Abstract
Partial hepatectomy (PHx) is a frequently used experimental model for the study of liver regeneration. Real-time quantitative PCR (qPCR) has become the one of the methods of choice for expression profiling of selected genes in order to elucidate the regulation of liver function and regeneration. The expression of five commonly used housekeeping genes (HKGs; Alb, UBC, Hprt, Ywhaz, and GAPDH) were evaluated by qPCR in 70% and 90% rat PHx model at 1, 2, and 7 d after PHx. We set up a closely controlled qPCR procedure validating each critical step and the gene expression stability was statistically evaluated by linear regression and analysis of variance. Our results showed the HKG best suited for the evaluation of gene expression in the extended 90% PHx model is Hprt. The amplification of an HKG can be omitted when the same amount of cDNA from all samples is introduced into the amplification reaction. Determination of cDNA concentration employing the bioanalyzer proved to be an easy and reproducible approach. Using this technique the potential regulation of the transcription level of the HKG in response to the experimental condition tested or the stability of a housekeeping gene becomes irrelevant.
Keywords: bioanalyzer, cDNA quantification, housekeeping gene, partial hepatectomy, real-time PCR
INTRODUCTION
Partial hepatectomy (PHx) is a frequently used experimental model for the study of liver regeneration. However, the molecular mechanism of resection associated liver damage and liver regeneration is not yet fully elucidated. Elucidating these questions often employs the determination of gene expression levels. Gene expression levels can be assessed using the quantitative polymerase chain reaction (qPCR) technique. This complex technique involves multiple experimental steps: tissue harvesting, handling and storage, RNA isolation and quantification plus cDNA synthesis and quantification. Only if all experimental steps are closely controlled, biological differences in the overall transcriptional activity of the tissues or cells can be detected reproducibly.
Expression levels of housekeeping genes (HKGs) are commonly used in the calculation of relative expression levels of target genes in different experimental samples employing qPCR.1 This approach is based on three assumptions: First, the HKG is unaffected by the experimental conditions; second, the expression level of the HKG and the target gene are in the same range2; and third, the efficiency of the PCR reaction for HKG and target gene is in similar range.
We hypothesized that the Ct value should reflect the amount of target-specific cDNA subjected to the PCR reaction when the same amount of total cDNA is used for the PCR reaction and PCR conditions are standardized. The aim of the present study is (1) to establish a standardized sample preparation and qPCR procedure, (2) to evaluate different HKG candidates with respect to their expression stability under different experimental conditions in the liver, and (3) to demonstrate that absolute normalization based on a quantified amount of cDNA entered into the PCR reaction is at least equal to relative normalization based on the use of HKGs.
MATERIALS AND METHODS
Rat Models of 70% and 90% PHx
Male inbred Lewis (LEW, RT1) rats (Charles River Wiga GmbH, Sulzfeld, Germany) with a body weight of 250–350 g (10–14 wk of age) were used in this study. Liver tissue from normal rats as well as from rats subjected to 70% and 90% PHx as previously described3 was used (n = 6/group). Animals were sacrificed 1, 2, and 7 d after operation (POD1, 2, 7) to obtain liver samples. Animals were housed under standard animal care conditions and fed with rat chow ad libitum before and after the operation. All procedures and housing of the animals were carried in accordance with German Animal Welfare Legislation.
Extraction of RNA
Total RNA from rat liver was prepared using the the RNeasy Mini kit and RNase-free DNase set (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations for animal tissue. Briefly, fresh or frozen (in liquid nitrogen) samples of rat liver (~25–30 mg) were homogenized in Buffer RLT through a 20-gauge needle with syringe and subsequently subjected to the QIAshredder (Qiagen). RNA of the lysed tissue was adsorbed to a silica matrix, DNase treated for 15 min, washed, and finally eluted with 30 μL RNase-free water by centrifugation. RNA samples were stored at −80°C.
