Abstract
We have identified a novel archaeal protein that apparently plays two distinct roles in ribosome metabolism. It is a polypeptide of about 18 kDa (termed Rbp18) that binds free cytosolic C/D box sRNAs in vivo and in vitro and behaves as a structural ribosomal protein, specifically a component of the 30S ribosomal subunit. As Rbp18 is selectively present in Crenarcheota and highly thermophilic Euryarchaeota, we propose that it serves to protect C/D box sRNAs from degradation and perhaps to stabilize thermophilic 30S subunits.
Keywords: C/D box sRNA, ribosomal protein, rRNA methylation, Sulfolobus solfataricus
Introduction
The importance of small RNA molecules (sRNAs) in the regulation of many cellular processes has become increasingly evident in recent years. Although most known cases of sRNA function are in eukaryotic cells, instances of fundamental roles for sRNAs are not lacking in the prokaryotic world, especially in archaea. A prominent example is the site-specific post-transcriptional modification of rRNA and tRNA, which in archaea, similar to eukarya, is carried out by ribonucleoproteic (RNP) complexes containing sRNA molecules (Kiss 2001, 2002, Bachellerie et al. 2002, Omer et al. 2002). In these complexes, proteins provide the catalytic activity, whereas the RNA molecule acts as the guide for target recognition. The sRNAs found in such complexes (termed snoRNAs in eukarya because of their nucleolar localization) are divided into two major classes: the C/D box snoRNAs, involved in 2′-OH ribose methylation; and the H/ACA snoRNAs, involved in site-specific pseudouridylation (Tollervey 1996, Kiss-Laszlo et al. 1996, Ganot et al. 1997, Ni et al. 1997). The members of each class contain characteristic conserved sequences and interact with core binding proteins. The C/D box snoRNAs guide site-specific modification by base-pairing with the pre-rRNA at the target sites. Specifically, C/D box snoRNAs contain one, or sometimes two, sequences of 10–21 nucleotides located immediately upstream of D and D′ boxes and are complementary to the target RNA. Methylation takes place on the ribose of the nucleotide located exactly five base pairs upstream of box D or D′ (Cavaille et al. 1996, Kiss-Laszlo et al. 1998, Omer et al. 2000).
In eukarya, four core proteins associate with all C/D box snoRNAs: Nop 56; Nop 58; fibrillarin; and a 15.5 kDa protein (termed Snu13p in yeast). All archaeal genomes sequenced to date contain genes encoding proteins homologous to the components of eukaryotic snoRNA complexes: fibrillarin; Nop56/58 (a single protein for NOP56 and NOP58); and L7Ae. The latter was already known as a protein of the large ribosomal subunit (Ban et al. 1999). In vitro experiments have provided evidence confirming that these three archaeal proteins associate with C/D box sRNAs and participate in post-transcriptional rRNA modification (Kuhn et al. 2002, Omer et al. 2002, Aittaleb et al. 2003, Ziesche et al. 2004).
To date, there has been a lack of in vivo studies aimed at the isolation and characterization of natural archaeal rRNA-modifying RNP complexes. Therefore, the possiblity cannot be excluded that there exist some proteins other than fibrillarin, NOP56/58 and L7Ae that have a role in rRNA modification and small RNA metabolism, or both. In this study, we have identified one such protein in the archaeon Sulfolobus solfataricus. We describe the properties of a novel protein, Rbp18, specific to Crenarcheota and Pyrococcales. Similar to L7Ae, Rbp18 is a structural ribosomal protein (of the small ribosomal subunit) that also binds the C/D box sRNA. However, unlike L7Ae, Rbp18 does not seem to interact with the other polypeptide components of the C/D box RNP complexes, but appears to be exclusively involved in binding free cytoplasmic C/D box RNAs. Being selectively present in hyperthermophilic species, Rbp18 probably functions as a stabilizer of the sRNAs at high temperature.
Materials and methods
Affinity chromatography with biotinylated oligonucleotides
The following biotinylated oligonucleotides were synthesized, three of which are complementary to the Sulfolobus solfataricus C/D box sRNAs sR1, sR4 and sR8: anti-sR1 (AACTACTCGATTTGAGGGTACCAGACTATAG); anti-sR4 (CGGTGCGGTCTTGACTCGGTCCTACTTGCCGAACCCTC); anti-sR8 (CTCCCAAGGTCTCGACTTCGCACTACTTACCAACTGTG); and oligo R1 (ACGCAGTTCACTTACGTGTGACTACTCTACTGCTTGCT). Anti-sR1, anti-sR4 and anti-sR8 are fully complementary to the corresponding sRNAs of S. solfataricus; all span the sequences complementary to the 16S rRNA. Anti-sR1 included the complement to the C′ and D box motifs, whereas anti-sR4 and anti-sR8 included the complement to the C′ and D′ box motifs (underlined). Oligo R1 was not complementary to any particular sRNA, but included sequences complementary to the C′ and D′ box motifs common to all of them (underlined). The 30-mer oligo T and oligo A oligonucleotides were used as the aspecific controls.
