Abstract
Alpha-glucan phosphorylase catalyzes the reversible cleavage of α-1-4-linked glucose polymers into α-D-glucose-1-phosphate. We report the recombinant production of an α-glucan/maltodextrin phosphorylase (PF1535) from a hyperthermophilic archaeon, Pyrococcus furiosus, and the first detailed biochemical characterization of this enzyme from any archaeal source using a mass-spectrometry-based assay. The apparent 98 kDa recombinant enzyme was active over a broad range of temperatures and pH, with optimal activity at 80 °C and pH 6.5–7. This archaeal protein retained its complete activity after 24 h at 80 °C in Tris-HCl buffer. Unlike other previously reported phosphorylases, the Ni-affinity column purified enzyme showed broad substrate specificity in both the synthesis and degradation of maltooligosaccharides. In the synthetic direction of the enzymatic reaction, the lowest oligosaccharide required for the chain elongation was maltose. In the degradative direction, the archaeal enzyme can produce glucose-1-phosphate from maltotriose or longer maltooligosaccharides including both glycogen and starch. The specific activity of the enzyme at 80 °C in the presence of 10 mM maltoheptaose and at 10 mg ml–1 glycogen concentration was 52 U mg–1 and 31 U mg–1, respectively. The apparent Michaelis constant and maximum velocity for inorganic phosphate were 31 ± 2 mM and 0.60 ± 0.02 mM min–1 µg–1, respectively. An initial velocity study of the enzymatic reaction indicated a sequential bi-bi catalytic mechanism. Unlike the more widely studied mammalian glycogen phosphorylase, the Pyrococcus enzyme is active in the absence of added AMP.
Keywords: electrospray ionization mass spectrometry (ESI-MS), thermostability
Introduction
Genome sequencing projects have generated hundreds of genes tentatively assigned biochemical functions in the synthesis and degradation of glucose polymers. In contrast, efforts to verify the functions of those gene products and probe their possible applications are just beginning. As part of our program to understand the origins of fidelity in carbohydrate biosynthesis, we have recently reported three promiscuous enzymes—glycogen synthase, UDP-glucose pyrophosphorylase and a bifunctional acetyltransferase/UDP-N-acetylglucosamine pyrophosphorylase—from a hyperthermophilic archaeon, Pyrococcus furiosus (Zea et al. 2003, Mizanur et al. 2004, 2005, Zea and Pohl 2005). The relatively broad substrate specificities and high thermostabilities of these enzymes hint that this small archaeal genome might harbor more enzymes with unusual and useful properties.
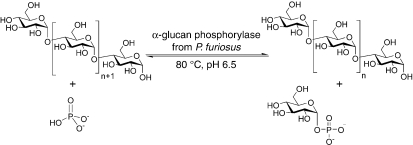
Alpha-glucan phosphorylases (EC 2.4.1.1) catalyze the reversible phosphorolytic cleavage of α-1-4-linked glucose oligosaccharides with formation of α-D-glucose-1-phosphate (G-1-P) and hence play a central role in the mobilization of storage polysaccharides (Figure 1). Although the enzyme catalyzes a reversible reaction, its major function in vivo is assumed to be phosphorolytic degradation of α-glucans. Significant progress has been made with the detailed structural and functional characterization of glycogen phosphorylases of mesophilic origins (Chen and Segel 1968, Hwang and Fletterick 1986, Browner et al. 1991, Rogers et al. 1992, Nidetzky et al. 1995, Weinhausel et al. 1995, Srivastava et al. 1996); however, little is known about those from hyperthermophilic organisms. Thermostable α-glucan phosphorylases reported to date have been isolated from the thermophilic bacteria Bacillus stearothermophilus, Thermus caldophilus, Thermus aquaticus, Thermus thermophilus, Thermotoga maritima and Aquifex aeolicus (Konig et al. 1982, Boeck and Schinzel 1996, Bibel et al. 1998, Takata et al. 1998, Takaha et al. 2001, Bhuiyan et al. 2003, Bae et al. 2005). Among archaea, the enzyme activity has been detected in Methanococcus maripaludis and Thermococcus litoralis (Yu et al. 1994, Xavier et al. 1999). A thin layer chromatography (TLC) study of an enzyme assigned as a maltodextrin phosphorylase (PF1535) from P. furiosus was shown to cleave maltotetraose as the smallest oligosaccharide for the formation G-1-P, although maltotriose may also have been a substrate (Lee et al. 2006). However, no detailed biochemical, stability and kinetic studies of any such archaeal enzymes have been made.
Figure 1.
Schematic diagram of the reaction catalyzed by α-glucan phosphorylase from Pyrococcus furiosus. The glucose molecule is liberated from the non-reducing end of the glucose polymer to form α-D-glucose-1-phosphate.
