Abstract
Very-long-chain fatty acids (VLCFAs) are important functional components of various lipid classes, including cuticular lipids in the higher plant epidermis and lipid-derived second messengers. Here, we report the characterization of transgenic Arabidopsis thaliana plants that epidermally express FATTY ACID ELONGATION1 (FAE1), the seed-specific β-ketoacyl-CoA synthase (KCS) catalyzing the first rate-limiting step in VLCFA biosynthesis. Misexpression of FAE1 changes the VLCFAs in different classes of lipids but surprisingly does not complement the KCS fiddlehead mutant. FAE1 misexpression plants are similar to the wild type but display an essentially glabrous phenotype, owing to the selective death of trichome cells. This cell death is accompanied by membrane damage, generation of reactive oxygen species, and callose deposition. We found that nuclei of arrested trichome cells in FAE1 misexpression plants cell-autonomously accumulate high levels of DNA damage, including double-strand breaks characteristic of lipoapoptosis. A chemical genetic screen revealed that inhibitors of KCS and phospholipase A2 (PLA2), but not inhibitors of de novo ceramide biosynthesis, rescue trichome cells from death. These results support the functional role of acyl chain length of fatty acids and PLA2 as determinants for programmed cell death, likely involving the exchange of VLCFAs between phospholipids and the acyl-CoA pool.
INTRODUCTION
In plants, very-long-chain fatty acids (VLCFAs; fatty acids with chain lengths >18 carbons) form important structural components of membranes and epidermal surfaces. VLCFAs are essential for the normal growth and development of Arabidopsis thaliana (Bach et al., 2008). They also appear to provide lipid signals involved in mediating rapid, localized death of plant cells at the site of pathogen invasion, a process known as hypersensitive response (HR), contributing to resistance (Raffaele et al., 2008; Wang et al., 2008).
VLCFAs are derived from shorter FAs by a sequential elongation process that occurs on the cytosolic face of microsomal membranes. FAs are initially activated by esterification with CoA, catalyzed by acyl-CoA synthase. The first step in fatty acid elongation is catalyzed by a β-ketoacyl-CoA synthase (KCS), condensing acyl-CoA and malonyl-CoA. It has also been proposed that FAs esterified to a glycerolipid or a phospholipid rather than CoA may also serve as substrates for elongation reactions (Hlousek-Radojcic et al., 1998). In yeast and animal species, the enzymes with KCS activity are encoded by the ELO family of genes. By contrast, in addition to encoding a small number of ELO genes, the genomes of higher plants contain a surprisingly large family of FATTY ACID ELONGATION (FAE) class KCSs (structurally unrelated to the ELOs). Based on sequence similarity to the seed-specific product of the FAE1 gene (Kunst et al., 1992; James et al., 1995; David et al., 1998), this family comprises 21 members in Arabidopsis (which, by contrast, has only four ELO genes), suggesting that the FAE genes take part in a number of plant-specific pathways. The KCS-catalyzed condensation is the rate-limiting step in microsomal fatty acid elongation, and current data suggest that each KCS catalyzes one or two individual condensing reactions in the progressive elongation of fatty acids and determines the VLCFAs produced. By contrast, the other three core enzyme activities, which are required for the elongation, play no direct role in the control of VLCFA synthesis (Millar and Kunst, 1997; Paul et al., 2006). Therefore, manipulation of KCSs via the use of mutants or overexpressor lines provides a means for deciphering the functional roles of VLCFAs in cellular responses and developmental processes.