Assessment of RNA Quantity and Quality
RNA quantity and purity were measured using the Nano-Drop ND-1000 UV-Vis Spectrophotometer (Nanodrop Technology, Wilmington, DE). In addition, RNA quantity and quality was monitored using Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) and RNA 6000 LabChip kit. For quality assessment the RNA integrity number (RIN) value by Agilent 2100 RIN Beta Version Software was used. All measurements were performed following the manufacturer’s instruction. Only high-quality RNA (RIN > 9; OD 260/280 and OD 260/230 > 2) was used for subsequent steps.4,5
Using the Nanodrop ND1000 spectrophotometer, we measured the same sample repeatedly (six times) with different sample volumes (1–5 μL) to determine the coefficient of variance (CV). Similarly, the same samples were also analyzed repeatedly (four samples in triplicates) using the Agilent System to assess the intrachip variability. In addition, interchip variability was determined. Furthermore, diluted samples were analyzed performing regression analysis of the respective curves.
cDNA Synthesis
The cDNA was amplified with the SuperScript III First-Strand Synthesis System for RT-PCR kit (Invitrogen GmbH, Karlsruhe, Germany) following the manufacturer’s protocol. Four to five micrograms of total RNA was mixed with oligo(dt) Primer and dNTP in a total volume of 10 μL. Samples were incubated at 65°C for 5 min. Then, RT Buffer, MgCl2, DTT, RNaseOUT and SuperScript III RT were added and incubated at 50°C for 50 min. The RT reaction was terminated by heating at 70°C for 15 min. One microliter of RNase H (2 U) was then added to the reaction mixture, and the mixture was incubated at 37°C for 20 min.
cDNA Quantification and Quality Control
One microliter of the first-strand reaction was used to measure the cDNA molecular distribution and quantity on Agilent 2100 bioanalyzer with RNA 6000 LabChip kit.
Reproducibility of this assay was assessed by subjecting triplicates of cDNA from four different reverse transcription reactions using the same RNA from the same animal to the assay. Linearity of the measurement was confirmed by quantifying a serial dilution of the sample in triplicates.
Primer and Probes
Primers were designed using Web-based software provided by Roche (https://www.roche-applied-science.com) and corresponding probes from the Universal ProbeLibrary were selected automatically (Table 1). Primers were synthesized by Microsynth (Balgach, Switzerland).
TABLE 1.
Characteristics of Primers and Probes of Selected Housekeeping Genes
| No. | Gene Symbol | Left Primer | Right Primer | Accession No. | Probea |
|---|---|---|---|---|---|
| 1 | Alb | aggatgacaaccccaacct | ggcaacttcatgcaaatagtgt | NM_134326.1 | R75 |
| 2 | Ubc | caggacaaggagggcatc | gccatcttccagctgctt | NM_017314 | R110 |
| 3 | Hprt | gaccggttctgtcatgtcg | acctggttcatcatcactaatcac | NM_012583 | R95 |
| 4 | Ywhaz | gctacttggctgaggttgct | tgctgtgactggtccacaat | NM_013011 | R9 |
| 5 | GAPDH | gcaagagagaggccctcag | tgtgagggagatgctcagtg | NM_017008 | R73 |
Universal ProbeLibrary probes.
Real-Time PCR
An Mx3000P QPCR System and Brilliant probe-based QPCR Master Mixes (Stratagene, La Jolla, CA) kit were used for qPCR following the manufacturer’s instruction. Amplifications were performed starting with a 10-min template denaturation step at 95°C, followed by 50 cycles of denaturation at 95°C for 30 s; combined primer annealing at 50°C for 30 s and extension at 72°C for 30 s. Fluorescence increase of FAM was automatically measured at the end of each PCR cycle. All samples were amplified at least in duplicate and the mean was obtained for further calculations. Replicates with differences of greater than 1 Ct value were not excluded from analysis. Ct values greater than 45 were excluded from further mathematical calculations.
Relative Amounts of Gene Expression Calculated by Standard Curve Method
A standard curve using a serial dilution of cDNA from normal rat liver was generated in each plate in order to calculate the fold changes of HKG expression in PHx samples compared with normal rat liver. Pooled total cDNA liver samples of 5 normal rats were serially diluted (1:1, 1:10, 1:100, 1:1000) and duplicated in each plate and then amplified simultaneously under the same condition together with the cDNA of PHx samples. One normal liver sample was included as internal control. The standard curve was generated using linear regression. Both coefficient of determination (r2) and PCR efficiency were calculated using the Stratagene program.
Interindividual Variation of HKG Expression in Normal Rats
In order to determine the variation of HKG expression in normal rats, samples from 5 normal Lewis rats were randomly chosen and analyzed within the same plate.
Statistical Analysis
We used one-way ANOVA (SigmaStat version 3.11), linear regression, and Pearson correlation (SigmaPlot version 9.0) analysis of Ct values to examine variation between samples in qPCR experiments.
