not known to play a role in either acetate or cobamide biosynthesis (Kim and Whitman 1999). Thus, it was possible that the auxotrophic phenotype of the mutant JJ117 was due to an effect on expression of a neighboring gene. In this work, experiments were performed to discover and characterize the gene directly responsible for the auxotrophic phenotype of JJ117.
Materials and methods
Strains, media and growth conditions
Methanococcus maripaludis JJ was obtained from W.J. Jones (Jones et al. 1983a). Escherichia coli strains SURE, XL-1 Blue MRF’ were obtained from Stratagene (La Jolla, CA) and TOP10 from Invitrogen (Carlsbad, CA). Methanococci were grown at 37 °C with 275 kPa of H2:CO2 (80:20) gas in the minimal medium McN (Whitman et al. 1986). Other media used were: McNA, McN medium + 10 mM acetate; McNAY, McNA medium + 2 g l–1 of yeast extract; McNV, McN medium + 1 µM of cobalamin; McNAV, McNA medium + 1 µM of cobalamin; McNAYV, McNAY medium + 1 µM of cobalamin. Plating was performed as described previously (Jones et al. 1983b). The inocula for the growth experiments were grown to an absorbance (at 600 nm) of 0.2 to 0.5 in McNYAV medium. For growth of methanococci with puromycin, a concentration of 2.5 µg ml–1 was used. Prior to inoculation, cultures were diluted in McN medium. Glassware for growth experiments was cleaned in 0.1 M HCl for at least 12 h at room temperature and autoclaved for 20 min. Stoppers for culture tubes were autoclaved with 0.2 N NaOH for 20 min. Glassware and stoppers were rinsed in tap and deionized water after autoclaving. Solid medium for M. maripaludis was prepared as described in Tumbula et al. (1995).
Molecular biology techniques
Ligation was performed at 4 or 16 °C with 1–3 units (Promega, Madison, WI) or 400 units (New England Biolabs, Beverly, MA) of T4 DNA ligase, respectively, as described by the manufacturer’s instructions. Plasmids from E. coli were purified with Wizard plus mini prep kits (Promega) or QIAprep spin miniprep kits (Qiagen, Hilden, Germany). The DNA was gel-purified with the QIAquick gel extraction kit (Qiagen). Restriction enzymes, alkaline phosphatase and Klenow fragments were purchased from New England Biolabs or Promega. The DNA was transformed into E. coli witha Gene Pulser Electroporator (Bio-Rad, Hercules, CA) set at 25 µF and 2.50 kV or with the One Shot kit (Invitrogen, Carlsbad, CA). Esherichia coli was screened on LB agar plates or LB broth medium with 50 µg ml–1 of ampicillin. Total genomic DNA of M. maripaludis was purified by a mini-prep method described previously (Ausubel et al. 1994). Plasmid DNA, 5 ng, or 50 ng of genomic DNA was used as a template for PCR amplifications. For PCR, 0.1 or 0.2 µM of primers were added. Unless specified differently, the PCR reactions were performed with the Ready-To-Go Kit (Amersham Pharmacia Biotech, Piscataway, NJ) with initial denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 2 min. Sequencing was carried out on an Applied Biosystems Model 373 automated sequencer in the Molecular Genetics Instrumentation Facility at the University of Georgia. Sequences were analyzed with the programs of GCG (University of Wisconsin), TIGR (http://www.tiger.org) and ERGO (http://wit. integratedgenomics.com/ERGO/).
Methanococcus maripaludis transformation
Polyethyleneglycol transformation procedures for M. maripaludis have been described previously (Tumbula et al. 1994). Transformations for gene replacement were performed with linear forms of plasmids, which were constructed by digestion with the specified restriction enzymes and subsequent gel purification.
Plasmid construction and identification of flanking sequences
Construction of pWDK117-11 was described previously (Table 1). JJ117-11, which was isolated from the transformation of the wild-type strain by pWDK117-11, was cultured in McNYAV + puromycin (Table 2). Genomic DNA was isolated and treated with 20 units of EcoRI overnight at 37 °C. The digested DNA was purified with a Wizard DNA clean-up kit (Promega), and 10 µl of the sample containing 0.14 µg of DNA was religated with T4 DNA ligase. The ligation mixture was electroporated into E. coli SURE after drop dialysis against distilled water on a VSWP filter paper (pore size 0.025 µm, Millipore, Billerica, MA). Plasmids that were purified from the E. coli transformants were screened by digestion with EcoRI and EcoRV. The presence of the flanking genomic DNA on the plasmid was confirmed by the increase in the size of digestion product to 2.0 kb. The plasmid containing the flanking sequence was named pWDK117-12. pWDK120 was constructed by ligating a 0.35 kb PCR amplification product from the new sequence (see below) into the NsiI site of pIJA03. For the PCR, 2.5 units of pfu polymerase (Stratagene) and the primers, 5′-TAATTTTAACCACCACAACA (positions +89–103 in Figure 1) and 5′-GGAAGTATCCTCACAA TCAG (positions +454–469) were employed. The standard PCR conditions were used except that the annealing temperature was 51.5 °C. The PCR product was gel-purified and ligated into the EcoRV site of p-ZErO. After isolation, the plasmid was digested with NsiI, and the resulting fragment was ligated into pIJA03, completing construction of pWDK120.
Table 1.