Among different Arabidopsis KCSs, FAE1 is the best-characterized example (Ghanevati and Jaworski, 2002). It directs two rounds of elongation of C18 (and probably C16) FAs to produce the C20 and C22 species that constitute 13.0 to 21.2% of total fatty acids in the triglycerides of Arabidopsis seed oil (O'Neill et al., 2003). These C20 and C22 species are absent in the seeds of fae1 mutants, which contain only C16 and C18 FAs in their oil (James et al., 1995). Although VLCFAs occur in sphingolipids in the plasma membrane, trans-Golgi network, and in recycling endosomes, C20 and longer fatty acids account for <1% of the total fatty acids in the leaves of wild-type Arabidopsis plants (Millar and Kunst, 1997), and most of them accumulate in the epidermis, as components of cuticular waxes and polyesters. Characterization of CaMV35S:FAE1 transgenic Arabidopsis plants (CaMV35S is a cauliflower mosaic virus 35S promoter sequence) revealed, however, that the plants are capable of accumulating high levels (>30%) of VLCFAs in leaf membrane lipids. The transgenic plants with relatively low levels of VLCFAs (less than ∼8.5% [w/w] of total fatty acids in 6-week-old plants) appeared wild-type but the transgenic plants with high levels of VLCFA (from ∼9.0 to ∼13.5%) exhibited a wide range of morphological changes and some failed to survive (Millar et al., 1998). To investigate the molecular mechanism by which VLCFA exert their effects, it can be helpful to use tissue-specific promoters to target the expression of a KCS, such as FAE1, accurately to appropriate cell types. The epidermis not only offers a model to study cell-type differentiation and provides the major physical barrier to invading pathogens and water permeation but also mediates a broad set of defense responses. The epidermis-specific FIDDLEHEAD (FDH) gene encodes a putative KCS (Yephremov et al., 1999; Pruitt et al., 2000), and its promoter shows strong expression in vegetative and floral meristems (Yephremov et al., 1999; Efremova et al., 2004). The fdh mutation has a deleterious effect on cuticle quality, plant morphology, and aspects of trichome differentiation. We reasoned that, if VLCFA biosynthesis affects these aspects of plant development, the FDH promoter is very suitable for driving expression of a well-characterized KCS.
In this article, we report the misexpression phenotype of FDH:FAE1 transgenic Arabidopsis plants. Most surprisingly, while the misexpression does not affect the overall morphology of the plants and cannot complement the fdh phenotype, it results in a glabrous phenotype, which is due to the specific execution of developing trichome cells. This phenotype contrasts sharply with the phenotypes of other glabrous mutants that are due to defects in trichome initiation, morphogenesis, or both. Phospholipase A2 (PLA2) inhibitors and chloracetamide herbicides that inhibit KCSs were found in pharmacological screening to rescue the development of trichome cells in the FDH:FAE1 plants. Cytochemical studies also provide support for the view that the death of trichomes in the transgenic plants may result from a process akin to lipoapoptosis in animal cells (Listenberger and Schaffer, 2002).
RESULTS
The FDH:FAE1 Transgenic Plants Show a Glabrous Phenotype
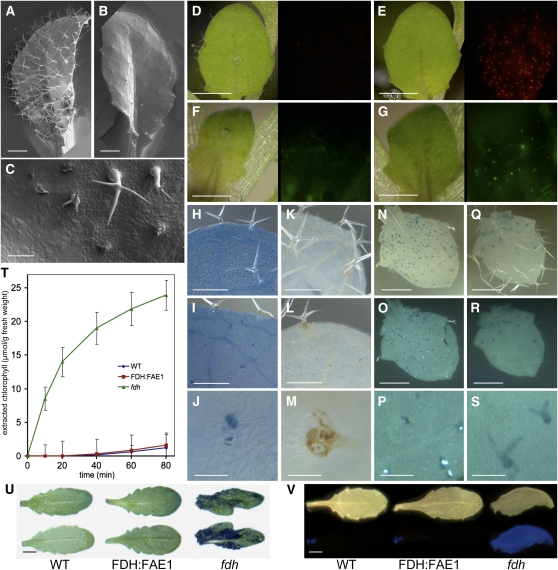
To investigate the effects of VLCFA biosynthesis in the epidermis, Arabidopsis (Columbia ecotype) plants were transformed with a construct expressing the FAE1 cDNA from the epidermis-specific FDH promoter (Efremova et al., 2004). All 27 independent FDH:FAE1 T1 transgenic plants were clearly distinct from the wild type in that they showed a glabrous phenotype in rosette and cauline leaves, stems, and sepals (Figures 1A to 1F). Trichome initials clearly formed in young developing leaves (∼0.5 cm), but no evidence of the presence of trichomes was observed in most areas of the surface of mature leaves. Infrequently (<1%), and mainly at leaf edges, rudimentary or mature trichomes were able to develop. In contrast with trichomes, the pavement cells in the epidermis, trichome subsidiary cells, and stomata were not noticeably affected. The glabrous phenotype of T1 plants segregated as a semidominant Mendelian trait in T2 and T3 generations, showing that one copy of the transgene was sufficient to confer the phenotype.
Figure 1.
Glabrous Phenotype of FDH:FAE1 Transgenic Plants.
