Abstract
Little is known about the methanogenic degradation of acetate, the fate of molecular hydrogen and formate or the ability of methanogens to grow and produce methane in cold, anoxic marine sediments. The microbes that produce methane were examined in permanently cold, anoxic marine sediments at Hydrate Ridge (44°35' N, 125°10' W, depth 800 m). Sediment samples (15 to 35 cm deep) were collected from areas of active methane ebullition or areas where methane hydrates occurred. The samples were diluted into enrichment medium with formate, acetate or trimethylamine as catabolic substrate. After 2 years of incubation at 4 °C to 15 °C, enrichment cultures produced methane. PCR amplification and sequencing of the rRNA genes from the highest dilutions with growth suggested that each enrichment culture contained a single strain of methanogen. The level of sequence similarity (91 to 98%) to previously characterized prokaryotes suggested that these methanogens belonged to novel genera or species within the orders Methanomicrobiales and Methanosarcinales. Analysis of the 16S rRNA gene libraries from DNA extracted directly from the sediment samples revealed phylotypes that were either distantly related to cultivated methanogens or possible anaerobic methane oxidizers related to the ANME-1 and ANME-2 groups of the Archaea. However, no methanogenic sequences were detected, suggesting that methanogens represented only a small proportion of the archaeal.
Keywords: cultivation, marine sediments, psychrotolerant
Introduction
Methane is an important greenhouse gas, with the potential to contribute 25 times per mole as much greenhouse warming as carbon dioxide (CO2) during the next century (Lashof and Ahuja 1990, Schlesinger 1997). In marine sediments, methane is either dissolved in the pore water, a free gas or trapped within crystalline ice-like cages called methane hydrates. Molecular and isotopic data suggest that most of the methane produced in marine sediments is of biogenic origin (Cragg et al. 1996, Kvenvolden and Lorenson 2001); however, little is known about the types of methanogens that produce methane there.
Generally, methanogenesis is limited in marine sediments when sulfate is present. Because of their higher affinity for hydrogen and acetate, sulfate-reducing bacteria (SRB) out-compete methanogens for these substrates (Oremland and Taylor 1978, Holmer and Kristensen 1994). However studies of deep ocean sediments and sediments at the Hydrate Ridge have demonstrated that methanogenesis does occur in zones with high sulfate concentrations as well as zones of anaerobic methane oxidation (Parkes et al. 1990, Cragg et al. 1996, Parkes et al. 2000, D’Hondt et al. 2004). The limited methane production that occurs in sulfate rich sediments may be due, in part, to the activity of methylotrophic methanogens that are able to utilize substrates not metabolized by the SRB (Oremland and Taylor 1978, Oremland and Polcin 1982, King 1984). Nevertheless, methane production from H2 and CO2 has been detected in at least some sediments with high sulfate concentrations (Parkes et al. 1990).
Hydrate Ridge is an area of intense scientific interest and numerous geological and geochemical (Bohrmann et al. 1998, Suess et al. 1999, Johnson et al. 2001, Tréhu and Flueh 2001, Torres et al. 2001, Grant and Whiticar 2002, Rehder et al. 2002, Torres et al. 2002, Tryon et al. 2002, Carson et al. 2003, Heeschen et al. 2003, Teichert et al. 2003) and biological studies (Cragg et al. 1996, Bidle et al. 1999, Boetius et al. 2000, Nauhaus et al. 2002, Sahling et al. 2002, Sommer et al. 2002, Knittel et al. 2003, Treude et al. 2003, Boetius and Suess 2004, Knittel et al. 2005, Lanoil et al. 2005, Nauhaus et al. 2005) have been conducted there. Uncultivated microbes belonging to the anaerobic methanotrophic groups ANME-1 and ANME-2 are the most abundant archaea in these sediments; whereas methanogens and uncultivated archaea belonging to the marine benthic groups B, C and D are typically found in lower numbers (Bidle et al. 1999, Boetius et al. 2000, Knittel et al. 2005, Lanoil et al. 2005). The microbial diversity at Hydrate Ridge is similar to that found at other methane seep and hydrate environments (Lanoil et al. 2001, Orphan et al. 2001a, Mills et al. 2003, Mills et al. 2005). However, only a few investigations have specifically addressed the abundance or activity of methanogens in these environments (Cragg et al. 1996, Bidle et al. 1999, Marchesi et al. 2001).
The main goal of the current study was to determine the types of methanogens that contribute to methane formation in cold marine sediments at Hydrate Ridge. Aceticlastic, CO2-reducing and methylotrophic methanogens were enriched in most probable number (MPN) dilutions to determine the relative abundance of each physiological type. The DNA was then extracted from the enrichment cultures to identify the methanogens. Additionally, DNA was extracted directly from sediment samples and rRNA gene libraries prepared to assess the overall diversity of archaea.