The bioseparation procedure required 40 µl of paramagnetic particles (PMP), washed thrice with 150 µl of 0.5× SSC. They were then incubated with 20 µl of a S. solfataricus cell lysate (corresponding to about 500 µg total proteins) and 230 µl 0.5× SSC for 30 min while being slowly rotated at room temperature. After incubation, the PMP were captured and discarded along with any nonspecifically bound proteins. After preclearing, the supernatant was supplemented with 200 pmol of a biotinylated oligonucleotide, and the mixtures were incubated for 7 min at 65 °C. Then, 60 µl of pre-washed fresh PMP was added, and the incubation was continued for 20 min at room temperature. The PMP were then captured and washed four times with 100 µl of 0.1× SSC, resuspended in 10 µl of 4× SDS Sample Buffer (60 mM, 25% Tris-HCl pH 6.8, 2% glycerol SDS, 14.4 mM 2-mercaptoethanol and 0.1% bromophenol blue) and heated at 95 °C for 3 min. The resin-bound proteins were resolved by SDS-PAGE and stained with Coomassie blue. For N-terminal sequencing by Edman degradation, Rbp18 was electrically transferred to PFVC paper.
Cloning and purification of Rbp18 and sRNP proteins
The genes encoding Rbp18 and the protein components of the rRNA-methylating sRNP (L7Ae, NOP56/58 and fibrillarin) were amplified by PCR from S. solfataricus genome using primers containing appropriate restriction sites at their 5′ and 3′ ends. The amplified fragments were cloned into the pRSETA vector (Invitrogen), which adds a 6× His tag at the N-terminal end of the recombinant proteins. The proteins were expressed in Escherichia coli BL21(DE3) cells after induction with 1 mM IPTG at 37 °C for 3 h. The cells were harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole), sonicated and heated for 10 min at 70 °C to remove most of the host’s mesophilic proteins. After centrifugation, the supernatant was loaded onto a Ni-NTA resin column (Qiagen). The column was washed, the recombinant proteins eluted according to the manufacturer’s protocol, then dialyzed overnight in a buffer containing 20 mM HEPES pH 7, 150 mM KCl, 1.5 mM MgCl2 and 20% glycerol). Mouse polyclonal antibodies were produced from the purified recombinant protein supplied to the firm Eurogentech.
Analysis of Rbp18/sR1 interaction by EMSA and affinity chromatography
The sR1 gene was cloned by RT-PCR into pBS-SK. The forward primer for amplification had a T7 promoter, and the reverse primer contained a PvuII site for linearization and run-off transcription. Radiolabeled sR1 was obtained by in vitro transcription with T7 RNA polymerase (Biolabs) in the presence of [32P] UTP. The control aspecific RNA was obtained by run-off transcription of the pBS S/K polylinker after digestion with PstI.
For electrophoretic mobility shift assays (EMSA), about 50 pmol of radiolabeled sR1 or unspecific control was incubated with increasing concentrations of recombinant Rbp18 (50, 80, 100, 150, 200, 250 and 300 pmol) in 20 µl (final volume) of a buffer containing 20 mM HEPES pH 7, 150 mM NaCl, 0.75 mM DTT, 1.5 mM MgCl2 and 0.1 mM EDTA. The samples were incubated at room temperature for 20 min. Then, 5 µl of loading buffer (0.8% bromophenol blue and 25% glycerol) was added, and the samples were loaded on a non-denaturing 8% polyacrylamide gel made in 0.5× TBE. At the end of the run, the gels were dried and autoradiographed.
For affinity chromatography assays, 40 µl Ni-NTA sepharose (Qiagen) was incubated for 12 h at 4 °C in 250 µl of lysis buffer containing 750 ng of Rbp18, or the same amount of control proteins. The latter were the proteins recovered from an Ni-NTA Sepharose column on which an E. coli lysate expressing the void pRSETA had been chromatographed. The resin coated with Rbp18 and the control resin were supplemented with 250 µl of 20 mM Tris-HCl pH 7, 200 mM KCl containing 80 µg of Torula yeast RNA (Sigma) and incubated for 1 h at room temperature in order to saturate any non-specific RNA binding sites. Finally, each sample was supplemented with increasing amounts (15–150 ng) of radiolabeled sR1 or control RNA and incubated for 1 h at room temperature. The resin was washed extensively, and bound radioactivity was measured with a β-counter.
In vitro immunoprecipitation of RNP complexes
Equimolar concentrations (about 30 pmol each) of recombinant L7Ae, NOP56/58, fibrillarin and Rbp18 were incubated in 1× TBS (0.5 M Tris-HCl pH 8 and 3 M NaCl ) + 0.5% NP40, for 30 min at 55 °C in the presence or absence of 30 pmol sR1 RNA. After incubation, the samples were supplemented with anti-fibrillarin antibodies and incubated for 1 h at room temperature with 50 µl of Sepharose A resin (Amersham) 50% (w/v). The resin containing the bound proteins was washed three times with buffer 1× TBS + 0.9% NP40, then supplemented with Laemmli buffer and heated for 5 min at 95 °C. The samples were run on a 12.5% SDS-polyacrylamide gel, blotted on Hybond C nitrocellulose filters and the immunoprecipitated proteins were visualized by Western Blot analysis using anti-His antibodies (Qiagen) at a 1:1000 dilution.