Plasmids used in this study.
| Plasmids | Characteristics | Reference |
| pIJA03 | Contains modified pac cassette (purR), developed from pMEB.2 | Stathopoulos et al. (2001) |
| pWLG40 + lacZ | Expression shuttle vector for methanococci, contains lacZ | Gardner and Whitman (1999) |
| pWDK200 | pIJA03 + S-layer promoter (Psla) | This work |
| pWDK117 | pMEB.2 + 1.2 kb of ppm from M. maripaludis JJ | Kim and Whitman (1999) |
| pWDK117-11 | pMEB.2 + 0.68 kb BglII-BamHI fragment of ppm | Kim and Whitman (1999) |
| pWDK117-12 | pWDK117-11 + 1.0 kb of DNA downstream of ppm | This work |
| pWDK117-13 | pWDK120 + 0.70 kb of DNA upstream of cbiJ | This work |
| pWDK117-14 | pMEB.2 + 0.55 kb of DNA downstream of ppm in strain JJ117 | This work |
| pWDK120 | pIJA03 + 0.38 kb of cbiJ | This work |
| pWDK121a | pIJA03 + 0.90 kb of DNA upstream of cbiJ | This work |
| pWDK121 | pWDK121a + 1.10 kb of DNA downstream of cbiJ | This work |
| pWDK201 | Inserts Psla upstream of ppm upon transformation | This work |
| pWDK202 | Inserts Psla upstream of cbiJ upon transformation | This work |
| pWDK220 | pWDK40 + plus 0.97 kb ppm | This work |
| pWDK240 | Inserts 2.10 kb DNA from ppm into pWDK40 | This work |
| pWDK241 | Inserts 1.20 kb DNA from ppm into pWDK40 | This work |
| pWDK242 | Inserts 1.10 kb DNA from ppm-cbiJ into pWDK40 | This work |
| pWDK243 | Inserts 0.20 kb DNA from ppm into pWDK40 | This work |
Table 2.
Strains used in this study.
| Strains | Characteristics | Reference |
| JJ | Wild-type strain of Methanococcus maripaludis | Jones et al. (1983) |
| JJ117 | Transformant of pWDK117, contains tandem insertions, cobalamin and acetate auxotroph | Kim and Whitman (1999) |
| JJ117-11 | Transformant of pWDK117-11 | Kim and Whitman (1999) |
| JJ117-12 | Transformant of pWDK117-12 | This work |
| JJ117-13 | Transformant of pWDK117-13 | This work |
| JJ117-14 | Transformant of pWDK117-14 | This work |
| JJ120 | Transformant of pWDK120, disruption of cbiJ | This work |
| JJ121 | Transformant of pWDK121, deletion of cbiJ | This work |
| JJ122 | Transformant of pWDK121, deletion of cbiJ | This work |
| JJ201 | Transformant of pWDK201, ppm under control of Psla | This work |
| JJ202 | Transformant of pWDK202, cbiJ under control of Psla | This work |
| JJ240 | Transformant of pWDK240, overexpress ppm and potential cbiJ antisense mRNA | This work |
| JJ241 | Transformant of pWDK241, overexpress ppm | This work |
| JJ242 | Transformant of pWDK242, overexpress 3′ portion of ppm and potential antisense cbiJ mRNA | This work |
| JJ243 | Transformant of pWDK243, overexpress 3′ portion of ppm | This work |
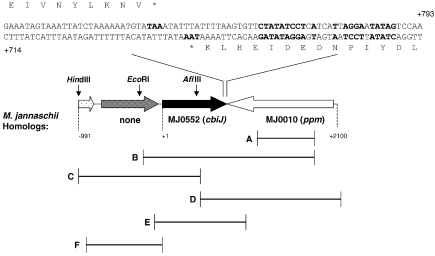
Figure 1.
Map of the genes downstream of the ppm homolog of Methanococcus maripaludis strain JJ. Numbering of the DNA sequence begins with the putative start codon of cbiJ. The nucleotide sequences of the termination regions of cbiJ and the ppm homolog are shown on the top. The bold characters in the sequences indicate potential termination codons and stem loop structures. The HindIII and EcoRIsites were used for identification of the flanking sequences of cbiJ. The AflIII site was used in the construction of pWDK117-14. A. The region of genomic DNA cloned in pWDK117-11. B. The region of genomic DNA cloned in pWDK117-12. C. The region of genomic DNA cloned in pWDK117-13. D. The region of genomic DNA from the auxotroph JJ117 cloned in pWDK117-14. E. The region deleted in JJ121 following gene replacement. F. The region used as a probe for the Southern blots of JJ121 and JJ122.
To identify sequences further upstream, pWDK120 was transformed into the wild type to form JJ120. Total DNA from JJ120, 1 µg, was treated with 30 units of HindIII overnight at 37 °C. The digested DNA was religated and electroporated as described above. The resulting plasmids were screened by restriction with XbaIand XhoI. An increase in size of the 0.7 kb fragment of the vector to 1.0 kb indicated the presence of the upstream sequences. This plasmid was named pWDK117-13.
To clone the region downstream of the ppm homolog in the mutant JJ117, a strategy similar to that of Hildebrandt and Nellen (1991) was used. Genomic DNA, 1 µg, was digested with 5 units of AflIII and 10 units of NruI. These sites were present in the vector portion of the tandem repeats immediately upstream of the cloned DNA. Therefore, restriction with two enzymes prevented ligation of the vector from tandem repeats. The only product expected would be formed by ligation between the AflIII sites in the vector and upstream in the genome. The resulting plasmid, pWDK117-14, contained an additional 0.55 kb of genomic DNA. To examine further the sequence of this region in JJ117 and JJ, genomic DNA was purified with the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). The primers cbiJ_for (5′-GGATAGTCCTGAAACGTCAAATCAG) and cbiJ_rev (5′-GGCCATTGAGATTAGAACACGATG) were used for PCR amplification of the region –190 bp upstream of the start of the cbiJ gene to +900 bp downstream and for DNA sequencing. PCR reactions were performed using 5 U of Herculase Enhanced DNA Polymerase (Stratagene) with the following PCR conditions: 92 °C for 2 min, 15 cycles of (92 °C for 30 s; 50 °C for 30 s, which increased 1 °C per cycle; 65 °C for 1 min), 25 cycles of 92 °C for 30 s, 52 °C for 30 s and 68 °C for 60 s, with an increase of 10 s per cycle. After electrophoresis, PCR products were extracted using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Sequencing was performed with an ABI Model 3730 sequencer at the University of Michigan DNA Sequencing Core (Ann Arbor, MI).