Determination of Expression Ratios
Quantification of gene expression was achieved employing the standard curve method. A reference gene was used for the comparative quantification(2−ΔΔCt).6 The ΔCt method is based on the assumption of perfect PCR efficiency of E = 1, which is rarely reached, even under optimized conditions.7 Thus the ΔCt method according to Pfaffl was employed.8
This mathematical model was established to standardize each reaction run with respect to RNA integrity, real-time RT-PCR primers, cDNA input, qPCR efficiency, and inter-PCR variations. It proved to be highly accurate and reproducible. Because of the similar inherent principle of this formula to our closely controlled procedure, it is applicable to our experimental condition for the purpose of comparison.
RESULTS
RNA Quantification
Quantification of total RNA was performed using the Nanodrop and the Agilent 2100 bioanalyzer. Using the Nanodrop, statistical analysis revealed the highest level of reproducibility using a sample volume of 2 μL (CV = 0.88%). Using the Agilent 2100 bioanalyzer and the corresponding RNA chip for quantitation, the CV was 0.87%–15.6%, a result inferior to the results obtained using the Nanodrop, albeit within specifications of the device.
Quantification of Reversely Transcribed cDNA
Quantification was performed employing the Agilent 2100 Bioanalyzer in combination with an RNA chip. A serial dilution (1, 2, 3, 4 mg) of the total RNA samples (the range of RT reagent kit was 5 mg) was retrotranscribed with oligo(dT) primer. The linear regression analysis showed high correlation between dilutions and their end products of cDNA (r2 = 0.987). When 4 mg total RNA from same sample was used for cDNA synthesis and measured in one chip and repeated in three chips, the concentration of cDNA product was 140.78 ± 12.50 (mean ± SD, n = 18), the intrachip variance (CV) was 5.27%, 7.40%, 9.80% in three chips, and the interchip CV was 2.11%–14.40%, still within the specification of the device.
Assessing Quality of cDNA
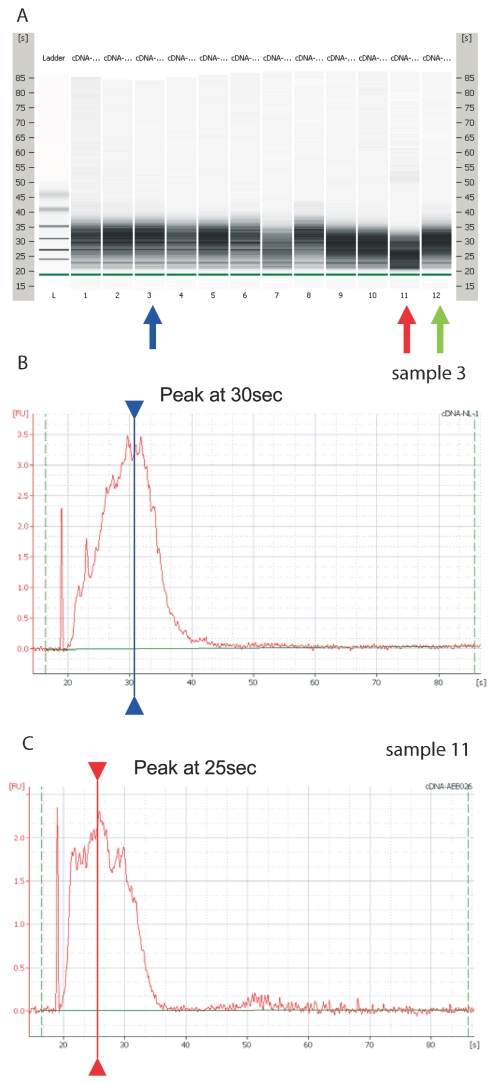
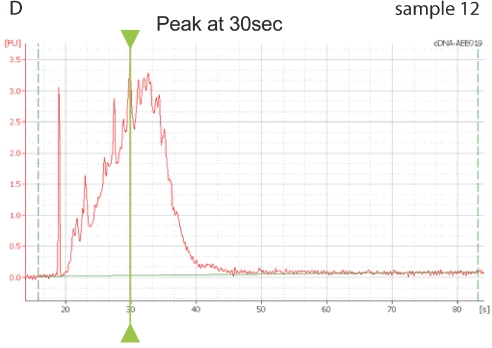
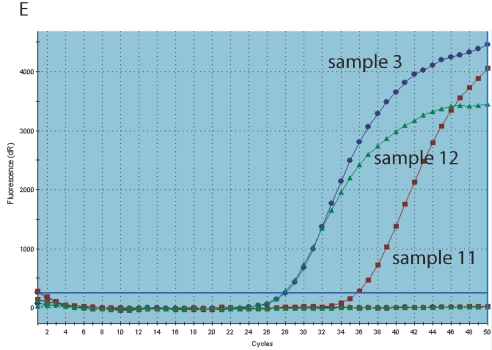
Using the Agilent bioanalyzer, the cDNA electropherogram was in general a bell-shaped curve with a peak occurring at 30 s or slightly thereafter. In single samples the peak occurred much earlier, around 25 s (Fig. 1A and B) and could even appear distorted. When using these cDNA samples (cDNA sample 11 versus 3 and 12, Fig. 1C), Ct values were much higher compared with cDNA samples synthesized starting from the same RNA sample. Using cDNA synthesized in independent runs (cDNA samples 3 and 12, Fig. 1), but fulfilling these criteria, the Ct values obtained in the same PCR run were very close.