Materials and methods
Study site
Hydrate Ridge is located about 90 km off the coast of Oregon, USA (44° N, 125° W). Hydrate Ridge is approximately 10 km in length and comprises a northern and southern summit. Methane hydrates occur in the shallow sediment as well as in deeper zones (Suess et al. 2001). Water depth (about 700 to 900 m) and temperatures (4 °C) place these seafloor gas hydrates close to their stability limit (Suess et al. 2001). The gas within the hydrates consists of methane (99%), hydrogen sulfide and ethane (Kastner et al. 1998). Diagenic carbonate is widespread throughout and areas with active methane ebullition occur along fractures in disrupted sediments (Suess et al. 2001). The seeps are associated with sediment-surface communities of clams, snails and microbial mats (Torres et al. 2002). The work described here was conducted solely at the southern summit of Hydrate Ridge.
Sample collection
The data reported are from sediment samples collected between July 26 and July 31, 2002, during cruise 7–18 of the R/V Atlantis. The submersible DSV Alvin used push-cores to collect sediment during a series of three dives (3811–3813) at depths of 700–900 m. Sediments associated with clam beds or microbial mats were collected. On board ship, the cores were immediately subsampled by inserting the barrel of a syringe into pre-drilled holes in the push cores. Parallel cores were processed for geochemistry, microbial abundance and other biological studies, and these results have been presented elsewhere (Hill et al. 2004, Valentine et al. 2005).
Culture techniques and media
We used the anaerobic techniques of Hungate (1969) with some modifications (Sowers and Noll 1995). The basal medium for characterization of these strains was MSH enrichment medium (Ni and Boone 1991), an anoxic, bicarbonate-buffered medium containing minerals (including 0.5 M NaCl), yeast extract, peptones and a catabolic substrate: 15 mM formate, 10 mM acetate or 20 mM trimethylamine (TMA). The medium was prepared at pH 6.7 with a gas phase of CO2.
Methanogenic enrichments were inoculated with dilutions of sediment taken from the deepest section (i.e., 31 cm below the sediment-seawater interface) of the core and incubated at 4 °C. On board ship, each of these suspensions was mixed, serially diluted and inoculated into MSH enrichment medium. Three sets of MPN enrichments from each site were prepared. In total, 54 tubes were inoculated (2 sites × 3 substrates × 3 dilutions × 3 replicates; Table 1). Upon returning to Portland State University, the enrichment cultures were taken into an anaerobic chamber and sealed into metal paint cans with an anoxic atmosphere. The double anoxic environment (anaerobic, sealed culture tubes placed inside sealed O2-free paint cans) was necessary to prevent diffusion of O2 through the stoppers during the long incubation times necessary for growth. When the cans were opened to sample the cultures, the cultures were placed in an ice bath to prevent warming. After sampling, the cans and cultures were returned to an anaerobic chamber where the tubes were resealed into the cans and returned to the incubator. All of the tubes were incubated initially at 4 °C to mimic in situ conditions and the cultures were screened periodically for methanogenesis by analyzing a sample of the headspace gas (Maestrojuán and Boone 1991).
Table 1.
Sites sampled in various experiments.
| Dive | Surface characteristic | Results presented |
| 3811 | Microbial mat | Methanogen enrichments |
| Clam field | Methanogen enrichments | |
| 3812 | Microbial mat | Methanogen enrichments |
| Culture-independent analysis | ||
| Clam field | Methanogen enrichments | |
| Culture-independent analysis | ||
| 3813 | Microbial mat | Methanogen enrichments |
| Clam field | Methanogen enrichments | |
Growth measurement
Growth was estimated from the accumulation of methane in the headspace gas of sealed culture vessels, as measured by gas chromatography (Maestrojuán and Boone 1991), taking into account the methane produced during growth of the inoculum (Powell 1983).
DNA isolation and amplification
Environmental DNA and DNA from cell pellets were extracted by a modified cetyltrimethylammonium bromide (CTAB) method (Ausubel et al. 1994). Initial samples were resuspended in TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA) for a final volume of 750 µl with 7.5% Chelex (Sigma), 0.05 EDTA (pH 7.0), 1% SDS and 200 µg of proteinase K. Samples were incubated with slow rotation for 1 h at 37 °C. Chelex was removed by centrifugation. Then 100 µl of 5 M NaCl and 80 µl of 10% (w/v) CTAB in 0.7 M NaCl were added, and the samples were incubated for 30 min at 65 °C. The DNA solution was extracted first with chloroform: isoamyl alcohol (24:1), then twice with phenol:chloroform: isoamyl alcohol (25:24:1). The DNA was precipitated with an equal volume of 2-propanol, washed with 70% ethanol and resuspended in 10 mM Tris buffer (pH 8.0).