Localization of RNP components in a cell lysate
The localization of L7Ae, fibrillarin and Rbp18 was determined as follows. Twenty-five µl of an S. solfataricus cell lysate (about 30 mg total protein ml–1) was fractionated on 10–30% sucrose density gradients (11.5 ml) made in 10 mM KCl, 20 mM Tris-HCl pH 7 and 20 mM MgCl2. The gradients were centrifuged for 4 h at 160,000 g in a Beckman SW41 rotor. The gradients were fractionated and the fractions assayed for absorbance at 260 nm to determine the ribosome position. Each fraction was then supplemented with 5 volumes of acetone to precipitate the proteins. Proteins were separated by SDS-PAGE and blotted on nitrocellulose. The presence of the proteins under study was revealed by Western blotting with antibodies anti-Fib, anti-L7Ae and anti-Rbp18.
High-salt treatment of 30S ribosomal subunits
Sulfolobus solfataricus 30S ribosomal subunits were obtained by zonal centrifugation of 500 mg of unfractionated ribosomes on a 7–38% sucrose gradient containing 500 mM NH4Cl, 20 mM Tris-HCl pH 7 and 10 mM MgCl2. About 175 pmol of 30S subunits were resuspended in 100 µl (final volume) of a buffer containing 20 mM Tris-HCl pH 7.5, 10 mM Mg(OAc) and either: 2 M NH4Cl; 2 M KCl; or 0.4 M LiCl. The samples were kept on ice for 1 h with occasional stirring. At the end of the incubation, the samples were loaded on 10–30% sucrose gradients made in 30 mM KCl, 20mM Tris-HCl pH 7.5 and 10 mM MgCl2, which were centrifuged for 4 h at 160,000 g. The centrifuged gradients were fractionated and the fractions assayed for absorbance at 260 nm. The fractions were then supplemented with 5 volumes of acetone to precipitate the proteins, which were resolved by electrophoresis on a 12.5% polyacrylamide-SDS gel. The presence of Rbp18 and of the control protein SUI1 was revealed by Western blotting with the specific polyclonal antibodies.
Results
Isolation of Rbp18 from cell lysates of Sulfolobus solfataricus
The proteins interacting in vivo with archaeal C/D box sRNAs were isolated from cell lysates of the hyperthermophilic archaeon Sulfolobus solfataricus by means of a bioseparation technique based on the use of specific oligonuclotides and PMP. Specifically, a set of 5′ biotinylated oligonucleotides complementary to three S. solfataricus C/D box sRNAs (sR1, sR4, sR7) was designed. All oligonucleotides spanned the guide sequences complementary to the rRNA and at least two of the box motifs. A control set of oligonucleotides of similar length was also designed: one of them (R1) contained only short tracts of sequence homology with the sRNAs, limited to the C′ and D′ box regions, whereas others consisted of oligo A or oligo T sequences. The biotinylated oligonucleotides were incubated with a cell extract of S. solfataricus to allow annealing with the sRNAs or the RNP complexes contained therein. The oligonucleotides and the cellular components bound to them were then selectively captured by streptavidin-coated PMP. After several washing steps, the proteins bound to the oligonucleotides were resolved by SDS-PAGE and visualized by Coomassie blue staining.
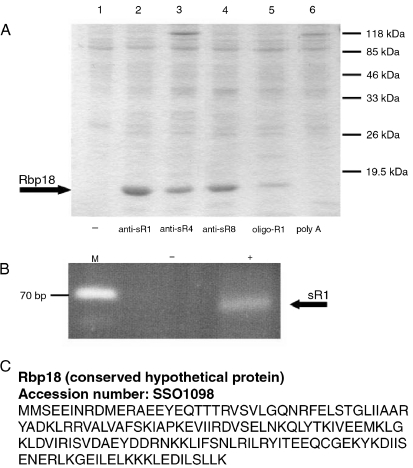
The results (Figure 1) showed that a polypeptide having an approximate molecular mass of 18 kDa was the principal component specifically recovered from the samples incubated with the anti-sRNA oligonucleotides. In the samples incubated with the R1 oligonucleotide, the amount of 18 kDa protein was much lower, whereas no 18 kDa protein was detected in the samples incubated with the aspecific oligonucleotides and in those lacking oligonucleotides. Several repetitions gave similar results, suggesting that the 18 kDa protein was specifically bound to sRNAs or sRNPs of S. solfataricus. That the sRNAs were present in the material captured with the antisense oligonucleotides was ascertained by means of RT-PCR using SR1-specific primers (Figure 1B).
Figure 1.