Glucan phosphorylases are valuable for the synthesis of G-1-P (Shin et al. 2000, Bae et al. 2005), a potential antibiotic or immunosuppressive drug (Takata et al. 1998), for the production of linear amylose and other related glucose polymers (Yanase et al. 2002, Fujii et al. 2003, Ohdan et al. 2007), for the synthesis of amylose-lipid complexes (Gelders et al. 2005) and for carbohydrate engineering (Yanase et al. 2006). The hyperthermostable glucan phosphorylase in combination with other thermostable sugar nucleotidyltransferases (Mizanur et al. 2004, 2005) in a reaction using starch as the starting polysaccharide for the subsequent synthesis of nucleotide sugars could be advantageous over the mesophilic counterpart, because the solubility of starch is increased at higher temperature. Because archaea contain a range of hyperthermophiles, these enzymes could be of particular use for their stability at higher temperatures. Here we report the first detailed biochemical and stability studies of a recombinant enzyme involved in glucan/starch metabolism from an archaeon, P. furiosus, from a gene assigned as an α-glucan/maltodextrin phosphorylase (locus tag PF1535).
Materials and methods
Materials
Enzymes and reagents used for the molecular biology procedures, DNA ladders and deoxynucleotide triphosphates (dNTPs) were purchased from Promega (Madison, WI) or New England Biolabs (Beverly, MA). Oligonucleotides for DNA amplification were synthesized by Integrated DNA Technologies (Coralville, IA). Isopropyl-β-thiogalactoside (IPTG) was obtained from Labscientific (Livingston, NJ). Protein molecular weight standards were obtained from BioRad (Hercules, CA). The QIAQuick gel extraction kit was obtained from Qiagen (Valencia, CA), and the Zero blunt PCR cloning kit was purchased from Invitrogen (Carlsbad, CA). Nucleotide triphosphates (NTPs), glycogen, maltooligosaccharides and all other chemicals were obtained from Sigma (St. Louis, MO) unless stated otherwise.
Bacterial strains and growth conditions
Genomic DNA of Pyrococcus furiosus DSM 3638 obtained from the American Type Culture Collection (Manassas, VA), was used as the source for the cloning experiments. Oneshot Top10 competent cells (Invitrogen, Carlsbad, CA), Escherichia coli XL-10Blue (Stratagene, La Jolla, CA) and PCR-Blunt vectors (Invitrogen) were used for direct cloning of PCR products. Escherichia coli BL21 Codon plus (DE3)-RIPL (Stratagene) was used in combination with the T7 expression system (pET21a vector; Novagen, Madison, WI) for expression of the α-glucan phosphorylase gene. Escherichia coli cells were grown on Luria Bertani (Sigma) medium at 37 °C in an incubator shaker at 225 rpm. When required, the antibiotics carbenicillin or kanamycin were added at 50 µg ml–1 to make the selective media.
General methods
The DNA procedures, including plasmid DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, DNA ligation and transformation of E. coli, were performed according to standard techniques (Sambrook et al. 1989) and manufacturers’ instructions. The PCR was carried out in an Eppendorf Mastercycler gradient thermocycler (Eppendorf Scientific, Westbury, NY). Protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Tris-HCl 10–20% gradients, Bio-Rad). The gels were stained with Coomassie brilliant blue. Protein concentrations were determined with the Bio-Rad protein assay kit according to the method of Bradford (1976), with bovine serum albumin as the standard.
PCR amplification, cloning, expression and purification of enzyme
Genomic DNA of P. furiosus DSM 3638 was amplified by PCR. The primers were designed to construct the α-glucan phosphorylase expression plasmid. The forward primer, 5′-AAACCATATGGTTAAAGTGGAGAATTCAATTAAG-3′ , containing an NdeI restriction site (in bold), and the reverse primer, 5′ -AAACTCGAGCTACTCTTTCTTTTCAATATCTTTCC-3′, containing an XhoI restriction site (in bold), were synthesized from the putative α-glucan phosphorylase of P. furiosus. The amplification reaction mixture contained standard Pfu DNA polymerase buffer, 375 µM of dNTPs, 3 ng of each primer, 4 ng of total genomic DNA and 2.5 units of Pfu DNA polymerase. The cycling parameters were as follows 94 °C for 2 min 40 s followed by 32 cycles of 94 °C for 30 s, 58 °C for 45 s and 72 °C for 2 min 15 s, with a final elongation step of 72 °C for 15 min. The amplified DNA was cloned in a pET21a vector and introduced into an E. coli BL21 codon plus (DE3)-RIPL strain following the method described by Mizanur et al. (2004). Isopropyl-β-thiogalactoside (1 mM) induced production of the recombinant enzyme, which was purified at 4 °C unless otherwise stated. The cells were disrupted by sonication (Fisher model 100 Sonic Dismembranator, Fisher Scientific, Pittsburgh, PA), after which unbroken cells and debris were removed by centrifugation (30 min at 10,000 g). The cleared lysate was then incubated at 100 °C for 5 min and centrifuged (10 min at 10,000 g) to remove most of the protein impurities. The supernatant was purified by metal chelate chromatography by following the recommended procedures provided by Novagen. The purified protein was concentrated and dialyzed against Tris-HCl buffer (50 mM, pH 7) using a Microcon Centrifugal Filter Device, MWCO 30 kDa (Millipore, Billerica, MA). The protein was analyzed by SDS-PAGE.