A portion of the 16S rDNA was amplified by PCR from genomic DNA by using the archaeal specific 4F (5′-TCCGGTTGATCCTGCCRG-3′) and the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′; Reysenbach et al. 1994). Reactions contained 1.5 mM MgCl2, 50 mM KCl, 30 mM Tris, 12.5 mM each dATP, dGTP, dCTP and dTTP, 1 U Taq Polymerase (USB), 20 pmol of each primer and DNA template. The reactions were incubated under the following conditions: initial denaturation at 94 °C for 5 min, 30 cycles of primer annealing at 53 °C for 30 s, extension at 72 °C for 90 s and denaturing at 94 °C for 30 s, and a final extension at 72 °C for 10 min. PCR products were purified with an UltraClean PCR Clean-up Kit (Mo Bio Laboratories, Carlsbad, CA).
Cloning and restriction fragment length polymorphism (RFLP) analysis
Clone libraries were prepared from sediments taken 15 to 35 cm below seep environments characterized by clam fields or microbial mats (Table 1). Samples for DNA extraction were either frozen directly or preserved in an ethanol/phosphate buffered saline solution before freezing. Samples were stored on board ship at –20 °C and were held at –20 °C during transportation to Portland State University for further processing.
The PCR products were cloned into the pCR2.1 vector using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). Plasmid DNA was prepared from isolated colonies by alkaline lysis (Sambrook and Russell 2001). The plasmid DNA was amplified with vector specific primers: M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′). The PCR-amplified inserts were digested with 1 U of the restriction endonucleases MspI and HindIII (Fermentas, Hanover, Maryland) for 3 hours at 37 °C. The resulting products were separated by gel electrophoresis on a 3.0 % GenePure 3:1 (ISC BioExpress, Kaysville, Utah) agarose gel in TAE buffer. Forty-five clones were analyzed and at least one phylotype from each group (determined by RFLP banding pattern and/or ≥ 97 sequence similarity) was subject to complete nucleotide sequencing.
Sequencing of the 16S rRNA genes
Sequencing reactions were performed with the ABI PRISM Big Dye Terminator Cycle Sequencing Kit and an ABI 3100 Avant Genetic Analyzer. The complete sequences of both strands were obtained with the primers 4F (described above), 515F (5′-GTGCCAGCMGCCGCGGTAA-3′), 1100F (5′-GGCAACGAGCGMGACCC-3′), 1492R (described above), 907R (5′-CCGTCAATTCCTTTRAGTTT-3′) and 340RA (5′-CCCCGTAGGGCCYGG-3′; Reysenbach et al. 1994) to produce an overlapping set of sequences that were assembled into one contiguous sequence by using the AutoAssembler Program (ABI).
Phylogenetic and statistical analysis
Nearly complete rRNA gene sequences from the enrichment cultures and clone libraries were manually aligned to related sequences obtain from GenBank. First, the sequences were automatically aligned with the program Clustal_X (Thompson et al. 1997) and then the alignments were checked manually in MacClade 4.0 (Maddison and Maddison 2003). Predicted secondary structures served as guides to ensure correct alignment. Chimeras were identified by the method described by Ashelford et al. (2005). Two chimeras were detected and excluded from further analysis. The phylogenetic relationships were determined by using neighbor-joining analysis in PAUP* (Swofford 2002). Ambiguous positions were excluded. The lengths of sequences used in the final analysis were 981 to 1415 bases. A bootstrap analysis was performed using 1000 trial replications. Rarefaction analysis was conducted using EstimateS version 7.5 (Colwell 1997).
Nucleotide sequence accession numbers
The rRNA gene sequences were submitted to GenBank and have been assigned Accession numbers DQ058802 to DQ058822 and DQ382335.
Results
Methanogen enrichments
Three 1-g samples (wet mass) from sediments taken at 31 to 35 cm depths were inoculated into MSH enrichment medium. Three other enrichment cultures were inoculated with 1 g of sediment collected from depths of 39 to 41 cm. In total, 54 tubes were inoculated (2 sites × 3 substrates × 3 dilutions × 3 replicates; Table 1). After one year of incubation at 4 °C, methane could be detected only in the 10–1 dilutions. The cultures were then shifted to 15 °C. This temperature was chosen to increase growth rates while still enriching for psychrophilic or psychrotolerant methanogens. At the end of the second year, methane could be detected from all 10–1 and 10–2 enrichments with formate or trimethylamine as catabolic substrate (a total of 24 cultures produced methane). These cultures were transferred and diluted into fresh medium and into roll-tubes to isolate potentially novel methanogens. No growth was detected in the aceticlastic enrichment cultures during the two years that this study was conducted.
The DNA was extracted and amplified by PCR with archaeal specific primers and the PCR products were sequenced from the 24 cultures that produced methane during the first 2 years of incubation. The presence of unambiguous bases in the sequencing chromatographs suggested that only a single phylotype was amplified from 20 of these cultures. The sequences from four cultures contained large numbers of ambiguous bases and were discarded from further analyses. Presumably, more than one phylotype was present in the PCR amplification products for these cultures.