pWDK121 was constructed in pIJA03 to make a gene replacement of cbiJ. The upstream fragment (0.9 kb) was obtained by digestion of pWDK117-13 with 20 units of NdeI, gel purification and ligation into the NdeI site of pIJA03. The resulting plasmids were screened to identify pWDK121a, which possessed the insert in the same transcriptional orientation as the puromycin transacetylase gene in the pac cassette. The second fragment was obtained by PCR amplification of the ppm homolog in pWDK117-12. The primers used were: 5′-CCCCCCGGTACCATCGTGTTCTAATCTCAATGG (positions +866–886 in Figure 1) and 5′-CCCCCCGCTAGCTTAGATTTTGACGGACTTTTG (positions +2000–2020 in Figure 1), where the underlined positions represent KpnI and NheI restriction sites, respectively. The PCR conditions were the same as those for the construction of pWDK120 except that 25 cycles were performed. The product was then digested with 10 units of KpnI and NheI for2 h and gel-purified. Thedevelopment of pWDK121 wascompleted byinsertion of the purified product into the KpnI and NheI sites of pWDK121a.
pWDK200 was constructed by ligating the strong S-layer promoter (Psla) into pIJA03. The promoter was obtained from pSla-vhcI (Kansy et al. 1994) by 25 cycles of PCR amplification with two primers: 5′-TAAAGTGACTAGTCAATTTCGAAAGTAATAAAAATAAT and 5′-ATTTGCATGCATATGCACCTTTTGTTTTTATTTT. The PCR conditions were denaturation at 94 °C for 1 min, annealing at 48.8 °C for 5 min, and extension at 65.0 °C for 2 min. The 0.25 kb product was then gel-purified and inserted into the EcoRV site of pZErO-2. To obtain the fragment containing the promoter sequence, the plasmid was digested with BamHI-NsiI. The digested fragment was then inserted into the BamHI-NsiI sites of pIJA03. pWDK201 was developed to insert Psla prior to the ppm homolog after integration of the plasmid into the genome (Gardner and Whitman 1999). Homologous recombination of the plasmid into the genome would yield a merodiploid where a truncated form of ppm is under the control of the native promoter and the full length gene is under the control of Psla. For the construction, the 5′ end of ppm was cloned next to Psla in pWDK200. First ppm was PCR amplified with two primers: 5′-GCGGTGGCATGCATGAAAACAATCGT (underline indicates NsiI site, positions +2021–2045 in Figure 1) and 5′-ATTACGGCGGCCTTTTCCACAG (positions +1080– 1101 in Figure1) from genomic DNA. The product was treated with NsiI and BamHI and cloned into the NsiI-BglII sites of pIJA200. This treatment was successful because the PCR product possessed an internal BamHI site at positions +1101– 1105. Similarly, pWDK202 was constructed to insert Psla prior to the genomic copy of cbiJ following transformation. Two primers 5′-CCGTAATAGAAGAAATGGATG (positions +1066–1086 in Figure 1) and 5′-TTGCATGCATAACATATGGATTCGTGGCGGA (underline indicates NsiI site, positions +4–24 in Figure 1) were used to amplify a 1.0 kb portion of cbiJ-ppm by PCR. Genomic DNA of the wild type was used as the template. The standard conditions were used for PCR amplification except that 25 cycles were performed, the annealing temperature was 59.5 °C, and extension was for 2 min and 30 s. The PCR product was digested with NsiI and ligated into the NsiI site of pIJA200. In addition to the NsiI site in the primer, the PCR product possessed another NsiI site in the amplified DNA (positions +1063–1068 in Figure 1). The orientations of the inserted fragments were determined by restriction of the plasmids with HindIII and BamHI.
pWDK240 and pWDK241 were derivatives of the expression shuttle vector pWLG40 + lacZ and were used for overexpression of the ppm and cbiJ antisense mRNA. Because ppm contained an internal NsiI site (at position +1065–1068 in Figure 1), pWDK220 was first constructed by ligating the 0.97 kb ppm fragment from the NsiI-XbaI digestion of pWDK201 into the NsiI -XbaI sites of pWLG40 + lac. This vector was then digested with BglII and XbaI. Because ppm contained an internal BglII site at positions +1785–1790, the product could then be used to clone the 3′-end of ppm without using NsiI. The remaining portions of ppm were obtained by PCR amplification. For pWDK240, the primers were 5′-AAGTTTAGTAGATAGGGCGG (positions +2042–2061) and 5′-CTAGTCTAGATAGCAAATACGACTTCGGGGA (underline indicates XbaI site, positions –93–73 in Figure 1). The PCR was performed under standard conditions except that the annealing was performed at 60 °C and the extension was performed for 3 min and 30 s. For pWDK241, the primers were 5′-TTAGATTTTGACGGACTTTTG (positions +2000–2019 in Figure 1) and 5′-CTAGTCTAGAGCTTTATCCATGTAATTTAATATTAA (underline indicates XbaI site, positions +828–853 in Figure 1). Standard conditions were used for the PCR except that only 25 cycles were performed and extension was for 2 min and 30 s. Both PCR products were digested with BglII and XbaI, ligated into the BglII-XbaI digested pWDK220. To make pWDK242 and pWDK243, the PCR products were digested with NsiI and XbaI to yield 0.96- and 0.2-kb gene fragments, respectively, and ligated into pWLG40 + lacZ. Because of the internal NsiI site at positions +1063–1068 in ppm, these constructions contained only the 3′ portion of ppm.
PCR confirmation of ΔcbiJ mutation
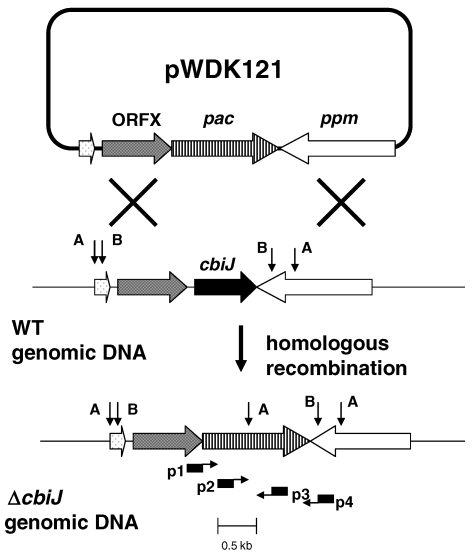
To confirm the replacement of the genomic cbiJ with the pac cassette, primer sequences for the PCR were selected from 100 bp upstream and 200 bp downstream of the cbiJ (Figure 2): p1 (5′-TAGCAAATACGACTTCGGGGA, positions –93–73 in Figure 1) and p4 (5′-GATGGGCGCGTTTGTAAATTC, positions +975–995 ). Other primers were complementary to the pac cassette: p3 (5′-TGCAAGAACTCTTCCTCACG) and p4 (5′-GTCTCTTTCACCAGCAGCTT). The reactions contained 50 ng of template DNA in 1 µl, 1 µl of each primer, 2.5 µl of DMSO (dimethylsulfoxide) and 19.5 µl of distilled water in the Ready-To-Go PCR tubes (Amersham Pharmacia Biotech). The standard PCR conditions were used except that the extension time was 3 min.