Mass spectrometry based enzyme assay
A Shimadzu LCMS 2010 quadruple mass spectrometer (Shimadzu Scientific Instruments, Columbia, MD) equipped with an electrospray ionization source was used in negative ion mode to develop an electrospray ionization mass spectrometry (ESI-MS) based enzyme assay according to the method described by Zea and Pohl (2004). Activity of the P. furiosus enzyme was determined in the phosphorolytic direction with glycogen as the substrate. The enzymatic reaction was initiated by the addition of glycogen to a reaction mixture of 50 µl containing α-glucan phosphorylase, 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (50 mM, pH 6.8), sodium phosphate (50 mM) and glycogen (7 mg ml–1). Before adding glycogen, the reaction mixture was incubated at 80 °C for 5 min. Reactions were carried out at 80 °C for 10 min, and 15 µl of the reaction mixture was quenched by the addition of 30 µl of 70% methanol containing AMP (3 mM) as an internal standard. The quenched solutions were centrifuged for 10 min at 10,000 g to precipitate the protein. Aliquots of the reaction mixtures were diluted with 135 µl of acetonitrile:water:triethylamine (35:65:0.2). These samples (5 µl) were subjected to analysis by ESI-MS to determine the amount of G-1-P formed and compared with a blank containing no enzyme. A control reaction contained all the reaction components except the enzyme. One unit of enzyme is defined as the amount of enzyme needed to produce one µmol of G-1-P per minute at specified temperature and conditions.
Optimal conditions for enzymatic activity
Optimal pH for the P. furiosus enzymatic activity was determined at 80 °C between pH 4 and pH 9 using 50 mM acetate, MOPS, N-[2-hydroxyethyl] piperazine-N′-[2-ethanesulfonic acid] (HEPES) and Tris-HCl buffer. The optimal temperature was measured at pH 6.5 between 0 and 99 °C. To check the stability of the enzyme at its optimal temperature, the purified enzyme in Tris-HCl buffer was incubated at 80 °C, and enzymatic activity determined for different times of incubation.
Substrate specificity
Substrate specificity of the enzyme in the phosphorolytic direction was determined with different maltooligosaccharides and glycogen/starch by monitoring the formation of G-1-P using the assay described above. Percent conversions were calculated as the amount of sugar phosphate remaining in the reaction after incubation at a specified time, divided by the amount of substrate used in the reaction, and then multiplied by 100. In the synthetic direction, the reactions were carried out in a mixture containing the appropriate amount of enzyme in the presence of G-1-P, various maltooligosaccharides and MOPS buffer (50 mM, pH 6.8). Chain elongation was confirmed by monitoring the disappearance of G-1-P and was expressed as percent relative intensity (RI), which was calculated as: (RI of G-1-P from oligosaccharide)/(RI of G-1-P from maltoheptaose) × 100. Each reaction was run in duplicate and the mean percent conversion was used. Controls for each set of reactions were run simultaneously containing all the components except enzyme.
Kinetic analysis
The values of the Michaelis constant (Km) and maximum velocity (Vmax) were derived from enzymatic reactions run in triplicate and determined from the initial rates of glycogen degradation by monitoring the rate of G-1-P formation using ESI-MS. The enzymatic reaction was initiated by the addition of α-glucan phosphorylase (0.05 units) to a reaction mixture containing MOPS buffer (50 mM, pH 6.5), glycogen (7 mg ml–1) and phosphate (2.5–200 mM). Reactions were carried out at 80 °C, and after 5 min, 20 µl of the reaction mixture was quenched by addition to 20 µl of 70% methanol containing AMP (3 mM) as an internal standard. The quenched solutions were centrifuged for 10 min at 10,000 g to precipitate the glycogen and protein. Aliquots (15 µl) of the reaction mixtures were diluted with 135 µl of acetonitrile:water:triethylamine (35:65:0.2). These samples (5 µl) were subjected to analysis by ESI-MS to determine the G-1-P concentrations. All kinetic data was fitted with the nonlinear regression algorithm in GraphPad Prism Version 4 (GraphPad Software, San Diego, CA). To determine the kinetic mechanism of the enzyme, the experiments were designed to provide data by varying the concentration of phosphate at several fixed concentrations of maltoheptaose and also varying the concentration of maltoheptaose at several fixed concentrations of phosphate. Additionally, an initial velocity study of the enzyme in the phosphorolytic direction with a maltotriose primer was carried out at 37 °C in a reaction mixture containing fixed concentrations of maltotriose (5 mM) and varying concentrations of phosphate (10–200 mM) using a coupled assay as described by Schinzel and Palm (1990).
Multiple sequence alignment
A multiple sequence alignment of the amino acids of α-glucan phosphorylase was performed using BLAST (www.ncbi.nih. gov). Two sequences were aligned using BLAST.