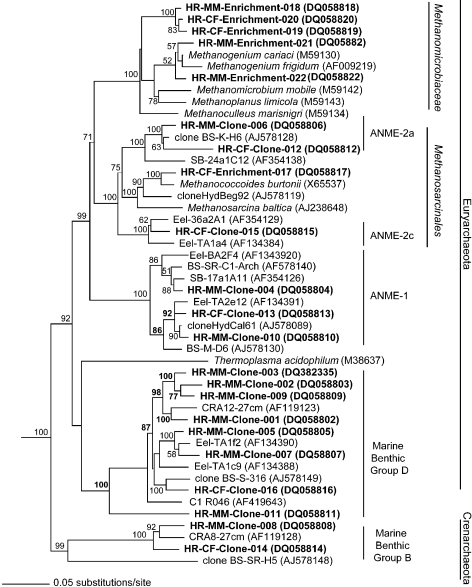
The CO2-reducing enrichments contained organisms that belonged to the order Methanomicrobiales(Figure 1; Table 2). Of these, seven cultures contained methanogens with 16S rDNA sequences that were ≥ 97% similar to each other and 96 to 98% similar to Methanogenium cariaci,a psychrotolerant methanogen isolated from marine sediments (Romesser et al. 1979). Five other sequences formed a monophyletic cladewithin the order Methanomicrobialesand seemed to represent a new genus. These sequences, represented by HR-MM-Enrichment-018 and related sequences in Figure 1, possessed only 91% similarity to Methanogenium cariaci, the most closely related described methanogen culture.
Figure 1.
Phylogenetic tree showing the affiliations of 16S rRNA gene sequences from the enrichment cultures and the clone sequences. Sequences from this study are indicated by bold-face type; sequences from enrichment cultures are identified. Sequences are labeled HR (Hydrate Ridge) and CF (clam field) or MM (microbial mat) indicate the source of the sequences. GenBank accession numbers are listed in parentheses. The tree was calculated by neighbor-joining analysis. The scale bar represents the number of fixed mutations per nucleotide position. The numbers at the branch nodes are bootstrap percentage values based on 1000 iterations and are shown for branches with more than 50% bootstrap support.
Table 2.
Phylogenetic affiliation of the enrichment cultures and phylotypes obtained from Hydrate Ridge sediments. Abbreviations: MM = microbial mat site; and CF = clam field site.
| Sample type | Phylogenetic group | Total | MM | CF |
| Enrichment | Methanomicrobiales | 17 | 9 | 8 |
| Enrichment | Methanosarcinales | 3 | 1 | 2 |
| Sediment | ANME-1 | 8 | 7 | 1 |
| Sediment | ANME-2 | 6 | 2 | 4 |
| Sediment | Marine benthic group D | 29 | 18 | 11 |
| (Thermoplasmales) | ||||
| Sediment | Marine benthic group B | 2 | 1 | 1 |
| (Crenarchaeota) | ||||
Three methylotrophic enrichment cultures contained methanogens that grouped within the Methanosarcinales. Of these, the enrichment culture HR-CF-Enrichment-017 (enriched with TMA) was the most distantly related to any cultivated methanogen (95% similar; Figure 1). A viable enrichment culture of HR-CF-Enrichment-017 has been maintained in mineral medium with trimethylamine. This strain appears to be an obligately psychrotolerant methanogen, as the enrichment culture produced methane at 15 °C but not at 20 °C. Two methylotrophic enrichments contained strains with rRNA genes that were 97–98% similar to those of Methanococcoides species and may represent new species within this genus (data not shown). Five of the methylotrophic enrichment cultures contained organisms that were closely related to CO2-reducing methanogens in the order Methanomicrobiales. These organisms might have been growing on hydrogen produced by heterotrophic bacteria in the cultures.
Phylogeny of environmental DNA sequences
Clone libraries were constructed from sediment samples taken at 15 to 35 cm below the sediment–water interface. A total of forty-five clones were analyzed (28 from the microbial mat site and 17 from the clam field site). Unique clones, or phylotypes, were initially distinguished according to RFLP banding patterns. At least one phylotype from each group was fully sequenced. A distance matrix was used to group the phylotypes into clades of 97% or greater sequence similarity and at least one representative phylotype from each clade was included in the final phylogenetic analysis. Rarefaction curves did not reach saturation for either the microbial mat or the clam field library. Additional libraries were not prepared because of the large amount of molecular data at this site and because the initial libraries were consistent with previously published reports (Bidle et al. 1999, Knittel et al. 2005, Lanoil et al. 2005).
Phylogenetic analyses indicated that the diversity of archaeadetected in the environmental samples was low, grouping only within four major clades (Figure 1; Table 2). The majority of the clones grouped within the Euryarchaeota (96%), and the majority of these (29/45) grouped in the Marine benthic group D (Thermoplasmales related). The remaining clones in the Euryarchaeota grouped in the ANME-1 (8/45) and ANME-2 (6/45) clades, which are associated with the anaerobic oxidation of methane (Hoehler and Alperin 1996, Hinrichs et al. 1999, Orphan et al. 2001a, Orphan et al. 2001b, Thomsen et al. 2001, Hallam et al. 2004). Clones related to marine benthic group B of the Crenarchaeota were also a minor component (4%) of the library. No methanogenic sequences were detected.