Figure 2.
Scheme for construction of the cbiJ deletion (ΔcbiJ) by gene replacement in M. maripaludis. A and B indicate locations of the restriction sites for AccI and BamHI, which were used during the Southern hybridization. Also, locations of primers p1–p4 for the PCR are indicated.
Southern hybridization
Genomic DNA, 2 µg, was digested with AccI (15 units) for 29 h or BamHI (20 units) for 44 h. The restricted DNA was separated on a 1.0 % (w/v) agarose gel and transferred to a positively charged nylon membrane (Micro Separations, Westborough, MA) by the downward capillary method (Sambrook and Russell 2001). Prehybridization and hybridization were performed at 65 °C overnight in a solution of 2× SSC, 0.01% (w/v) SDS, 0.1% (w/v) N-lauroylsarcosine and 1% blocking reagent (nonfat milk). Membranes were washed twice in solutions of 2× SSC plus 0.1% SDS for 5 min at room temperature and 2× SSC plus 1% SDS for 15 min at 65 °C. The probe was radiolabeled with [α-P32] dATP (ICN, Aurora, OH) according to a random priming method (Sambrook and Russell 2001). The template for the random priming reaction was the 0.96 kb NdeIfragment (Positions –951 to +6 in Figure 1) from pWDK121. Approximately 2.5 × 107 dpm of labeled probe was added to the hybridization. Prior to visualization with ImageQuant 1.1 software, the membranes were exposed in a Phosphoimager cassette for 36 h. The molecular weight markers were constructed by digestion of the PCR product of the wild-type genomic DNA and primers p1–p4 with AccI, BamHI and AccI plus PvuII.
Measurement of cobamide concentration
Cobamides were extracted from cells as described by Gorris et al. (1988). In this experiment, 100 ml of mid-exponential phase cultures (A600 = 0.45–0.55) were harvested by centrifugation at 4 °C. After the extraction, the total volume was 25 ml. Of this, 4 ml was concentrated to dryness in a high speed vac (Uniequip, Martinsried, Germany) and resuspended in 50 µl of distilled water. The cobamide concentration was estimated as described by Maggio-Hall and Escalante-Semerena (1999) and Thomas and Escalante-Semerena (2000). For this experiment, Vogel-Bonner medium overlaid by 3 ml of medium containing Salmonella enterica serovar Typhimurium TR6583 [metE205 ara-9] or JE1299 [metE205 ara-9 btuB7::MudJ] were spotted with 2.5 or 5 µl of various dilutions of the methanococcal extracts. The standards were spots of cobalamin solutions containing 0.0025 to 20 pmol of cobalamin in 2.5 or 5 µl, depending on the experiment. The quantity of the cobamide in the methanogens was determined by comparing the diameters of the confluent growth between the standard and samples. For the control JE1299, 2.5 or 5 µl of methionine (0.4 to 3 µmol) solutions was also spotted.
Accession number
The GenBank accession number for the M. maripaludis cbiJ and surrounding sequence is AF402610.
Results
Identification of the flanking sequence of the ppm homolog
In previous studies, the insertion of tandem repeats of the plasmid pWDK117 into the M. maripaludis homolog of the Methanocaldococcus jannaschii gene MJ0010 yielded an acetate and cobamide auxotroph (Kim and Whitman 1999). However, inactivation of the MJ0010 homolog did not appear to be responsible for the observed phenotype for the following reasons. First, the insertion was only expected to produce a small truncation of the ORF, and it seemed unlikely that this would be sufficient to inactivate the gene product. Second, mutants bearing a single insertion, which would also truncate the ORF, were not auxotrophic, and their growth properties were indistinguishable from wild type. Therefore, the phenotype depended on the presence of the tandem repeats. Third, the MJ0010 homolog possessed sequence similarity to ppm, and it was difficult to rationalize a role for this gene product in cobamide or acetate biosynthesis (Kim and Whitman 1999). Thus, it was hypothesized that inactivation of a downstream gene might be responsible for the auxotrophic phenotype.
The downstream genes were cloned by transforming M. maripaludis with pWDK117-11 to yield JJ117-11 (Table 1). The plasmid pWDK117-11 contained a 0.68 kb gene fragment internal to the ppm homolog and was expected to integrate within this gene by homologous recombination. The genomic DNA of JJ117-11 was then digested with EcoRI, ligated and transformed into E. coli. In this fashion, pWDK117-12 was isolated (Figure 1). Sequencing of pWDK117-12 revealed that it contained genomic DNA, including 0.3 kb of the ppm homolog and 1.0 kb of a downstream ORF. To obtain additional flanking genes, the original strategy was repeated. First, a portion of the downstream ORF was cloned into the insertion plasmid pIJA03 to form pWDK120. After transformation of the wild type strain, pWDK117-13 was isolated following HindIII digestion and transformation into E. coli. The sequencing of this plasmid revealed additional downstream genes (Figure 1).
Analysis of the sequences of these clones identified the 3′ region of the ppm homolog, which had been missing in previous clones (Kim and Whitman 1999). Analysis of the ORF immediately downstream of the ppm homolog showed that it possessed 52% sequence similarity to the cbiJ homolog of Methanocaldococcus jannaschii (Figure 1). Downstream of the cbiJ homolog was a hypothetical ORF lacking similarity to any known gene and the 3′-end of an ORF that was only 180 bp and too short to identify.