Results and discussion
Amino acid sequence alignment
To compare the P. furiosus α-glucan phosphorylase with other known phosphorylases from different thermostable organisms, we initiated a BLAST search with the amino acid sequence of the enzyme. The enzyme showed significant alignments with three different conserved domains: (1) carbohydrate phosphorylases (Residues 273 to 617) that catalyze the formation of G-1-P from glycogen, starch, glucan or maltodextrin; (2) GlgP (Residues 21 to 698) that corresponds to glucan phosphorylase; and (3) GlgA (Residues 214 to 583) that corresponds to glycogen synthase. Next, we carried out a domain search of the enzyme and found it to have two different domains: a carbohydrate phosphorylase domain (Residues 91 to 797) and a phosphorylase pyridoxal phosphate attachment site (Residues 638 to 650; Figure 2). The pyridoxal phosphate (PLP) domain is common to all α-glucan phosphorylases. Catalysis by phosphorylases is dependent on the ionizable phosphate residue of the enzyme-bound PLP (Johnson 1992, Boeck and Schinzel 1996). The amino acid sequence alignment of the P. furiosus a-glucan phosphorylase with the E. coli maltodextrin phosphorylase (Palm et al. 1982) showed only 23% identity (Figure 2). However, the PLP domain is conserved in both enzymes and is almost 70% identical (Figure 2). The important AMP binding residues of glycogen phosphorylase of rabbit muscle (Cori and Green 1943) are not conserved in the E. coli and P. furiosus enzymes. The P. furiosus enzyme is not likely regulated by AMP nor is AMP required for enzyme activity, although no such biochemical studies have yet been reported with any archaeal enzymes. When the P. furiosus enzyme was compared with putative enzymes from other thermophilic organisms, it showed a high degree of sequence identity with the glucan phosphorylases from P. horikoshii (locus tag PH1512), P. abyssi (locus tag PAB2414) and Thermococcus litoralis (locus AF115479) sequences with 78, 80 and 70% identity, respectively (90, 90 and 83% similarities, respectively; Table 1). The P. furiosus enzyme also has significant sequence alignment with putative proteins from the thermophilic bacterium Thermotoga maritima (locus CAA04523). All enzymes shown in Table 1 have nearly the same molecular mass, except the protein from Aquifex aeolicus. Taken as a whole, the α-glucan phosphorylases have 41–80% identity and 57–99% similarity with each other. The putative α-glucan phosphorylase from another closely related organism, Thermococcus kodakarensis (locus tag TK 1406), is 69% identical to the enzyme from P. furiosus. Two neighboring genes (locus tags PF1534 and PF1536) of the putative a-glucan phosphorylase (PF1535) have no sequence similarity with any of the known proteins and were assigned as hypothetical proteins. To study the biochemical function of the α-glucan phosphorylase from P. furiosus and test its temperature stability, we cloned and expressed the enzyme in an E. coli strain.
Figure 2.
Amino acid sequence alignment of Pyrococcus furiosus (pfu) α-glucan phosphorylase with an Escherichia coli (eco) maltodextrin phosphorylase. Identical amino acids are indicated in bold. The conserved pyridoxal phosphate domain residues are highlighted.
Table 1.
Similarity matrix of amino acids pair wise identity for α-glucan phosphorylases from different thermophiles. Amino acid identity among organisms specifies the number of amino acid residues between each pair of organisms which are identical, with the percent of identical and similar residues given in parentheses. Abbreviations: Accn. no., Accession number; and amino acids, number of amino acids.
| Organism | Accn. no. | Amino acids | Amino acid identity among organisms | ||||||
| Pyrococcushorikoshii | Thermococcuslitoralis | Thermococcusmaritima | Thermuscaldophilus | Aquifexaeolicus | Thermusthermophilus | Pyrococcusabyssi | |||
| Pyrococcus furiosus | NC003413.1 | 839 | 78 (90) | 70 (83) | 62 (77) | 41 (57) | 43 (62) | 44 (62) | 80 (90) |
| Pyrococcus horikoshii | NC000961.1 | 837 | 68 (83) | 59 (76) | 39 (58) | 43 (63) | 39 (58) | 86 (94) | |
| Thermococcus litoralis | AF115479.1 | 831 | 62 (77) | 45 (63) | 45 (63) | 44 (63) | 69 (82) | ||
| Thermotoga maritima | AJ001088 | 822 | 44 (61) | 43 (63) | 44 (60) | 59 (75) | |||
| Thermus caldophilus | AY804131.1 | 819 | 39 (58) | 99 (99) | 44 (62) | ||||
| Aquifex aeolicus | NC000918.1 | 692 | 39 (57) | 43 (63) | |||||
| Thermus thermophilus | NC006461.1 | 819 | 44 (62) | ||||||
| Pyrococcus abyssi | NC000868.1 | 835 | |||||||
Cloning and purification of the PF1535 protein
We successfully expressed recombinant protein in E. coli harboring a heterologous expression vector containing the PF1535 gene. The open reading frame encoding the P. furiosus α-glucan phosphorylase (2517 bp) was PCR amplified and cloned into a pET21a vector with a hexahistidine tag at the C-terminus to provide plasmid pGPH1. To confirm the presence of a functional recombinant α-glucan phosphorylase, the expression construct pGPH1 was introduced into E. coli BL21 codon plus (DE3)-RIPL cells, which were then grown in Luria Bertani broth for IPTG-initiated induction of the enzyme. To purify the enzyme, the cells were disrupted by sonication and centrifuged to collect the soluble fraction. The crude protein extract was heat treated at 100 °C for 5 min, the temperature at which most native E. coli proteins are precipitated, and the supernatant was collected for further purification by column chromatography. The resulting cell-free extract was passed through an Ni-affinity column and eluted with imidazole buffer. The purity, of the recombinant protein was confirmed by SDS-PAGE, which revealed a single band with an apparent molecular mass of 98 kDa that is consistent with the molecular mass expected from the nucleotide sequence (data not shown). It has been observed that most known phosphorylases have a subunit molecular mass of around 90 kDa and usually exist as homodimers or tetramers (Palm et al. 1985). At 25 °C the thermostable α-glucan phosphorylases from Thermus thermophilus, Thermus caldophilus and Thermococcus litoralis form octamers, hexamers and trimers, respectively (Boeck and Schinzel 1996, Xavier et al 1999, Bae et al 2005).