Discussion
We combined molecular and cultivation techniques to assess the diversity of methanogens at Hydrate Ridge. The clone libraries detected no sequences that were closely related to any cultivated methanogens; in fact, the environmental sequences from the clone libraries were most closely related to environmental sequences from this and other marine sediment environments (Figure 1). Although the number of clones screened in this study was small, similar results have been reported previously at Hydrate Ridge and other methane seep environments (Bidle et al. 1999, Lanoil et al. 2001, Orphan et al. 2001a, Reed et al. 2002, Knittel et al. 2005, Lanoil et al. 2005). Presumably, the reason that methanogens were not detected is because they are a small fraction of the archaea present. For instance, the ANME-1 and ANME-2 groups of anaerobic methane oxidizing organisms constitute about 94% of the archaeal population (3 × 109 archaeal cells ml–1) at Hydrate Ridge (Boetius et al. 2000). In contrast, our MPN data indicated that the number of methanogens could be as small as 101 to 102 cells ml–1.
The CO2-reducing enrichments contained organisms belonging to the order Methanomicrobiales (Figure 1; Table 2). Methanogens belonging to this order grow by reducing CO2 and oxidizing H2, CO or formate (Boone et al. 2001). Based on studies of carbon and hydrogen stable isotopes, Whiticar et al. (1986, 1999) have suggested that CO2-reduction is the most important pathway of methanogenesis in marine sediments. Additionally, in deep marine sediments at the Nankai Trough, methane formation was dominated by CO2-reducing methanogens (Newberry et al. 2004). Our cultivation data are consistent with these results.
All methanogens that reduce CO2 can use H2 as an electron donor and many (but not all) CO2-reducing methanogens can also use formate. Thus, our enrichments may have selected against obligate H2-oxidizing methanogens that may have been present in these marine sediments. Formate was used as the electron donor because the long incubation times required to cultivate these slow growing, psychrotolerant methanogens precluded the use of H2, which would have diffused from the tubes during these incubations.
Methanogens that belong to the order Methanosarcinaleshave the broadest substrate range of methanogens: many can grow by reducing CO2 with H2, by dismutating methyl compounds or by the splitting of acetate (Kendall and Boone 2004). Methanogens closely related to Methanococcoides spp. were cultivated from these sediments. Many of the organisms in this group catabolize methylated compounds such as methylamines and methanol and have a limited capacity to reduce CO2 (Kendall and Boone 2004). Many osmolytes are methylated and contribute a significant fraction of the organic matter to marine sedimentary environments (Fiebig and Gottschalk 1983, King 1984, Vairavamurthy et al. 1985). Thus, these compounds could be available to support methanogenesis at Hydrate Ridge.
Although we have isolated an aceticlastic methanogen from cold marine sediments in Skan Bay, Alaska (unpublished data), we were unable to cultivate aceticlastic methanogens from sediments collected at the Hydrate Ridge. In fact, of all described psychrotolerant methanogens, only Methanosarcina baltica, which was isolated from the Gotland deep of the Baltic Sea, is able to catabolize acetate (von Klein et al. 2002). Isotopic data have suggested that acetate is only a minor precursor to methanogenesis in marine sediments, most likely because sulfate-reducing bacteria out-compete methanogens for this substrate (Whiticar et al. 1986).
By maintaining long-term (2 years) methanogenic enrichment cultures, we assessed the diversity of very slow growing, psychrotolerant methanogens at Hydrate Ridge. In addition to allowing for the detection and identification of methanogens that are present in the sediment in very low numbers, this approach provided inferences of physiological characteristics about the methanogens (e.g., catabolic substrates, response to temperature and pH) that cannot necessarily be inferred only from sequence data. The results of this study suggest that CO2-reducing methanogens have an important role in methane production in marine sediments. Further studies are being conducted at non-seep and non-hydrate marine environments to determine if these results are consistent with other marine sediment environments.
Acknowledgments
We thank Miriam Kastner, Doug Bartlett, Dave Valentine and the crews of the R/V Atlantis and the DSV Alvin for their assistance in sample collection. We also thank Yitai Liu, Karl Rusterholtz, Melissa DeYoung and Adam Bonin for their technical assistance, and Susan Masta for her guidance with the phylogenetic analyses. We are grateful to Ken Stedman, members of the Valentine lab and Dave Rasko for their helpful suggestions in the preparation of this manuscript. This research was supported by a grant from the National Science Foundation’s LexEn program (prime Contract OCE-0085607 via a subcontract from the University of California at Irvine [L00OCE0085607]), and by a NSF IGERT Earth Subsurface Biosphere fellowship to MMK.