FIGURE 1.
Amplification plots. Assessing quality of cDNA with bioanalyzer. A: Gel demonstrating the different electropherograms obtained with three cDNAs (lanes 3, 11, and 12) synthesized in three runs from the same tissue sample and analyzed in the same RNA nano6000 chip with mRNA nano assay. B: Electropherogram of sample 3 (blue arrow). C: Electropherogram of sample 11 (red arrow). D: Electropherogram of sample 12 (green arrow). Note time of peak in different cDNA samples E: Amplification plots of Hprt qPCR with cDNA sample 3 (blue), 11 (red), and 12 (green).
Interindividual Variation of HKG Expression Assessed in Normal Rats
The same amount of cDNA from 5 normal rats was subjected to qPCR in the same plate in quadruplicate. The CV of each sample within the respective quadruplicate ranged from 0.35% to 2.85%; the CV of each rat within one run was 1.15%–2.78%; and the CV within the group of 5 rats was 1.45%, all showing the high reproducibility of the assay.
HKG Expression in Classical 70% and Extended 90% Hepatectomy Model after 24 Hours
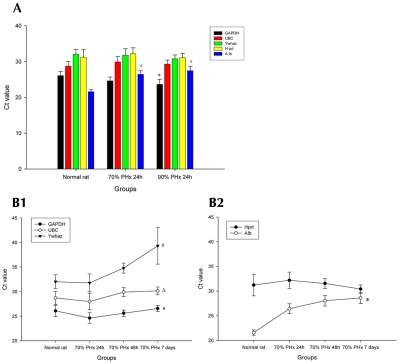
Alb, UBC, Hprt, Ywhaz, and GAPDH mRNA expression levels were employed in the qPCR in rat liver samples obtained 24 h after 70% and 90% PHx (Fig. 2A). A significant upregulation (lower Ct value, 3.19-fold) of mRNA level of GAPDH was found in the 90% PHx model (P = 0.004) but not in the 70% PHx model as compared with the normal group. In contrast, downregulation of mRNA level of Alb was observed after both 70% (13.13-fold) and 90% PHx (22.44-fold) as compared with the normal group (P = 0.002). However, mRNA expression of UBC, Hprt, and Ywhaz was not regulated in any direction at 24 h after 70% or 90% PHx.
FIGURE 2.
Evaluation of HKG in rat PHx model. A: GAPDH, UBC, Ywhaz, Hprt, and Alb mRNA expression level were measured by real-time RT-PCR in liver samples obtained from rats 24 h after 70% and 90% PHx. A significant upregulation (lower Ct value, 3.19-fold) of mRNA level of GAPDH was found in the 90% PHx model (*P = 0.004) but not in the 70% PHx model as compared with the normal group. A downregulation (higher Ct value) of the mRNA level of Alb was found in both 70% (13.13-fold) and 90% PHx (22.44-fold) as compared with the normal group (#P = 0.002). B1, B2: The mRNA levels of five HKGs were measured at different time points (24 h, 48 h, and 7 d) after 70% PHx. B1: mRNA levels of three HKGs increased at 24 h after 70% PHx, and decreased continuously thereafter (48 h; 7 d): GAPDH 2.52-fold, *P = 0.008; UBC 2.85-fold, P = 0.019; and Ywhaz 17.42-fold, #P < 0.001. B2: mRNA level of Alb decreased significantly at POD1 (13.13-fold) and slightly more thereafter: 0.88-fold at 48 h and 41.60-fold at POD7, *P < 0.001. Hprt expression was fluctuant but without statistical significance (P = 0.270).