Isolation of Rbp18 from Sulfolobus solfataricus cell lysates. (A) Proteins isolated from S. solfataricus lysates by the bioseparation technique, resolved by SDS-PAGE and stained with Coomassie blue. Each lane contains the proteins recovered after bioseparation using the indicated anti C/D box sRNAs (anti-sR1, anti-sR4 and anti-sR8), an oligonucleotide with limited specificity (oligo-R1) and a nonspecific oligonucleotide (poly A). The numbers on the right indicate the positions of the molecular mass markers. (B) Products of RT-PCR reactions performed on the material recovered from a bioseparation with the anti-sR1 oligonucleotide (from Lane 2 of (A)). Abbreviations: M = molecular mass marker; (–) = negative control (no proteins added); and (+) = addition of anti-sR1 isolated material. (C) The Accession number and amino acid sequence of Rbp18.
The 18-kDa protein isolated by the procedure described above did not correspond to any of the known core proteins of the C/D box sRNPs. No band that could be interpreted as fibrillarin, NOP 56/58 or L7Ae was discernible in any sample. The 18 kDa polypeptide stood out as the sole protein specifically captured with the sRNAs antisense oligonucleotides, at least the only one present in sufficient abundance to be detected by Coomassie blue staining. To get more insight into the features of this protein, we set out to identify it by subjecting the band isolated from the gels to N-terminal sequencing by Edman degradation.
Identification of the gene encoding Rbp18 in the Sulfolobus solfataricus genome
N-Terminal sequencing of the 18 kDa protein yielded an unambiguous sequence of 15 amino acids that was compared with the products of translation of all the ORFs identified in the complete genome of S. solfataricus. It corresponded to a polypeptide of 18,157 Da annotated as a hypothetical protein of unknown function (Figure 1C). A more extended comparison of the full-length S. solfataricus polypeptide with comprehensive protein data banks revealed that the unknown protein had a highly selective distribution, as it was present exclusively in certain hyperthermophilic archaeal species (Figure 2). In particular, it was found in all Crenarchaeota whose genomic sequences are available and in the four completely sequenced genomes of the family Pyrococcales (Pyrococcus abyssi, P. furiosus, P. horikoshii and Thermococcus kodakaraensis). In all cases the protein was annotated as a polypeptide of unknown function. The rest of the Euryarcheota, including thermophilic species such as Methanococcus jannaschii and M. thermoautrophicum, possessed no obvious homologues of this protein. Only weak, and probably meaningless, homologies were detected with eukaryotic or bacterial proteins. Because of its putative interaction with C/D box RNAs, we termed the protein Rbp18 (18 kDa RNA binding protein). A search of the Rbp18 sequence for known functional domains failed to identify obvious RNA binding motifs.
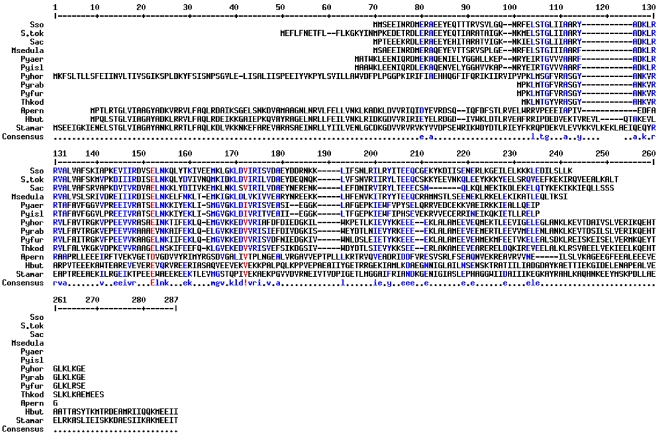
Figure 2.
Alignment of the archaeal Rbp18 proteins. Amino acids that are identical or similar in most, but not all, sequences are shown in blue; amino acids that are identical or similar in all sequences are shown in red.
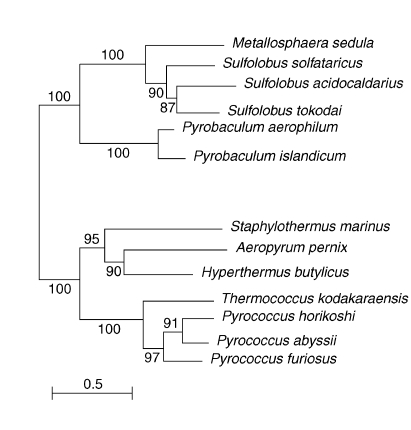
The available sequences of all Rbp18 homologues were compared to draw a phylogenetic tree and to reconstruct the evolutionary history of the polypeptide. The unrooted distance tree shown in Figure 3 revealed that all the Pyrococcales sequences grouped together, whereas the crenarchaeal proteins clustered in two groups. The group including Hyperthermus butylicus, Aeropyrum pernix and Staphylothermus marinus appeared to be more closely related to the Pyrococcales (Figures 2 and 3).
Figure 3.
Distance tree showing the phylogenetic distribution of Rbp18 proteins. The scale bar gives the number of amino acid substitutions per site.