References
- R1.Ashelford K.E., Chuzhanova N., Fry J.C., Jones A.J., Weightman A.J. At least one in twenty 16S rRNA sequence records currently held in public repositories estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. New York: Greene Publishing Associates Wiley Interscience; 1994. Current protocols in molecular biology. Units 2.3.1–2.4.5. [Google Scholar]
- R3.Bidle K.A., Kastner M., Bartlett D.H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B) FEMS Microbiol. Lett. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- R4.Boetius A., Suess E. Hydrate Ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem. Geol. 2004;205:291–310. [Google Scholar]
- R5.Boetius A., Ravenschlag K., Schubert C.J., et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- R6.Bohrmann G., Greinert J., Suess E., Torres M.E. Authigenic carbonates from the Cascadia subduction zone and their relation to gas hydrate stability. Geology. 1998;26:647–650. [Google Scholar]
- R7.Boone D.R., Whitman W.B., Koga Y. Order II: Methanomicrobiales. In: Boone D.R., Castenholz R.W., editors. New York: Springer-Verlag; 2001. pp. 246–247. [Google Scholar]
- R8.Carson B., Kastner M., Bartlett D.H., Jaeger J., Jannasch H.W., Weinstein Y. Implications of carbon flux from the Cascadia accretionary prism: results from long-term, in situ measurements at ODP Site 892B. Mar. Geol. 2003;198:159–180. [Google Scholar]
- R9.Colwell R.K. EstimateS: statistical estimation of species richness and shared species from samples. 1997 Version 7.5. Persistent URL <purl.oclc.org/estimates>. [Google Scholar]
- R10.Cragg B.A., Parkes R.J., Fry J.C., Weightman A.J., Rochelle P.A., Maxwell J.R. Bacterial populations and processes in sediments containing gas hydrates (ODP Leg 146: Cascadia Margin) Earth Planet. Sci. Lett. 1996;139:497–507. [Google Scholar]
- R11.D’Hondt S., Jorgensen B.B., Miller D.J., et al. Distributions of microbial activities in deep subseafloor sediments. Science. 2004;306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- R12.Fiebig K., Gottschalk G. Methanogenesis from choline by a coculture of Desulfovibrio sp. and Methanosarcina barkeri. Appl. Environ. Microbiol. 1983;45:161–168. doi: 10.1128/aem.45.1.161-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Grant N.J., Whiticar M.J. Stable carbon isotopic evidence for methane oxidation in plumes above Hydrate Ridge, Cascadia Oregon Margin. Global Biogeochem. Cycles. 2002;16:1124. [Google Scholar]
- R14.Hallam S.J., Putnam N., Preston C.M., Detter J.C., Rokhsar D., Richardson P.M., Delong E.F. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- R15.Heeschen K., Tréhu A.M., Collier R.W., Suess E., Rehder G. Distribution and height of methane bubble plumes on the Cascadia Margin characterized by acoustic imaging. Geophys. Res. Lett. 2003;30:1643. [Google Scholar]
- R16.Hill T.M., Kennett J.P., Valentine D.L. Isotopic evidence for the incorporation of methane-derived carbon into foraminifera from modern methane seeps, Hydrate Ridge, Northeast Pacific. Geochim. Cosmochim. Acta. 2004;68:4619–4627. [Google Scholar]
- R17.Hinrichs K.-U., Hayes J.M., Sylva S.P., Brewer P.G., Delong E.F. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- R18.Hoehler T.M., Alperin M.J. Anaerobic methane oxidation by a methanogen-sulfate reducer consortium: geochemical evidence and biochemical considerations. In: Lidstrom M.E., Tabitha F.R., editors. Microbial Growth on C1 Compounds. Dordrecht: Kluwer Academic Publishers; 1996. pp. 326–331. [Google Scholar]
- R19.Holmer M., Kristensen E. Coexistence of sulfate reduction and methane production in an organic-rich sediment. Mar. Ecol. Prog. Ser. 1994;107:117–184. [Google Scholar]
- R20.Hungate R.E. A roll tube method for cultivation of strict anaerobes. In: Norris J.R., Ribbons D.W., editors. New York: Academic Press; 1969. pp. 117–132. [Google Scholar]
- R21.Johnson J.E., Goldfinger C., Suess E. Geophysical constraints on the surface distribution of authigenic carbonates across the Hydrate Ridge region, Cascadia margin. Mar. Geol. 2001;202:79–110. [Google Scholar]
- R22.Kastner M., Kvenvolden K.A., Lorenson T.D. Chemistry, isotopic composition, and origin of a methane-hydrogen sulfide hydrate at the Cascadia subduction zone. Earth Planet. Sci. Lett. 1998;156:173–183. [Google Scholar]
- R23.Kendall M.M., Boone D.R. The order Methanosarcinales. In: Dworkin M., editor. New York: Springer-Verlag; 2004. http://link.springer-ny.co m/link/service/books/10125/ [Google Scholar]