Activity of the recombinant protein
The activity of both the crude extract and purified enzyme was determined at 80 °C with glycogen as substrate in the phosphorolytic direction in the presence of other appropriate reaction components. Formation of G-1-P both in the crude extract and with purified protein was monitored by an ESI-MS-based detection method (Zea and Pohl 2004) and confirmed the presence of a functional PF1535 protein. Enzymatic activity was also detected in the direction of chain elongation using glycogen and G-1-P by monitoring the disappearance of the phosphate sugar. Consequently, time- and concentration-dependent formation and accumulation of G-1-P were observed when various concentrations of glycogen and maltoheptaose were incubated in separate reactions with the purified recombinant protein at 80 °C for different time periods (data not shown). The purified enzyme was used directly for further biochemical and kinetic characterization or stored at –20 °C as a 25% glycerol stock solution for future use with little loss of activity after 180 days. Although heat treatment should have denatured all native proteins, an additional control experiment was run with extracts of E. coli carrying a pET21a vector without an inserted gene. No additional protein band corresponding to 98 kDa was detected by SDS-PAGE of the crude extract after passing through the Ni-affinity column, and no phosphorylase activity was detected at 80 °C (data not shown). No product formation was observed in the absence of the enzyme and glycogen. Unlike rabbit muscle glycogen phosphorylase, the P. furiosus enzyme is not likely regulated by AMP, as it is completely active in the absence of added AMP. This result is consistent with the finding that no conserved amino acid residues for AMP binding are present in the P. furiosus enzyme (Figure 2). The activity of the enzyme is unaffected by the presence of ADP or ATP in the reaction mixture. As expected, the enzyme does not require divalent cation for its activity nor do added divalent cations show any inhibitory activity.
Optimization of pH and temperature conditions and thermostability
The effect of pH on the phosphorolytic reaction of the enzyme was analyzed using four different buffer systems. The recombinant protein exhibited relatively high activity around pH 6.0 and 7.5, with maximum activity at pH 6.5 (data not shown). The shape and maxima of the pH velocity curves, however, varied somewhat with the nature of the buffer ion present. Because the recombinant protein was originally cloned from the hyperthermophilic archaeon P. furiosus, which grows optimally at 100 °C, the temperature dependence of the enzymatic activity was analyzed between 0 and 100 °C. The enzyme had relatively high activity between 60 and 100 °C with a maximum at 80 °C. To select a suitable temperature for enzyme storage for short periods of time, we checked enzyme stability after incubation with MOPS (pH 6.5) buffer for 30 min at different temperatures ranging from 0 to 100 °C. The enzyme remained stable at all temperatures tested with no loss of residual activity. The purified recombinant protein at 0.05 mg ml–1 concentration was incubated at 100 °C for over 24 h, and aliquots were removed at 3-h intervals and stored at –20 °C. The activity of these aliquots was then checked at 80 °C for 10 min. No loss of activity was detected after incubation at 100 °C for 24 h (data not shown), suggesting that the protein encoded by PF1535 is an extremely thermostable enzyme. Maximum enzymatic activity was detected between 70 and 90 °C, but over 50% of activity still remained at 50 and 60 °C, indicating that the enzyme can function over a broad temperature range. The hyperthermostability as well as broad temperature range for activity could be beneficial for industrial uses.
Substrate specificity
The specificity of the PF1535 recombinant protein for various maltooligosaccharides (7 mM) and glycogen (7 mg ml–1) was examined in both directions under our standard assay conditions. Initially, the specificity for various oligosaccharides was examined. When G-1-P was utilized in the synthetic direction, maltose and larger oligosaccharides were shown as effective acceptors for oligosaccharide synthesis. The highest activity was detected with the smallest oligosaccharide, maltose, which was comparable with the value observed in the presence of glycogen (Table 2).
Table 2.