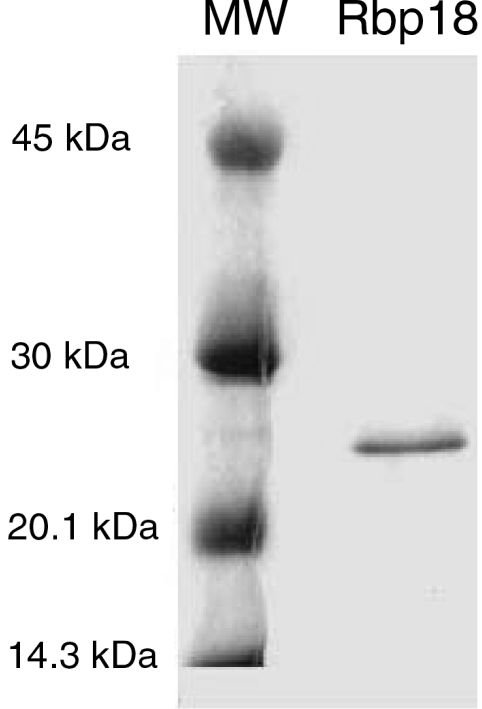
To obtain information about the function of Rbp18, we cloned the S. solfataricus gene encoding it and produced a recombinant protein. The gene was amplified from total genomic DNA by standard PCR techniques and inserted in the bacterial expression vector pRSETB. The recombinant protein was expressed in E. coli and purified. The final purity of Rbp18 as verified by SDS-PAGE and Coomassie blue staining appeared to be over 90% (Figure 4).
Figure 4.
Purification of recombinant Rbp18 by affinity chromatography.
Before analyzing the RNA-binding capacity of Rbp18, we wished to exclude the possibility that it could interact generically with single-stranded nucleic acids. To this end, the recombinant Rbp18 was incubated with the antisense biotinylated oligonucleotides, and the purification procedure was carried out as previously described for the whole cell lysate. No detectable amounts of Rbp18 were retained on the paramagnetic resin under these conditions, indicating that the protein did not bind to the sRNA-complementary oligonucleotides (not shown). We conclude that the Rbp18 in the cell lysates was captured by the antisense oligonucleotides because of its direct or indirect interaction with the sRNAs. Consistent with the conclusion, the sRNA sR1 was found among the cytoplasmic components retained by the resin (Figure 1B). To learn more about the properties of the new protein, we began by testing the possibility that Rbp18 interacts directly with S. solfataricus sRNAs.
Rbp18 binds the sR1 sRNA of Sulfolobus solfataricus
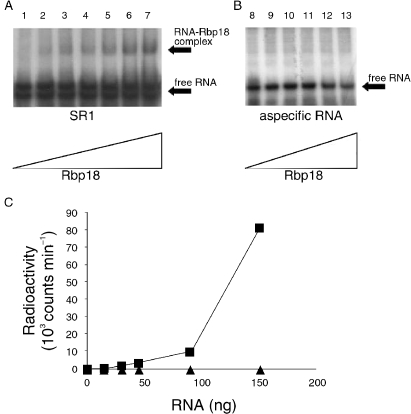
To verify if Rbp18 interacts directly with the sRNAs, the recombinant protein was tested in the electrophoretic mobility shift assay (EMSA) with the sR1 RNA uniformly labeled with [32P]. Sulfolobus solfataricus sR1 was obtained by in vitro transcription of the corresponding gene cloned by RT-PCR; this was inserted downstream of a T7 promoter to obtain a transcript exactly corresponding in size and sequence to the native sR1. Increasing amounts of recombinant Rbp18 were incubated at room temperature with the in vitro labeled sR1, and the samples subjected to electrophoresis on non-denaturing polyacrylamide gels. The results, shown in Figure 5A, revealed that Rbp18 retarded the mobility of sR1, indicating that an RNA–protein complex was being formed. Control experiments with an aspecific RNA of length and composition comparable to sR1 demonstrated the specificity of the Rbp18–sR1 interaction (Figure 5B).
Figure 5.
Rbp18 interacts specifically with C/D box sRNAs. (A) electrophoretic mobility shift assay showing the interaction of increasing amounts of recombinant Rbp18 (0, 50, 80, 100, 150, 200, 250 pmol; Lanes 1–7) with 50 pmol of sR1. (B) 50 pmol of a nonspecific oligonucleotide having length and composition similar to sR1 were incubated with increasing amounts of recombinant Rbp18 (50, 80, 100, 200, 250 pmol; Lanes 8–13). (C) Increasing amounts of sR1 (■) or a nonspecific oligonucleotide (▲) labeled in vitro with [32P]UTP were mixed to the Ni-NTA resin pre-bound to the recombinant Rbp18 and pretreated with Torula yeast RNA to saturate nonspecific RNA binding sites. After extensive washing, the radioactivity reatained by the resin was measured by liquid scintillation spectrometry.
The results obtained by EMSA were confirmed by a different experimental technique. An aliquot of the Ni-NTA resin was saturated with the recombinant Rbp18 and incubated with an excess of cold non-specific RNA (yeast tRNA) to block aspecific RNA binding sites. Control samples were prepared in a similar way, except that the resin was incubated with the final eluate of an affinity column on which E. coli cell lysates transformed with the void PRSET plasmid had been fractionated. This control was made to rule out the possibility that the RNA binding protein was not Rbp18 but some E. coli polypeptide contaminating the S. solfataricus protein. The Rbp18-coated and control resins were finally incubated with increasing amounts of radioactively labeled sR1 RNA, or with the control nonspecific RNA.