- R24.King G.M. Utilization of hydrogen, acetate, and “noncompetitive” substrates by methanogenic bacteria in marine sediments. Geomicrobiol. J. 1984;3:275–306. [Google Scholar]
- R25.Knittel K., Boetius A., Lemke A., Eilers H., Lochte K., Pfannkuche O., Linke P., Amann R. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon) Geomicrobiol. J. 2003;20:269–294. [Google Scholar]
- R26.Knittel K., Losekann T., Boetius A., Kort R., Amann R. Diversity and distribution of methanotrophic Archaea at cold seeps. Appl. Environ. Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Kvenvolden K.A., Lorenson T.D. The global occurrence of natural gas hydrate. In: Paull C.K., Dillon W.P., editors. Washington D.C.: American Geophysical Union; 2001. pp. 3–18. [Google Scholar]
- R28.Lanoil B., Sassen R., Laduc M.T., Sweet S.T., Nealson K.H. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 2001;67:5143–5153. doi: 10.1128/AEM.67.11.5143-5153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Lanoil B.D., Laduc M.T., Wright M., Kastner M., Nealson K.H., Bartlett D.H. Archaeal diversity in ODP legacy borehole 892b and associated seawater and sediments of the Cascadia Margin. FEMS Microbiol. Ecol. 2005;54:167–177. doi: 10.1016/j.femsec.2005.03.015. [DOI] [PubMed] [Google Scholar]
- R30.Lashof D.A., Ahuja D.R. Relative contributions of greenhouse gas emissions to global warming. Nature. 1990;344:529–531. [Google Scholar]
- R31.Maddison D.L., Maddison W.P. Sunderland, MA: 2003. MacClade 4: analysis of phylogeny and character evolution: Sinauer Associaters. [Google Scholar]
- R32.Maestrojuán G.M., Boone D.R. Characterization of Methanosarcina barkeri MST and 227. Methanosarcina mazei S-6T and Methanosarcina vacuolata Z-761T. Int. J. Syst. Bacteriol. 1991;41:267–274. [Google Scholar]
- R33.Marchesi J.R., Weightman A.J., Cragg B.A., Parkes R.J., Fry J.C. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 2001;34:221–228. doi: 10.1111/j.1574-6941.2001.tb00773.x. [DOI] [PubMed] [Google Scholar]
- R34.Mills H.J., Hodges C., Wilson K., Macdonald I.R., Sobecky P.A. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 2003;46:39–52. doi: 10.1016/S0168-6496(03)00191-0. [DOI] [PubMed] [Google Scholar]
- R35.Mills H.J., Martinez R.J., Story S., Sobecky P.A. Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl. Environ. Microbiol. 2005;71:3235–3247. doi: 10.1128/AEM.71.6.3235-3247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Nauhaus K., Boetius A., Kruger M., Widdel F. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 2002;4:296–305. doi: 10.1046/j.1462-2920.2002.00299.x. [DOI] [PubMed] [Google Scholar]
- R37.Nauhaus K., Treude T., Boetius A., Kruger M. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-1 and ANME-2 communities. Environ. Microbiol. 2005;7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- R38.Newberry C.J., Webster G., Cragg B.A., Parkes R.J., Weightman A.J., Fry J.C. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 2004;6:274–287. doi: 10.1111/j.1462-2920.2004.00568.x. [DOI] [PubMed] [Google Scholar]
- R39.Ni S., Boone D.R. Isolation and characterization of a dimethyl sulfide-degrading methanogen, Methanolobus siciliae HI350, from an oil well, characterization of M. siciliae T4/MT, and emendation of M. siciliae . Int. J. Syst. Bacteriol. 1991;41:410–416. doi: 10.1099/00207713-41-3-410. [DOI] [PubMed] [Google Scholar]
- R40.Oremland R.S., Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl. Environ. Microbiol. 1982;44:1270–1276. doi: 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Oremland R.S., Taylor B.F. Sulfate reduction and methanogenesis in marine sediments. Geochim. Cosmochim. Acta. 1978;42:209–214. [Google Scholar]
- R42.Orphan V.J., Hinrichs K.-U., Iii W.U., Paull C.K., Taylor L.T., Sylva S.P., Hayes J.M., Delong E.F. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R43.Orphan V.J., House C.H., Hinrichs K.-U., Mckeegan K.D., Delong E.F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- R44.Parkes R.J., Cragg B.A., Fry J.C., Herbert R.A., Wimpenny J.W.T. Bacterial biomass and activity in deep sediment layers from the Peru margin. Philos. Trans. R. Soc. Lond. A. 1990;331:139–153. [Google Scholar]
- R45.Parkes R.J., Cragg B.A., Wellsbury P. Recent studies on bacterial publications and processes in subseafloor sediments: a review. Hydrogeol. J. 2000;8:11–28. [Google Scholar]
- R46.Powell G.E. Interpreting gas kinetics of batch culture. Biotechnol. Lett. 1983;5:437–440. [Google Scholar]