Mutagenesis of cbiJ in M. maripaludis
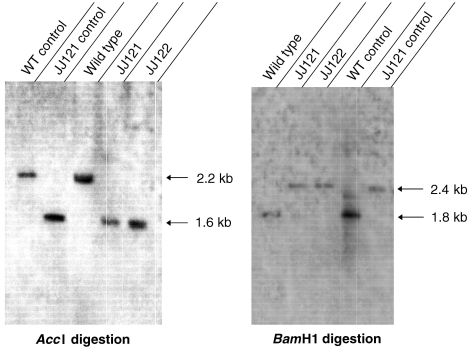
If the original phenotype of JJ117 was due to an effect on expression of cbiJ, deletion of cbiJ would be expected to yield a mutant with the same phenotype. Therefore, the mutants JJ121 and JJ122 were constructed by replacing cbiJ with the pac cassette (Figure 2). The genotypes of these mutants were confirmed by PCR amplification and Southern hybridization (Figure 3 and data not shown). First, PCR amplification of the genomic DNA was performed to verify the gene replacement of cbiJ in JJ121 and JJ122. Although the primer pair p1–p4 failed to yield a product with JJ121 (data not shown), the expected 1.1 kb product was found with the wild type. However, amplification of JJ121 DNA with primer pair p2–p3 confirmed the placement of the pac cassette in the genome, and amplification with the primer pair p2–p4 confirmed the replacement downstream of ppm. Because primer pair p1–p3 also failed to produce a product in either JJ121 genomic DNA or the plasmid control, it seemed likely that the high GC content of the puromycin transacetylase gene prevented amplification across the 5′ boundary of the cassette (Henke et al. 1997). Thus, it was not possible to demonstrate the 5′-end of the gene replacement and the absence of tandem insertions by this method. Southern hybridization was then used to confirm the gene replacement of cbiJ in the mutants. In the first hybridization, AccI-digested genomic DNA produced a 2.2 kb band for wild type and 1.6 kb band for the mutants JJ121 and JJ122 (Figure 3). These were the sizes expected for a replacement of cbiJ with the pac cassette. Similarly, hybridization of BamHI digested genomic DNA produced a 1.8 kb band for wild type and a 2.4 kb band for the mutants (Figure 3). These results confirmed the gene replacement of cbiJ by a single copy of the pac cassette.
Figure 3.
Southern hybridizations of the wild type and ΔcbiJ mutants, JJ121 and JJ122, with a probe to the cbiJ locus. Genomic DNA was restricted by either AccI or BamHI. The probe was constructed by random priming of a linear portion of pWDK121 corresponding to positions –951 to +6 (Figure 1). Controls were PCR amplicands of genomic DNA that had been digested with the restriction enzymes.
Phenotype of the ΔcbiJ mutants
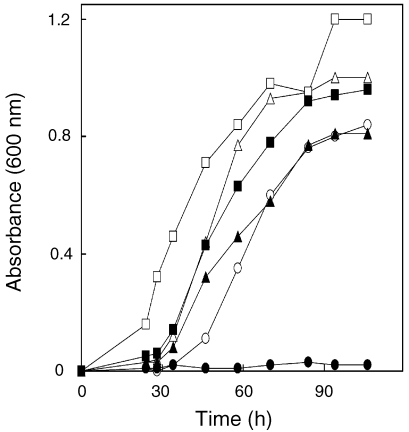
The original mutant JJ117 was an auxotroph that required either acetate or cobamide for growth (Kim and Whitman 1999). In the presence of both compounds, growth was comparable to that of the wild type. For the ΔcbiJ mutant JJ121, a similar phenotype was observed. Addition of acetate, cobamide or acetate plus cobamide allowed growth in minimal medium (Figure 4 and data not shown). To determine if JJ117 and JJ121 required similar concentrations of cobamide for growth, the cultures were incubated in minimal medium plus 0.05, 0.3 and 1.0 µM of cobamide. Based on a cobamide content of 500 nmol per gram of protein in methanococci (see below), the concentration necessary to support full growth in minimal medium was expected to be 0.3 µM. Both JJ117 and JJ121 grew in medium containing 0.3 and 1.0 µM of cobalamin. However, no growth was observed with 0.05 µM of cobalamin. These results confirmed the similarity of their phenotypes.
Figure 4.
Stimulation of growth of the ΔcbiJ mutant JJ121 by acetate and cobalamin. Growth of wild type in McN (○), McNA (△) and McNV (□) medium. Growth of the mutant JJ121 in McN (●), McNA (▲) and McNV medium (■). The inoculum was 4.6 × 105 cells that had been grown in McNAYV medium.
In experiments where large inocula were used, JJ121 grew in minimal medium without acetate or cobalamin after a long lag. Upon subsequent transfers to minimal medium, a lag was no longer observed. Therefore, the occasional growth observed in minimal medium appeared to be due to selection for revertants. Because the revertants remained puromycin-resistant, the mutation presumably occurred at a second site.
Mechanism of cbiJ inactivation in JJ117
Although the original mutant JJ117, with an insertion of tandem repeats, possessed a severe auxotrophic phenotype, other transformants formed by integration of single copies of other vectors at the same site were not auxotrophs (Kim and Whitman 1999). For this reason, it seemed possible that the cobamide auxotrophy of JJ117 may have been due to a second mutation in cbiJ, possibly caused by the formation of tandem repeats. To test this hypothesis, the 3′-end of cbiJ was cloned and sequenced from JJ117 (positions +394–+945 in Figure 1), but no differences were observed between the sequence of the gene from the wild type and that from the mutant. The entire cbiJ gene was then PCR amplified from JJ117 and the wild type and sequenced. These experiments confirmed the identity of the entire cbiJ gene in the mutant and wild type. Thus, the phenotype of JJ117 did not appear to be due to deletions or other mutations in cbiJ.