As shown in Figure 5C, sR1 was retained by the Rbp18-coated resin in amounts proportional to its concentration. By contrast, essentially no binding of the aspecific RNA was observed under the same conditions. These results confirm those obtained by EMSA, showing that S. solfataricus sR1 RNA could bind specifically to Rbp18. Altogether, the experiments of sR1–Rbp18 interaction supported the hypothesis that Rbp18 is a novel archaeal RNA-binding protein, interacting specifically with C/D box small RNAs. It remained to be determined if Rbp18 also interacted with the other members of C/D box RNA complexes.
Rbp18 is not a stable component of the C/D box RNPs
Since Rbp18 was able to bind the C/D box RNA sR1, we investigated the relationship of Rbp18 with the polypeptides characterized as components of the archaeal C/D box sRNPs; namely, fibrillarin, L7Ae and Nop 56/58. The S. solfataricus sRNP proteins were obtained in recombinant form by cloning and expression in E. coli of the corresponding genes, as described for Rbp18. All contained an N-terminal His-tag. Polyclonal antibodies against fibrillarin and L7Ae were also produced.
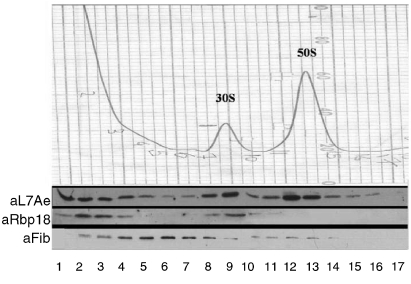
First, we determined the cellular localization of the proteins in question. For this purpose, S. solfataricus whole-cell lysates were fractionated on density gradients and the gradient fractions were probed in turn with anti-fibrillarin, anti-L7Ae and anti-Rbp18 antibodies. The position of Nop56/58 was not analyzed because we lacked efficient antibodies against this polypeptide.
The results (Figure 6) revealed that the three proteins had different localizations. Fibrillarin was scarce in the topmost fractions (containing the components of lowest molecular mass), peaked in the fractions corresponding to 10–20S material and trailed off to the 50S region. However, neither L7Ae nor Rbp18 was present in significant amounts in the fractions where fibrillarin was most abundant. L7Ae was found either in the top fractions of the gradients or on both ribosomal subunits. Rbp18 had a similar distribution except that it was only selectively associated with the 30S subunits.
Figure 6.
Localization of fibrillarin, Rbp18 and L7Ae. Sulfolobus solfataricus cell lysates were fractionated on sucrose density gradients; the fractions were checked for absorbance at 260 nm (top) to determine the position of the ribosomal subunits and for the presence of the different proteins using the specific antibodies against each of them as indicated (bottom). Abbreviations: aL7Ae, anti-L7Ae; aRbp18, anti-Rbp18; and aFib, anti-fibrillarin.
Given its presence in relatively heavy fractions, fibrillarin is likely to be complexed with some other cellular component (possibly pre-ribosomal particles). L7Ae, however, had the distribution expected of ribosomal proteins, partly in a free cytosolic pool and partly ribosome-bound. Notably, L7Ae, although known as a structural protein of the 50S particle, was present on both ribosomal subunits, consistent with its other role as a component of the rRNA-methylating RNP.
These results indicate that the C/D box sRNP proteins fibrillarin and L7Ae are not quantitatively associated in S. solfataricus cytosol. This may indicate that either the C/D box RNP particles are unstable (at least under the experimental conditions employed), or they assemble only transiently during ribosome biogenesis. However, the archaeal C/D box RNP proteins have been shown to interact in vitro (Omer et al. 2002). Therefore, we investigated whether Rbp18 was able to interact in vitro with fibrillarin, Nop 56/58 and L7Ae, individually or in the various combinations, in the presence of the C/D box RNA sR1.
For this purpose, recombinant fibrillarin, Nop56/58 and L7Ae were incubated with or without Rbp18 in the presence of sR1 RNA at 55 °C for 30 min. At the end of incubation, the samples were immunoprecipitated with polyclonal antibodies specific for S. solfataricus fibrillarin. The other proteins were therefore expected to be found in the final precipitate only if they had interacted with fibrillarin. The proteins retained by the resin were resolved by SDS-PAGE and visualized by Western blotting with anti-His antibodies, which recognized all four recombinant His-tagged proteins at the same time.
The results (not shown) revealed that the three sRNP core proteins (fibrillarin, Nop 56/58 and L7Ae) formed a complex stable enough to withstand immunoprecipitation. Rbp18, however, was not detected in the immunoprecipitates. These results indicated that Rbp18 is not an integral component of the C/D box RNPs, although a weak interaction with this particle cannot be excluded.