- R47.Reed D.W., Fujita Y., Delwiche M.E., Blackwelder D.B., Sheridan P.P., Uchida T., Colwell F.S. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 2002;68:3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R48.Rehder G., Collier R.W., Heeschen K., Kosro P.M., Barth J., Suess E. Enhanced marine CH4 emissions to the atmosphere off Oregon caused by coastal upwelling. Global Biogeochem. Cycles. 2002;16:1081. [Google Scholar]
- R49.Reysenbach A.-L., Wickham G.S., Pace N.R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R50.Sahling H., Rickert D., Lee R.W., Linke P., Suess E. Macrofaunal community structure and sulfide flux at gas hydrate deposits from the Cascadia convergent margin, NE Pacific. Mar. Ecol. Prog. Ser. 2002;231:121–138. [Google Scholar]
- R51.Sambrook J., Russell D.W. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: a laboratory manual 3rd Edn. Units 1.38–1.39. [Google Scholar]
- R52.Schlesinger W.H. San Diego: Academic Press; 1997. Biogeochemistry: an analysis of global change; pp. 358–382. [Google Scholar]
- R53.Sommer S., Pfannkuche O., Rickert D., Kahler A. Ecological implications of surficial marine gas hydrates for the associate small-sized benthic biota at the Hydrate Ridge (Cascadia Convergent Margin, NE Pacific) Mar. Ecol. Prog. Ser. 2002;243:25–38. [Google Scholar]
- R54.Sowers K.R., Noll K.M. Techniques for anaerobic growth. In: Robb F.T., Sowers K.R., Schreier H.J., Dassarma S., Fleischmann E.M., editors. New York: Cold Spring Harbor Laboratory Press; 1995. pp. 15–47. [Google Scholar]
- R55.Suess E., Torres M.E., Bohrmann G., et al. Gas hydrate destabilization: enhanced dewatering, benthic material turnover and large methane plumes at the Cascadia convergent margin. Earth Planet. Sci. Lett. 1999;170:1–15. [Google Scholar]
- R56.Suess E., Torres M.E., Bohrmann G., et al. Sea floor methane hydrates at Hydrate Ridge, Cascadia Margin. In: Paull C.K., Dillon W.P., et al., editors. Washington D.C.: American Geophysical Union; 2001. pp. 87–98. [Google Scholar]
- R57.Swofford D.L. Sunderland, MA: Sinauer Associates; 2002. PAUP*. Phylogenetic analysis using parsimony and other methods. [Google Scholar]
- R58.Teichert B.M.A., Eisenhauer G., Bohrmann G. U/Th systematics and ages of authigenic carbonates from Hydrate Ridge, Cascadia Margin: recorders of fluid flow variations. Geochim. Cosmochim. Acta. 2003;67:3845–3857. [Google Scholar]
- R59.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R60.Thomsen T.R., Finster K., Ramsing N.B. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 2001;67:1646–1656. doi: 10.1128/AEM.67.4.1646-1656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R61.Torres M.E., Barry J.P., Hubbard D.A., Suess E. Reconstructing the history of fluid flow at cold seep sites from Ba/Ca ratios in vesicomyid clam shells. Limnol. Oceanogr. 2001;46:1701–1708. [Google Scholar]
- R62.Torres M.E., Mcmanus J., Hammond D.E., De angelis M.A., Heeschen K.U., Colbert S.L., Tryon M.D., Brown K.M., Suess E. Fluid and chemical fluxes in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, I: hydrological provinces. Earth Planet. Sci. Lett. 2002;201:525–540. [Google Scholar]
- R63.Treude T., Boetius A., Knittel K., Wallmann K., Jorgensen B.B. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 2003;264:1–14. [Google Scholar]
- R64.Tryon M.D., Brown K.M., Torres M.E. Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, II: hydrological processes. Earth Planet. Sci. Lett. 2002;201:541–557. [Google Scholar]
- R65.Tréhu A.M., Flueh E.R. Estimating the thickness of the free gas zone beneath Hydrate Ridge, Oregon continental margin, from seismic velocities and attenuation. J. Geophys. Res. 2001;106:2035–2045. [Google Scholar]
- R66.Vairavamurthy A., Andreae M.O., Iverson R.L. Biosynthesis of dimethylsulfide and dimethylpropiothetin by Hymenomonas carterae in relation to sulfur source and salinity variations. Limnol. Oceanogr. 1985;30:59–70. [Google Scholar]
- R67.Valentine D.L., Kastner M., Wardlaw G.D., Wang X., Purdy A., Bartlett D.H. Biogeochemical investigations of marine methane seeps, Hydrate Ridge, Oregon. J. Geophys. Res. 2005;110:GO2005. [Google Scholar]
- R68.von Klein D., Arab H., Volker H., Thomm M. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles. 2002;6:103–110. doi: 10.1007/s007920100234. [DOI] [PubMed] [Google Scholar]
- R69.Whiticar M.J. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 1999;161:291–314. [Google Scholar]
- R70.Whiticar M.J., Faber E., Schoell M. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta. 1986;50:693–709. [Google Scholar]