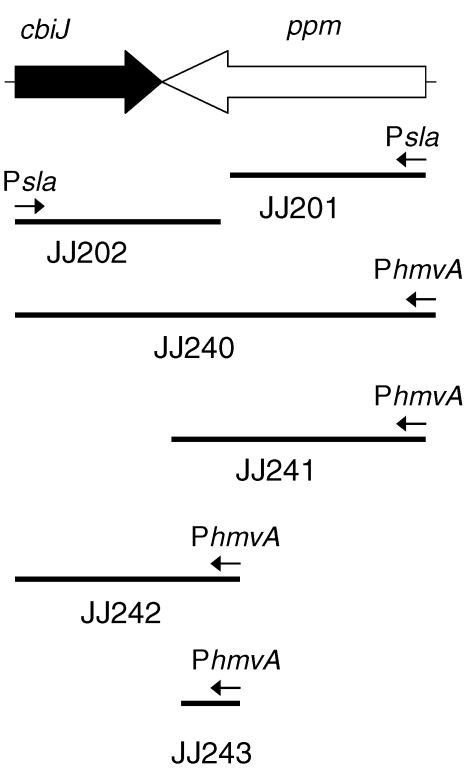
An alternative explanation for the phenotype of JJ117 was that overexpression of ppm from tandem repeats interfered with the expression of cbiJ. For instance, the production of cbiJ antisense mRNA might inhibit either translation or the stability of the mRNA. Therefore, integration and expression vectors containing different regions of ppm and cbiJ were transformed into wild-type M. maripaludis to determine if overexpression of ppm caused cobamide auxotrophy (Figure 5). The transformation efficiencies of the expression vectors were comparable to controls to insure that the efficiencies were high. Thus, the transformants were not formed by unusual or rare genetic events. In JJ201, the very strong Psla promoter was integrated upstream of the genomic copy of ppm (Figure 5). This construction was expected to result in a merodiploid with very high expression of the copy of ppm adjacent to cbiJ (Gardner and Whitman 1999). For the characterization of the transformants, growth experiments were performed in McN, McNA and McNV broth media after isolating the transformants from McNAYV agar plates. In these experiments, significantly slower growth of the transformants was not observed in the McN medium (data not shown). These results suggested that the transformants were not auxotrophs. However, because merodiploids are unstable in M. maripaludis, it is possible that JJ201 was overgrown by revertants. Therefore, other constructions were attempted where ppm was overexpressed from the shuttle vectors, JJ240–JJ243 (Figure 5). In these constructions, JJ240 and JJ242 were expected to contain high amounts of cbiJ antisense RNA produced from the expression vector. JJ241 and JJ243 were designed as negative controls. However, these transformants also failed to show the auxotrophic phenotype. Lastly, it was possible that the auxotrophic phenotype in JJ117 resulted from overexpression of cbiJ. In JJ202, cbiJ was expected to be overexpressed by the very strong histone promoter Psla (Figure 5). However, it was also not auxotrophic. Therefore, another model of gene regulation must be required to explain the effect of tandem repeats on cbiJ expression.
Figure 5.
Overexpression of ppm-cbiJ in Methanococcus maripaludis. The lines indicate the DNA fragments cloned into the vectors for overexpression. JJ201 and JJ202 were transformed by expression vectors designed to integrate into the genome, placing the promoters upstream of ppm and cbiJ, respectively. JJ240–JJ243 were transformed by shuttle vectors containing the indicated cloned DNA. Psla and PhmvA represent promoter sequences for the S-layer gene and a histone gene in Methanococcus voltae, respectively. Both promoters are believed to be highly expressed in M. maripaludis.
Quantification of cobamide in the wild type and JJ117
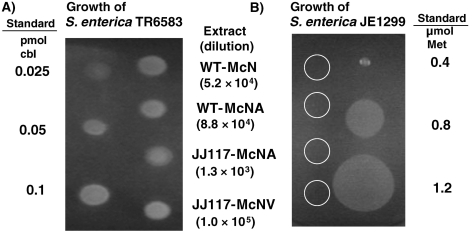
To understand the stimulation of growth of JJ117 with acetate in minimal medium, the cobamide content of the strain was measured. The stimulation with acetate was unexpected because acetate is unlikely to be specifically involved in the biosynthesis of cobamide. Moreover, cobamides are components of the N5-methyltetrahydromethanopterin: CoM methyltransferase, an essential enzyme in the pathway of methanogenesis. Therefore, cobamides were expected to be essential for growth. Thus, it was hypothesized that a small amount of cobamide was synthesized in JJ117, which was sufficient to allow for growth in the presence of acetate. In M. maripaludis, acetate can be converted into acetyl–CoA (Shieh and Whitman 1987). In the absence of acetate, acetyl-CoA decarbonylase/ synthase (ACDS) is required for growth. This enzyme complex also requires cobamide. Therefore, the addition of acetate to minimal medium might reduce the total cobamide demand and allow for growth on reduced concentrations of cobamide. To verify this hypothesis, the concentrations of cobamides in extracts of wild type and JJ117 were determined with the indicator strain S. enterica serovar Typhimurium TR6583 (Maggio-Hall and Escalante-Semerena 1999, Thomas and Escalante-Semerena 2000). Detectible but low amounts of cobamides were found in extracts of the mutant JJ117 (Figure 6A). Because S. enterica serovar Typhimurium TR 6583 also responds to methionine, S. enterica serovar Typhimurium JE1299 was utilized as an indicator for this amino acid. The concentrations of methionine in the methanococcal extracts were too low to produce a response in the assay (Figure 6B). This result indicates that a small amount of the cobamide was synthesized in the mutant JJ117. From other experiments, the cobamide concentrations were estimated from the diameter of confluent growth from strain TR6583. For the wild type, the concentrations of cobamide were 800–1100 nmol g–1 protein following growth in McN and McNA medium. For JJ117, only 15 nmol g–1 protein was found following growth in McA medium. Following growth of JJ117 in McV medium, which contained cobalamin, the concentrations were 1200 nmol g–1 protein or comparable to wild-type cells. The concentrations of cobamide determined by this bioassay were comparable to the concentrations of 145–620 nmol g–1 protein determined in methanococci by liquid chromatography (Stupperich and Kräutler 1988, Gorris and van der Drift 1994).
Figure 6.
Bioassay for cobamides in cell extracts of the wild type and JJ117. All spots were 2.5 µl, and extracts were diluted by the indicated amounts to produce zones of confluent growth. Salmonella enterica serovar Typhimurium TR6583 is dependent upon either cobamide (cbl) or methionine (Met). Salmonella enterica serovar Typhimurium JE1299 requires methionine for growth and does not respond to cobamides. For this control, undiluted methanococcal extracts were used, and the locations of the spots are indicated by the circles.
