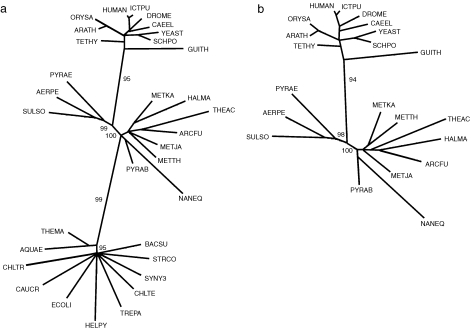
Figure 4.
Maximum-likelihood unrooted trees constructed from the positional variation among aligned sets of ribosomal proteins. (a) Tree constructed from the concatenated set of alignment blocks common in the 34 universal ribosomal proteins from the three phylodomains: Bacteria, Archaea and Eukarya, labeled by orange bars in Figure 3. (b) Tree constructed from the concatenated set of alignment blocks common to both the eukaryotic and archaeal representatives and labeled by purple bars in Figures 1 and 2. The trees were constructed by the maximum likelihood approach with the TREE-PUZZLE procedure, Quartet Puzzling, 1000 steps and the JTT model of substitution. Abbreviations are Swiss-Prot codes: Eukarya (top set): ARATH = Arabidopsis thaliana, CAEEL = Caenorhabditis elegans, DROME = Drosophila melanogaster, GUITH = Guillardia theta, HUMAN = Homo sapiens, ICTPU = Ictalurus punctatus, ORYSA = Oryza sativa, SCHPO = Schizosaccharomyces pombe, TETHY = Tetrahymena thermophila and YEAST = Saccharomyces cerevisiae; Archaea (middle set): AERPE = Aeropyrum pernix, ARCFU = Archaeoglobus fulgidus, HALMA = Haloarcula marismortui, METJA = Methanococcus jannischii, METKA = Methanopyrus kandleri, METTH = Methanobacterium thermoautotrophicum, NANEQ = Nanoarchaeum equitans, PYRAB = Pyrococcus abyssi, PYRAE = Pyrobaculum aerophilum, SULSO = Sulfolobolus solfataricus and THEAC = Thermoplasma acidophilum; Bacteria (bottom set): AQUAE = Aquifex aeolicus, BACSU = Bacillus subtilis, CAUCR = Caulobacter crescentus, CHLTR = Chlamydia trachomatis, CHLTE = Chlorobium tepidum, ECOLI = Escherichia coli, HELPY = Helicobacter pylori, STRCO = Streptomyces coelicolor, SYNY3 = Synechocystis sp. (strain PCC 6803), THEMA = Thermotoga maritima and TREPA = Treponema pallidum.