HKG Expression in 70% Hepatectomy Model at Different Observation Time Points
In a separate experiment we analyzed mRNA levels of the five HKGs at three different time points throughout the first postoperative week after 70% PHx. As in the previous experiment, GAPDH was slightly, but not significantly, upregulated at POD1, but significantly down-regulated by POD7 (2.52-fold, P = 0.007).
Similar results were observed for the initially unregulated genes UBC and Ywhaz when prolonging the observation time to POD7 (UBC, 2.85-fold, P = 0.019 and Ywhaz, 17.42-fold, P < 0.001, see Fig. 2, B1). In contrast, the mRNA level of Alb decreased significantly at POD1 (13.13-fold) and slightly more thereafter (30.88-fold at 48 h and 41.60-fold at POD7, P < 0.001). Hprt mRNA expression levels fluctuated but did not show any significant changes throughout the observation period (P = 0.270) (Fig. 2, B2).
Comparison Between Normalization Based on the Closely Controlled Sample Preparation Procedure and the HKG
As shown in Table 2, the ΔΔCt method can overestimate the expression ratio up to 15-fold if the real efficiency of the PCR run is neglected. The results were statistically different from what we obtained in our closely controlled procedure (P = 0.022). When adjusting the calculation according to the efficiency, the expression ratios obtained in either method were statistically similar (P = 0.759).
TABLE 2.
Comparison of Methods Determining Gene Expression Ratio Between Normalized Standard Curve Method and ΔΔCt Method
| Gene | Group | Regulation Trend |
Expression Ratio |
||
|---|---|---|---|---|---|
| Standard Curve |
ΔΔCt Method |
||||
| Corrected with E* | 2−Δ ΔCt | ||||
| Normal Rat: 70% PHx 24 h and 90% PHx 24 h | |||||
| GAPDH | 90% PHx 24 h vs. Normal | Upregulated | 3.19 | 3.12 | 5.35 |
| Alb | 70% PHx 24 h vs. Normal | Downregulated | 13.13 | 13.38 | 29.62 |
| 90% PHx 24 h vs. Normal | Downregulated | 22.44 | 22.72 | 59.14 | |
| Normal Rat: 70% PHx 24 h, 48 h and 7 d | |||||
| GAPDH | 7 d vs. 24 h | Downregulated | 2.52 | 2.69 | 4.23 |
| UBC | 7 d vs. 24 h | Downregulated | 2.85 | 2.97 | 4.96 |
| Ywhaz | 7 d vs. Normal | Downregulated | 17.42 | 18.01 | 270.60 |
| Alb | 7 d vs. Normal | Downregulated | 41.60 | 39.22 | 224.41 |
| 48 h vs. Normal | Downregulated | 30.88 | 32.86 | 172.45 | |
| 24 h vs. Normal | Downregulated | 13.13 | 13.25 | 45.25 | |
| P Value | 0.759 | 0.022 | |||
Corrected with qPCR efficiency (PFAFFL Method8).
DISCUSSION
The qPCR technique is widely used in biomedical research.9,10 Quantification is often performed employing the ΔΔCt method.1 This approach requires the use of at least one HKG to normalize for the amount of cDNA used in different samples.11,12
RNA levels of HKGs in tissues should be stable across the different experimental conditions. However, the level of expression of HKGs is often subject to local or systematic modulation depending on the biological function they are involved in.9 In the PHx model, differentially expressed HKGs reflect the shift in the transcriptional program from genes involved in biosynthesis (e.g., albumin) in the quiescent liver to those contributing to DNA and structural protein synthesis (e.g., Ywhaz, UBC) in the proliferating liver.