Discussion
In this project, we sought to explain the cobamide and acetate auxotrophy of the mutant JJ117, which contained tandem repeats of an insertion vector and portions of ppm, a homolog of phosphopentose mutase. The discovery of a homolog of cbiJ immediately downstream of ppm suggested that inhibition of expression of this gene was the cause of cobamide auxotrophy. In bacteria, the gene cbiJ encodes the cobalt-precorrin 6-X reductase, which catalyzes the reduction of the C18/C19 double bond of the tetrapyrrole “D” ring and the formation of cobalt-precorrin 6-Y during an early step in the anaerobic pathway of cobamide biosynthesis. In Paracoccus,mutants of cobK (a homolog of cbiJ that catalyzes the analogous step in the aerobic pathway) are cobamide auxotrophs (Blanche et al. 1992, Rondon et al. 1997, Shearer et al. 1999). However, the amino acid sequence of the cbiJ homolog in M. maripaludis and the other archaea possesses only 24–33% identity to the Paracoccus and other bacterial genes, so it was necessary to demonstrate its function in methanococci. Although the enzymatic specificity was not demonstrated directly, the cobamide auxotrophy of the cbiJ deletion mutant JJ121 confirmed the requirement of this gene for cobamide synthesis in M. maripaludis.
Although good growth of the original mutant JJ117 was found only in the presence of both acetate and cobamide, some growth was also observed in the presence of either acetate or cobamide alone. This phenotype could have resulted from two independent leaky mutants, one each in the pathways of acetate and cobamide biosynthesis. The isolation of a second mutant with a similar phenotype and genotype as JJ117 suggested that the phenotype was not due to a rare double mutation (Kim and Whitman 1999). The discovery of acetate auxotrophy in the ΔcbiJ mutants further eliminated this possibility. Cobamides are required for methyltetrahydromethanopterin: CoM methyltransferase as well as the acetyl–CoA decarbonylase/synthase reactions in methanococci. Because the first reaction is a required step in methanogens, mutations in cobamide biosynthesis were expected to be lethal. Growth of JJ121 on acetate alone suggested that the mutants contained small amounts of cobamide synthesized by an alternative reaction that bypasses cbiJ. Although the genome sequence of the related organism Methanococcus maripaludis strain S2 does not contain a detectable paralog to cbiJ (Hendrickson et al. 2004), the oxidation and reduction of C-C double bonds are common biosynthetic reactions and low levels of a nonspecific activity are not unexpected. Moreover, the possibility of nonspecific reductions have also been demonstrated for the conversion of Co(III) to Co(I) in a later step of cobamide biosynthesis (Fonseca and Escalante-Semerena 2001). In this hypothesis, the amount of the cobamide biosynthesized by the bypass reaction would be insufficient to maintain normal growth in minimal medium. However, acetate would spare the cobamide requirement because it is no longer required by the acetyl-CoA decarbonylase/synthase, and the small amount of remaining cobamide would then be sufficient for the methyltransferase. This conclusion was supported by showing that JJ117 in fact contained 1–2% of the cobamide of the wild type. The characteristics of these mutants suggest that the cells possess a hierarchy for cobamide utilization. When the amount of coenzyme is limited, it is used preferentially for essential functions such as methanogenesis.
The cbiJ and the ppm homologs in M. maripaludis are convergently transcribed. The cbiJ homologs (named cobK) are also convergently transcribed with other genes in Rhodococcus sp., Pseudomonas denitrificans and Paracoccus denitrificans (De Mot et al. 1994, Shearer et al. 1999). In M. maripaludis, the cbiJ and ppm homologs were separated by four bases and potential termination structures of stem loop and oligo thymidines overlapped in 3′ regions of the messages. Thus, it seemed possible that mutation in ppm could affect the expression of cbiJ through interactions of their mRNAs. For instance, overexpression of ppm could produce a large quantity of antisense cbiJ mRNA that might inhibit translation of cbiJ. However, overexpression of the ppm homolog in either cis or trans failed to generate the auxotrophic phenotype in minimal medium. Therefore, the mechanism of the effect of the original mutation in JJ117 on cbiJ expression is still not understood.
Acknowledgments
This work was supported by a DOE grant to WBW. We thank J.C. Escalante-Semerena for helpful discussions and providing the Salmonella strains and M.J. Warren for helpful comments.