Substrate specificity of α-glucan phosphorylase from Pyrococcus furiosus in the synthetic and phosphorolytic directions in the presence of glucose-1-phosphate (G-1-P) after 30 min. Percent conversions were calculated as the amount of sugar phosphate remaining in the reaction after incubation at a specified time, divided by the amount of substrate used in the reaction, and then multiplied by 100. The percent G-1-P produced was expressed as percent relative intensity (RI) which was calculated as: (RI of G-1-P from oligosaccharide)/(RI of G-1-P from maltoheptaose) × 100. Substrate concentration was 7 mg ml–1 for glycogen and 7 mM for maltose through maltoheptaose.
| Substrate | G-1-P consumed (Synthetic) | G-1-P produced (Phosphorolytic) |
| Maltose | 100 | 0 |
| Maltotriose | 95 | 35 |
| Maltotetraose | 90 | 61 |
| Maltopentaose | 86 | 78 |
| Maltohexaose | 82 | 85 |
| Maltoheptaose | 80 | 100 |
| Glycogen | 75 | 64 |
In the presence of inorganic phosphate, the enzyme degraded maltotriose and larger oligosaccharides effectively with relative activity ranging from 34 to 100% (Table 2). In a previous study, it was concluded from TLC analysis that maltotetraose is the smallest substrate for the P. furiosus glucan/maltodextrin phosphorylase (Lee et al. 2006). However, with our mass spectrometry assay, we demonstrated that the enzyme can also cleave maltotriose to form G-1-P. Inspection of the earlier reported TLC data also indicates the possibility of cleavage of this smaller substrate, although those data were not clear. To clarify if maltotriose can indeed be used as a substrate in the phosphorolytic reaction, we carried out an additional experiment to check the initial rate by using a coupled enzyme assay as described by Schinzel and Palm (1990). Nonlinear regression of the data yielded KM and Vmax values of 70 ± 5 mM and 0.67 ± 0.02 mM min–1 µg–1, respectively. These data indicate that maltotriose is also a substrate of the enzyme in the phosphorolytic direction. Maltodextrin phosphorylase from T. litoralis, a close relative of P. furiosus, accepts maltotetraose as a substrate in the synthetic direction, and glycogen phosphorylase from A. aeolicus accepts maltotriose in the direction of chain elongation (Xavier et al. 1999, Bhuiyan et al. 2003). Studies have shown that P. furiosus hydrolyses starch to maltodextrins using extracellular amylopullulanase (PF1935*; Brown and Kelly 1993, Lee et al. 2006). Maltodextrin thus produced in the extracellular medium is then taken up into the cell by the maltooligosaccharide transporter Mal-II (Lee et al. 2006). Lee and coworkers proposed that intracellular maltodextrins are converted to G-1-P by this glucan/maltodextrin phosphorylase (PF1535). Our in-vitro studies have shown that, in addition to maltooligosaccharides, the enzyme degrades larger glucose-based polysaccharides to form G-1-P, an essential component for the biosynthesis of various nucleotide-glucoses. In extreme environments with nutrient scarcity, the α-glucan phosphorylase-catalyzed synthesis of G-1-P might serve as the donor substrate for the synthesis of nucleoside diphospho-glucoses—essential components for various glycosyltransferase reactions. In addition to maltodextrins, the phosphorolytic cleavage of starch at a comparable rate also hints that the enzyme might play a significant role in mobilization and storage of glucose polymers.
Kinetic properties
To obtain the kinetic properties of the enzyme in the phosphorolytic direction, phosphate ion concentrations were varied from 2.5 to 250 mM. Traditional enzymatic analysis such as a Michaelis–Menten plot of the velocity versus phosphate ion concentration was determined using the ESI-MS technique. The enzymatic reactions were run in triplicate. Nonlinear regression of the data yielded KM and Vmax values of 31 ± 2 mM and 0.60 ± 0.02 mM min–1 µg–1, respectively, which are comparable with values obtained from homologous enzymes. To determine the kinetic mechanism of the enzyme, double reciprocal plots of the initial velocity were obtained by varying the concentration of one substrate at fixed concentrations of the other. The kinetic data were obtained from studies in the direction of phosphorolysis utilizing maltoheptaose as a substrate. All the double reciprocal plots were linear and showed converging patterns. A clear intersecting pattern left of the 1/V axis was indicative of a sequential bi-bi mechanism, which is similar to that of other bacterial and eukaryotic phosphorylases (Chao et al. 1969, Engers et al. 1969, Gold et al. 1971).
This first detailed biochemical and thermostability study of a recombinant α-glucan/maltodextrin pyrophosphorylase from any archaeal source has uncovered an unusually heat-stable enzyme for the chemoenzymatic synthesis of a range of glucose biopolymer fragments. Hyperthermostability and activity over a broad range of temperatures make this archaeal enzyme potentially attractive for biotechnological applications that involve degradation reactions of α-glucose polymers. We are now investigating the one-pot synthesis of activated glucose nucleotides from starch using a mix of archaeal enzymes including this glucan/maltodextrin phosphorylase and our recently reported sugar nucleotidyltransferases (Mizanur et al. 2004, 2005).