The downregulation of Alb after PHx in both the classical and the extended resection model is consistent with the report from Tygstrup et al.13 in which slot-blot hybridization was used to evaluate mRNA level. This decrease of Alb expression undoubtedly correlates with the deprivation of hepatocytes after resection. GAPDH was only upregulated in the 90% PHx model but not after 70% PHx, suggesting that the fluctuation of GAPDH level might be related to the lost liver mass and the subsequently reduced liver function. The downregulation of UBC in PHx might reflect attenuation of UBC-dependent protein degradation,14 which in turn may promote the proliferation of remnant hepatocytes. In our hands, Hprt was the only relatively stable HKG in the PHx model. This observation is supported by reported results that were also obtained using strictly normalized methods.15–17 In the case where a closely controlled procedure cannot be thoroughly implemented because of a lack of equipment or inferior sample quality, Hprt can be regarded as the candidate of choice as the reference gene for comparative quantitative RT-PCR when using models of extended hepatectomy in the rat.
Normalizing the cDNA based on an HKG in real time PCR can be omitted, when the cDNA is quantified otherwise and an equal amount of cDNA is entered into the amplification reaction. Quantification using the Agilent lab-on-a-chip assay turned out to be an easy and reproducible approach. Employing this technique the potential regulation of the transcription level or the stability of an HKG is not going to affect the qPCR results. Furthermore, using this approach, the potentially time-comsuming step of identifying a truly not regulated HKG can be omitted when establishing the qPCR. In addition this new technique allows for a quality assessment of the cDNA.
cDNA samples can be assessed quantitatively and qualitatively employing the bioanalyzer. Only cDNA leading to a bell-shaped electropherogram with the peak occurring at 30 sec or slightly thereafter should be used for the qPCR. Samples not fulfilling these quality criteria should not be used in the qPCR reaction, but the cDNA synthesis step should be repeated. The use of samples not fulfilling the quality criteria might lead to falsely high Ct values for the given tissue sample and thereby an underestimated level of gene expression.
In summary, employing the Ct method for the quantitation of qPCR results requires the use of at least one HKG to normalize for the amount of cDNA of different samples. The HKG best suited for the evaluation of gene expression in the extended 90% PHx model is Hprt. The amplification of a HKG can be omitted, when the same amount of cDNA from all samples is introduced into the amplification reaction. Determination of cDNA concentration employing the Bioanalyzer proved to be an easy and reproducible approach. Using this technique the potential regulation of the transcription level of the HKG in response to the experimental condition tested or the stability of an HKG becomes irrelevant.
Acknowledgments
Research supported by DFG-Grant KFO117, BII and BIII.
REFERENCES
- 1.Radonic A, Thulke S, Mackay IM, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 2.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 3.Madrahimov N, Dirsch O, Broelsch C, Dahmen U. Marginal hepatectomy in the rat: From anatomy to surgery. Ann Surg. 2006;244:89–98. doi: 10.1097/01.sla.0000218093.12408.0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 5.Imbeaud S, Graudens E, Boulanger V, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s CT difference” formula. J Mol Med. 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- 7.Yuan JS, Wang D, Stewart CN., Jr Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol J. 2008;3:112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]
- 8.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 10.Ginzinger DG. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp Hematol. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 11.Tricarico C, Pinzani P, Bianchi S, et al. Quantitative real-time reverse transcription polymerase chain reaction: Normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 12.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tygstrup N, Jensen SA, Krog B, Pietrangelo A, Shafritz DA. Expression of messenger RNA for liver functions following 70% and 90% hepatectomy. J Hepatol. 1996;25:72–78. doi: 10.1016/s0168-8278(96)80330-8. [DOI] [PubMed] [Google Scholar]
- 14.Kim DJ, Akiyama TE, Harman FS, et al. Peroxisome proliferator-activated receptor beta (delta)-dependent regulation of ubiquitin C expression contributes to attenuation of skin car-cinogenesis. J Biol Chem. 2004;279:23719–23727. doi: 10.1074/jbc.M312063200. [DOI] [PubMed] [Google Scholar]
- 15.de Kok JB, Roelofs RW, Giesendorf BA, et al. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- 16.Fischer M, Skowron M, Berthold F. Reliable transcript quantification by real-time reverse transcriptase-polymerase chain reaction in primary neuroblastoma using normalization to averaged expression levels of the control genes HPRT1 and SDHA. J Mol Diagn. 2005;7:89–96. doi: 10.1016/S1525-1578(10)60013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meller M, Vadachkoria S, Luthy DA, Williams MA. Evaluation of housekeeping genes in placental comparative expression studies. Placenta. 2005;26:601–607. doi: 10.1016/j.placenta.2004.09.009. [DOI] [PubMed] [Google Scholar]