Rbp18 is a small ribosomal subunit protein in Sulfolobus solfataricus
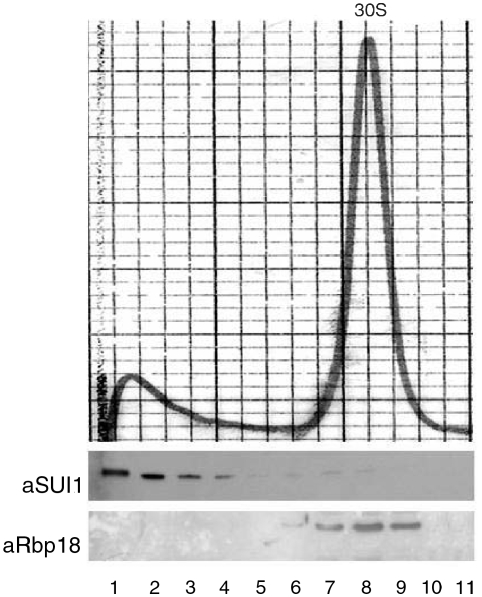
The localization experiments revealed that Rbp18 was in part associated with the 30S ribosomal subunit, thus suggesting that it could resemble L7Ae in being a ribosomal protein. To test this possibility, we determined whether Rbp18 could be detached from the 30S subunits by treatments designed to wash away extrinsic ribosome-bound proteins. The ribosomes were exposed to high concentrations of monovalent cations to weaken the electrostatic interactions whereby most extrinsic factors interact with ribosomal particles. The washing experiments were done with S. solfataricus 30S subunits that had been purified by zonal centrifugation. These particles were already devoid of most extrinsic proteins, but still retained Rbp18 and certain translation factors, including the small polypeptide aSUI1, a homologue of a eukaryotic translation initiation factor.
To strip 30S particles of remaining extrinsic proteins, they were treated with high concentrations of either of three salts: 2 M NH4Cl; 2 M KCl; and 0.5 M LiCl. Subsequently, the 30S particles were run on sucrose density gradients to assess their integrity and to determine the localization of Rbp18. Before gradient analysis, the salt-treated ribosomes were assayed for activity in a poly-U directed cell-free protein synthesis system in the presence of untreated 50S subunits. This was especially important for LiCl-treated particles, since it is known that this salt causes the partial disassembly and inactivation of E. coli ribosomes at a concentration of 0.4 M (Nierhaus and Montejo 1973). Sulfolobus solfataricus 30S particles, however, remained active in poly-U translation after washing with the various salts, including LiCl (results not shown). They also appeared structurally intact, as shown by the density gradient profiles in Figure 7, relative to LiCl-treated subunits. The salt-washed 30S subunits formed a sharp peak indistinguishable from untreated particles in its sedimentation behavior. Probing the gradient fractions with anti-Rbp18 antibodies showed that all of the protein remained concentrated in the 30S-subunit-containing fractions, thus demonstrating that it was firmly associated with the ribosomal particles. The same results were obtained with particles treated with either 2 M NH4Cl or 2 M KCl (not shown). In contrast, all salt treatments detached most of the translation factor SUI1 from the ribosomes (Figure 7).
Figure 7.
Rbp18 is a small subunit protein. Purified Sulfolobus solfataricus 30S subunits were treated with 0.4 M LiCl and fractionated on sucrose density gradients. Absorbance profile of the LiCl-washed subunits (top). Immunostaining of the gradient fractions with anti-Rbp18 (aRbp18) and anti-SUI1 (aSUI1) antibodies (bottom).
These results strongly suggest that Rbp18 is a structural protein of S. solfataricus ribosomes. Therefore, it appears to be similar to L7Ae in simultaneously performing the roles of ribosomal protein and of RNA-binding protein in the cytoplasm.
Discussion
We identified an archaea-specific component of the rRNA post-transcriptional modification machinery. A technique devised to specifically capture free cytosolic C/D box sRNPs resulted in the isolation of a protein having an apparent molecular mass of 18 kDa that we termed Rbp18 (18 kDa RNA-binding protein). Following N-terminal sequencing, Rbp18 was cloned and identified as a polypeptide of 155 amino acids of unknown function.
Notably, the protein is selectively present only in archaea. Furthermore, it is restricted to all the Crenarchaeota whose complete genome is known, whereas among the Euryarchaeota, Rbp18 seems to be present only in the order Pyrococcales.
An unrooted phylogenetic tree of the available Rbp18 sequences (Figure 3) showed that the crenarcheal and pyrococcal Rbp18 cluster into distinct clades. Specifically, the crenarchaeal Rbp18 sequences form two groups, one of which, including Hyperthermus butylicus, Aeropyrum pernix and Staphylothermus marinus, appears to be the closest to the clade formed by the Pyrococcales. This distribution is compatible with a crenarchaeal origin of Rbp18 and its later acquisition by the Pyrococcales by horizontal gene transfer, most likely from one of the species forming the nearest crenarcheal clade. Another possibility, though less probable because it requires several repeated events of gene transfer, is that Rbp18 was given by the Pyrococcales to the Crenarcheota. Since the archaea possessing Rbp18 are all thermophiles, in particular members of the Pyrococcales, which include the most thermophilic archaeal species known, the protein likely represents a specific adaptation to high temperatures.