Acknowledgments
This report is based in part on work supported by the National Science Foundation under CAREER Grant No. 0349139. The authors thank the Department of Chemistry, the Caldwell Trust, the Plant Sciences Institute, a Shimadzu University Research Grant and the Herman Frasch Foundation (American Chemical Society) for partial support of this research. NLP is a Cottrell Scholar of Research Corporation and an Alfred P. Sloan Foundation Research Fellow.
References
- R1.Bae J., Lee D.H., Kim D. Facile synthesis of glucose-1-phosphate from starch by Thermus caldophilus GK24 alpha-glucan phosphorylase. Process Biochem. 2005;40:3707–3713. [Google Scholar]
- R2.Bhuiyan S.H., Rus'd A.A., Kitaoka M., Hayashi K.J. Characterization of a hyperthermostable glycogen phosphorylase from Aquifex aeolicus expressed in Escherichia coli . J. Mol. Catal. B: Enz. 2003;22:173–180. [Google Scholar]
- R3.Bibel M., Brettle C., Gosslar U., Kriegshauser G., Liebl W. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima . FEMS Microbiol. Lett. 1998;158:9–15. doi: 10.1111/j.1574-6968.1998.tb12793.x. [DOI] [PubMed] [Google Scholar]
- R4.Boeck B., Schinzel R. Purification and characterization of an alpha-glucan pyrophosphorylase from the thermophilic bacterium Thermus thermohilus. Eur. J. Biochem. 1996;239:150–155. doi: 10.1111/j.1432-1033.1996.0150u.x. [DOI] [PubMed] [Google Scholar]
- R5.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- R6.Brown S.H., Kelly R.M. Characterization of amylolytic enzymes, having both α-1,4 and α-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis . Appl. Environ. Microbiol. 1993;59:2614–2621. doi: 10.1128/aem.59.8.2614-2621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Browner M.F., Rasor P., Tugendreich S., Fletterick R.J. Temperature-sensitive production of rabbit muscle glycogen phosphorylase in Escherichia coli . Protein Eng. 1991;4:351–357. doi: 10.1093/protein/4.3.351. [DOI] [PubMed] [Google Scholar]
- R8.Chao J., Johnson G.F., Gravest D.J. Kinetic mechanism of maltodextrin phosphorylase. Biochemistry . 1969;8:1459–1466. doi: 10.1021/bi00832a022. [DOI] [PubMed] [Google Scholar]
- R9.Chen G.S., Segel I.H. Purification and properties of glycogen phosphorylase from Escherichia coli . Arch. Biochem. Biophys. 1968;127:175–186. doi: 10.1016/0003-9861(68)90214-2. [DOI] [PubMed] [Google Scholar]
- R10.Cori G.T., Green A.A. Crystalline muscle phosphorylase. II. Prosthetic group. J. Biol. Chem. 1943;151:31–38. [Google Scholar]
- R11.Engers H.D., Bridger W.A., Madsen N.B. Kinetic mechanism of phosphorylase b. Rates of initial velocities and of isotope exchange at equilibrium. J. Biol. Chem. 1969;244:5936–5942. [PubMed] [Google Scholar]
- R12.Fujii K., Takata H., Yanase M., Terada Y., Ohdan K., Takaha T., Okada S., Kuriki T. Bioengineering and application of novel glucose polymer. Biocatal. Biotransform. 2003;21:167–172. [Google Scholar]
- R13.Gelders G.G., Goesaert H., Declour J.A. Potato phosphorylase catalyzed synthesis of amylose-lipid complexes. Biomacromolecules. 2005;6:2622–2629. doi: 10.1021/bm0502011. [DOI] [PubMed] [Google Scholar]
- R14.Gold A.M., Johnson R.M., Sanchez G.R. Kinetic mechanism of potato phosphorylase. J. Biol. Chem. 1971;246:3444–3450. [PubMed] [Google Scholar]
- R15.Hwang P.K., Fletterick R.J. Convergent and divergent evolution of regulatory sites in eukaryotic phosphorylases. . Nature. 1986;324:80–84. doi: 10.1038/324080a0. [DOI] [PubMed] [Google Scholar]
- R16.Johnson L.N. Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J. . 1992;6:2274–2282. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- R17.Konig H., Skorko R., Zilling W., Reiter W.D. Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus and Thermococcus. Arch. Microbiol. 1982;132:297–303. [Google Scholar]
- R18.Lee H.-S., Shockley K.R., Schut G.J., et al. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 2006;188:2115–2125. doi: 10.1128/JB.188.6.2115-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Mizanur R.M., Zea C.J., Pohl N.L. Unusually broad substrate tolerance of a heat stable archaeal sugar nucleotidyltransferase for the synthesis of sugar nucleotides. J. Am. Chem. Soc. 2004;126:15993–15998. doi: 10.1021/ja046070d. [DOI] [PubMed] [Google Scholar]