References
- R1.Abbanat D.R., Ferry J.G. Resolution of component proteins in an enzyme complex from Methanosarcina thermophila catalyzing the synthesis or cleavage of acetyl-CoA. Proc. Natl. Acad. Sci. USA. 1991;88:3272–3276. doi: 10.1073/pnas.88.8.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Smith J.A., Seidman J.G., Struhl K. New York, Unit 2.4: Green Publishing Associates and Wiley-Interscience,; 1994. Current protocols in molecular biology; p. 4. [Google Scholar]
- R3.Blanche F., Robin C., Couder M., Faucher D., Cauchois L., Cameron B., Crouzet J. Purification, characterization, and molecular cloning of S-adenosyl-L-methionine:uroporphyrinogen III methyltransferase from Methanobacterium ivanovii . J. Bacteriol. 1991;173:4637–4645. doi: 10.1128/jb.173.15.4637-4645.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Blanche F., Thibaut D., Famechon A., Debussche L., Cameron B., Crouzet J. Precorrin-6x reductase from Pseudomonas denitrificans: purification and characterization of the enzyme and identification of the structural gene. J. Bacteriol. 1992;174:1036–1042. doi: 10.1128/jb.174.3.1036-1042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5.De Mot R., Nagy I., Schoofs G., Vanderleyden J. Sequences of the cobalamin biosynthetic genes cobK, cobL and cobM from Rhodococcus sp. NI86/21. Gene. 1994;143:91–93. doi: 10.1016/0378-1119(94)90610-6. [DOI] [PubMed] [Google Scholar]
- R6.Eisenreich W., Bacher A. Biosynthesis of 5-hydroxybenzimidazolylcobamid (factor III) in Methanobacterium thermoautotrophicum . J. Biol. Chem. 1991;266:23840–23849. [PubMed] [Google Scholar]
- R7.Fonseca M.V., Escalante-Semerena J.C. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J. Biol. Chem. 2001;276:32101–32108. doi: 10.1074/jbc.M102510200. [DOI] [PubMed] [Google Scholar]
- R8.Gardner W.L., Whitman W.B. Expression vectors for Methanococcusmaripaludis: overexpression of acetohydroxyacid synthase and α−galactosidase. Genetics. 1999;152:1439–1447. doi: 10.1093/genetics/152.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Gorris L.G.M., van der Drift C. Cofactor contents of methanogenic bacteria reviewed. Biofactors. 1994;4:139–145. [PubMed] [Google Scholar]
- R10.Gorris L.G.M., Van Der Drift C., Vogels G.D. Separation and quantification of cofactors from methanogenic bacteria by high–performance liquid chromatography: optimum and routine analyses. J. Microbiol. Methods. 1988;8:175–190. [Google Scholar]
- R11.Hendrickson E.L., Kaul R., Zhou Y., et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Henke W., Herdel K., Jung K., Schnorr D., Loening S.A. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Hildebrandt M., Nellen W. Library-independent cloning of genomic fragments adjacent to vector integration sites—isolation of the EB4-PSV gene from a Dictyostelium gene disruption transformant. Biochem. Biophys. Res. Commun. 1991;181:884–888. doi: 10.1016/0006-291x(91)91273-f. [DOI] [PubMed] [Google Scholar]
- R14.Jones W.J., Paynter M.J.B., Gupta R. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 1983;135:91–97. [Google Scholar]
- R15.Jones W.J., Whitman W.B., Fields R.D., Wolfe R.S. Growth and plating efficiency of methanococci on agar media. Appl. Environ. Microbiol. 1983;46:220–226. doi: 10.1128/aem.46.1.220-226.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Kansy J.W., Carinato M.E., Monteggia L.M., Konisky J. In vivo transcripts of the S-layer-encoding structural gene of the archaeon Methanococcus voltae . Gene. 1994;148:131–135. doi: 10.1016/0378-1119(94)90246-1. [DOI] [PubMed] [Google Scholar]
- R17.Kengen S.W.M., Daas P.J.H., Duits E.F.G., Keltjens J.T., Van Der Drift C., Vogels G.D. Isolation of a 5-hydroxylbenzimidazoyl cobamide-containing enzyme involved in the methyltetrahydromethanopterin:coenzyme M methyltransferase reaction in Methanobacterium thermoautotrophicum . Biochim. Biophys. Acta. 1992;1118:249–260. doi: 10.1016/0167-4838(92)90282-i. [DOI] [PubMed] [Google Scholar]
- R18.Kim W., Whitman W.B. Isolation of acetate auxotrophs of the methane-producing archaeon Methanococcus maripaludis by random insertional mutagenesis. Genetics. 1999;152:1429–1437. doi: 10.1093/genetics/152.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Maggio-Hall L.A., Escalante-Semerena J.C. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. USA. 1999;96:11798–11803. doi: 10.1073/pnas.96.21.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Rondon M.R., Trzebiatowski J.R., Escalante-Semerena J.C. Biochemistry and molecular genetics of cobalamin biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:347–384. doi: 10.1016/s0079-6603(08)61010-7. [DOI] [PubMed] [Google Scholar]
- R21.Sambrook J., Russell D.W. 3rd Edn. . Cold Spring Harbor, NY, Units 6.50–6.55 and A9.38–9.: Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: a laboratory manual; p. 39. [Google Scholar]
- R22.Scherer P., Höllriegel V., Krug C., Bokel M., Renz P. On the biosynthesis of 5-hydroxybenzimidazoylcobamide (vitamin B12–factor III) in Methanosarcina barkeri . Arch. Microbiol. 1984;148:354–359. [Google Scholar]
- R23.Selkov E., Maltsev N., Olsen G.J., Overbeek R., Whitman W.B. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- R24.Shearer N., Hinsley A.P., Van Spanning R.J.M., Spiro S. Anaerobic growth of Paracoccus denitrificans requires cobalamin: characterization of cobK and cobJ genes. J. Bacteriol. 1999;181:6907–6913. doi: 10.1128/jb.181.22.6907-6913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Shieh J., Whitman W.B. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 1987;169:5327–5329. doi: 10.1128/jb.169.11.5327-5329.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Stathopoulos C., Kim W., Li T., Anderson I., Deutsch B., Palioura S., Whitman W.B., Söll D. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis . Proc. Natl. Acad. Sci. USA. 2001;98:14292–14297. doi: 10.1073/pnas.201540498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Stupperich E., Kräutler B. Pseudo vitamin B12 or 5-hydroxybenzimidazoyl-cobamide are the corrinoids found in methanogenic bacteria. Arch. Microbiol. 1988;149:268–271. [Google Scholar]
- R28.Thomas M.G., Escalante-Semerena J.C. Identification of an alternative nucleoside triphosphate: 5′-deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 2000;182:4227–4233. doi: 10.1128/jb.182.15.4227-4233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Tumbula D.L., Makula R.A., Whitman W.B. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 1994;121:309–314. [Google Scholar]
- R30.Tumbula D.L., Bowen T.L., Whitman W.B. Growth of methanogens on solidified medium. In: Sowers K.R., Schreier H.J., editors. Archaea: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. pp. 49–55. [Google Scholar]
- R31.Whitman W.B., Shieh J., Sohn S., Caras D.S., Premachandran U. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 1986;7:235–240. [Google Scholar]
- R32.Woodson J.D., Escalante-Semerena J.C. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc. Natl. Acad. Sci. USA. 2004;101:3591–3596. doi: 10.1073/pnas.0305939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Woodson J.D., Peck R.F., Krebs M.P., Escalante-Semerena J.C. The cobY gene of the archaeon Halobacterium sp. Strain NRC-1 is required for de novo cobamide synthesis. J. Bacteriol. 2003;185:311–316. doi: 10.1128/JB.185.1.311-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]