We show here that Rbp18 is an RNA-binding protein that interacts with C/D box sRNAs in vivo and in vitro. In vivo, S. solfataricus Rbp18 seems to be the principal, if not the only, component associated with free C/D box sRNAs in the cytoplasm. Our technique captures only free sRNAs (as opposed to those associated with nascent ribosomes), since the oligonucleotides employed were complementary to the sequences expected to pair with rRNA. The proteins that are known components of the C/D box sRNPs, namely fibrillarin, L7Ae and Nop56/58, were not recovered by the bioseparation technique employed in this work. Although possibly an artifact, this may indicate that the C/D box sRNPs are not permanently, or stably, assembled in the cytoplasm. This possibility is also suggested by the observation that two of the rRNA-methylating sRNP components, L7Ae and fibrillarin, are differently distributed in a density-fractionated cell lysate. Although L7Ae exists either in a free or ribosome-bound state, fibrillarin is found mainly in pre-ribosomal fractions, especially in the 10–20S range, indicating that the majority of it is bound to some other cellular component of relatively high molecular mass, possibly pre-ribosomal particles. However, the protein components of the C/D box sRNP interacted in vitro, whereas Rbp18 does not belong in this complex and appears to associate principally with free C/D box sRNA.
Because Rbp18 is specific to highly thermophilic archaea, we propose that its main role is in stabilizing the sRNAs during the life of the cell. It is conceivable that Rbp18 also helps the assembly of rRNA-modifying RNPs, possibly by carrying sRNAs to the pre-ribosomal particle and promoting its annealing with the target sequence. The fact that Rbp18 is not ubiquitous even in thermophilic archaea (many thermophilic euryarchaea lack it) suggests that it is not an essential protein. However, some selective advantage may accrue to organisms that have it, by conferring additional stability to the sRNAs that participate in rRNA post-transcriptional modification, and perhaps by facilitating the assembly of the methylation complex.
Remarkably, our results show that, in addition to its sRNA binding capacity, Rbp18 is a structural protein of the small ribosomal subunit. This is borne out by the fact that the ribosome-bound Rbp18 cannot be removed from the 30S subunits even by harsh, high-salt treatments. Therefore, Rbp18 would be a further example of a multifunctional ribosomal protein, and would resemble L7Ae in being implicated in the rRNA processing machinery in addition to serving as a ribosomal constituent. In this latter capacity, Rbp18 may also promote survival at high-temperatures; for instance, it might confer additional heat-stability to the small ribosomal subunits. However, further studies are needed to elucidate details of Rbp18 function.
Acknowledgments
This work was supported in part by funds from the MIUR PRIN 2002 and 2005 projects: "Evolution of the gene expression machinery: characterization of a set of universally conserved factors modulating translational initiation" and "Dynamics of ribosome-ligand interactions during translation initiation in bacteria and archaea" and by funds from Progetti di Ateneo 2002–2005 of Bari University. We thank Dr. L. Nicolini of Istituto Superiore di Sanità for growing S. solfataricus cells and Prof. P. Cammarano of Rome University Sapienza for drawing the phylogenetic tree shown in Figure 3.
References
- R1.Aittaleb M., Rashid R., Chen Q., Palmer J.R., Daniels C.J., Li H. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol.Nat. Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- R2.Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. BiochemieBiochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- R3.Ban N., Nissen P., Hansen J., Capel M., Moore P.B., Steitz T.A. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- R4.Cavaille J., Nicoloso M., Bachellerie J.-P. Targeted ribose methylationof RNAin vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- R5.Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small mucleolar RNAs. CellCell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- R6.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Kiss T. Small nucleolar RNAs: an abundant group of non-coding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- R8.Kiss-Laszlo Z., Henry Y., Bachellerie J.-P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nuclear RNAs. CellCell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- R9.Kiss-Laszlo Z., Henry Y., Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-RNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Kuhn J.F., Tran E.J., Maxwell E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p sno RNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Ni J., Tien A.L., Fournier M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- R12.Neirhaus K.H., Montejo V. A protein involved in the peptidyltransferase activity of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USAProc. Natl. Acad. Sci. USA. 1973;70:1931–1935. doi: 10.1073/pnas.70.7.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Omer A.D., Lowe T.M., Russell A.G., Ebhardt H., Eddy S.R., Dennis P.P. Homologs of small nucleolar RNAs in Archaea. ScienceScience. 2002;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- R14.Omer A.D., Ziesche S., Ebhardt H., Dennis P.P. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Tollervey D. Small nucleolar RNAs guide ribosomal RNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- R16.Zeische S.M., Omer A.D., Dennis P.P. RNA-guided nuclotide modification of ribosomal and non-ribosomal RNA in archaea. Mol. Microbiol. 2004;54:980–993. doi: 10.1111/j.1365-2958.2004.04319.x. [DOI] [PubMed] [Google Scholar]