- R20.Mizanur R.M., Jaipuri F.A., Pohl N.L. One-pot synthesis of labeled sugar nucleotides by archaeal sugar nucleotidyltransferases. J. Am. Chem. Soc. 2005;127:836–837. doi: 10.1021/ja044117p. [DOI] [PubMed] [Google Scholar]
- R21.Nidetzky B., Weinhausel A., Gryssler R., Kulbe K.D. Enzymatic synthesis of α-glucose-1-phosphate: a study employing a new α-1,4-glucan phosphorylase from Corynebacterium callunae. J. Carbohydr. Chem. 1995;14:1017–1028. [Google Scholar]
- R22.Ohdan K., Fujii K., Yanase M., Takaha T., Kuruki T. Phosphorylase coupling as a tool to convert cellobiose into amylose. J. Biotechnol. 2007;127:496–502. doi: 10.1016/j.jbiotec.2006.07.023. [DOI] [PubMed] [Google Scholar]
- R23.Palm D., Goerl R., Schaechtele K.H. Comparison of structural properties and primer requirements of Escherichia coli maltodextrin phosphorylase and rabbit muscle glycogen phosphorylase. Ann. Microbiol. 1982;133A:55–58. [PubMed] [Google Scholar]
- R24.Palm D., Goerl R., Burger K.J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985;313:500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- R25.Rogers P.V., Luo S., Sucic J.F., Rutherford C.L. Characterization and cloning of glycogen phosphorylase 1 from Dictyostelium discoideum . Biochim. Biophys. Acta. 1992;1129:262–272. doi: 10.1016/0167-4781(92)90502-q. [DOI] [PubMed] [Google Scholar]
- R26.Sambrook J., Fritsch E.F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual, 2nd edn.1659 [Google Scholar]
- R27.Schinzel R., Palm D. Escherichia coli maltodextrin phosphorylase: contribution of active site residues glutamate-637 and tyrosine-538 to the phosphorolytic cleavage of α-glucans. Biochemistry. 1990;29:9956–9962. doi: 10.1021/bi00494a028. [DOI] [PubMed] [Google Scholar]
- R28.Shin H.-J., Shin Y., Lee D.-S. Formation of α-D-glucose-1-phosphate by thermophilic α-1,4-D-glucan phosphorylase. J. Ind. Microbiol. Biotechnol. 2000;24:89–93. [Google Scholar]
- R29.Srivastava S., Nighojkar A., Kumar A. . Immobilization of Cuscuta reflexa starch phosphorylase: production of glucose-1-phosphate using biorectors. J. Ferment. Bioengineer. 1996;81:355–357. [Google Scholar]
- R30.Takaha T., Yanase M., Takata H., Okada S. Structure and properties of Thermus aquaticus α-glucan phosphorylae expressed in Escherichia coli. J. Appl. Glycosci. 2001;48:71–78. [Google Scholar]
- R31.Takata H., Takaha T., Okada S., Takagi M., Imanaka T. Purification and characterization of α-glucan phosphorylase from Bacillus stearothermophilus. J. Ferment. Bioengineer. 1998;85:156–161. [Google Scholar]
- R32.Weinhausel A., Nidetzky B., Kysela C., Kulbe K.D. Application of Escherichia coli maltodextrin phosphorylase for the continuous production of glucose-1-phosphate. Enzyme Microb. Technol. 1995;17:140–146. [Google Scholar]
- R33.Xavier K.B., Peist R., Kossmann M., Boos W., Santos H. Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J. Bacteriol. 1999;181:3358–3367. doi: 10.1128/jb.181.11.3358-3367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Yanase M., Takata H., Takaha T., Kuriki T., Smith S.M., Okada S. Cyclization reaction catalyzed by glycogen debranching enzyme (EC 2.4.1.25/EC 3.2.1.33) and its potential for cycloamylose production. Appl. Environ. Microbiol. 2002;68:4233–4239. doi: 10.1128/AEM.68.9.4233-4239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Yanase M., Takaha T., Kuruki T. α-Glucan phosphrylase and its use in carbohydrate engineering. J. Sci. Food Agric. 2006;86:1631–1635. [Google Scholar]
- R36.Yu J.P., Ladapo J., Whitman W.B. Pathway of glycogen metabolism in Methanococcus maripaludis . J. Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R37.Zea C.J., Pohl N.L. Kinetic and substrate binding analysis of phosphorylase b via electrospray ionization mass spectrometry: a model for chemical proteomics of sugar phosphorylases. Anal. Biochem. 2004;327:107–113. doi: 10.1016/j.ab.2003.12.022. [DOI] [PubMed] [Google Scholar]
- R38.Zea C.J., Pohl N.L. Unusual sugar nucleotide recognition elements of mesophilic vs. thermophilic glycogen synthases. Biopolymers. 2005;79:106–113. doi: 10.1002/bip.20338. [DOI] [PubMed] [Google Scholar]
- R39.Zea C.J., MacDonell S.W., Pohl N.L. Discovery of the archaeal chemical link between glycogen (starch) synthase families using a new mass spectrometry assay. J. Am. Chem. Soc. 2003;125:13666–13667. doi: 10.1021/ja037298o. [DOI] [PubMed] [Google Scholar]