(A) to (F) Comparison of 3.5-week-old wild-type (A) and FDH:FAE1 (B) rosettes (grown under short-day conditions), wild-type (C) and FDH:FAE1 (D) inflorescences, and stems from 5-week-old wild-type (E) and FDH:FAE1 (F) plants carrying cauline leaves and axillary shoots.
(G) and (H) Expression of the GL2:GUS reporter in developing leaves of 2-week-old wild-type (G) and FDH:FAE1 (H) seedlings. Note that most trichomes remained undeveloped and were not histochemically stained for GUS in the double transgenics (arrows). Bars = 500 μm.
(I) and (J) Chemical rescue of trichome development with 0.005% alachlor. (I) and (J) show the same plant before and 1 week after exposure to alachlor, a potent inhibitor of KCSs. The inhibitor was applied to 3-week-old FDH:FAE1 seedlings by spraying, to inactivate FAE1 (I). Leaves were numbered from the base to the apex of the plant. Young leaves, which developed 1 week after spraying, exhibit trichomes (J).
With respect to plant architecture, organ morphology, flowering time, and other visible phenotypes, including those affected in the most extreme examples of CaMV35S:FAE1 transgenic plants described by Millar et al. (1998), all FDH:FAE1 transgenic plants appeared to be normal in the T1 and subsequent generations.
Closer examination of the FDH:FAE1 plants showed that trichome differentiation was often arrested at the initiation stage or soon afterwards. Several transcription factors are known to play essential roles during trichome differentiation. GL1, GL3, and TTG constitute the primary transcription protein complex and regulate GLABRA2 (GL2), which is also involved in trichome initiation. GL2 is required for trichome morphogenesis, and its expression marks developing trichomes throughout development (Szymanski et al., 1998; Ohashi et al., 2002). Histochemical staining for β-glucuronidase (GUS) activity in double transgenic plants expressing both FDH:FAE1 and GL2:GUS reporter fusions showed that, in most trichomes, which were arrested at very early stages of development in these plants, GUS expression was not maintained. Only a few trichomes that had reached the stage at which they formed branches (three, as in the wild-type plants, or fewer) were stained (Figures 1G and 1H). GL2 is a homeodomain transcription factor, but it has a so-called StAR-related lipid transfer (START) domain, raising the possibility that the activity or cell localization of GL2 may be regulated by a lipid ligand. Therefore, we also used the native GL2 promoter to drive expression of a GL2-green fluorescent protein (GFP) fusion in transgenic GL2-GFP plants and double transgenic FDH:FAE1 GL2-GFP plants. However, the GFP fluorescent signal, when present, was localized to nuclei (see Supplemental Figure 1 and Supplemental Movie 1 online), and no evidence for mislocalization of the GL2-GFP protein was observed in the FDH:FAE1 plants. These results suggest that either FDH:FAE1 acts on trichome differentiation upstream of GL2 or that newly initiated trichomes undergo rapid cell death.
Enzymic Activity of FAE1 Is Required to Suppress Trichome Development
Ectopic expression of the FAE1gene in the epidermis could affect fatty acid elongation and cell differentiation in two alternative ways. The enzymic activity of FAE1, by elevating levels of longer fatty acids (and accordingly depleting levels of shorter fatty acids) in the epidermis, might account for the suppression of cell differentiation. Alternatively, FAE1 transcripts might reduce the efficiency of FA elongation by inducing cosuppression of homologous elongase genes expressed in the epidermis or at the protein level by competing for interacting proteins (i.e., other components of the elongase), cofactors, and substrates.
To study whether the fatty acid elongation activity of FAE1 is responsible for the suppression of trichome differentiation, we sprayed FDH:FAE1 transgenic plants with sublethal dosages of herbicidal chloroacetamides and oxyacetamides (alachlor and flufenacet, respectively) that are known to inhibit KCSs through the covalent modification of the active site (Boger et al., 2000; Trenkamp et al., 2004). Rescue of trichome development became apparent within several days after multiple applications of these chemicals (Figures 1I and 1J). These results confirm that enzymatically active FAE1 is required to suppress trichome development in the transgenic plants. Some rescued trichomes were arrested at intermediate stages, but most developed three lateral branches and ultimately reached the normal cell size. However, they typically showed a flattened appearance, suggesting that the trichome morphogenesis program constitutively depends on the elongation of fatty acids in FDH:FAE1 plants.
FAE1 Misexpression Differentially Affects Various Fatty Acid Classes in Acyl-CoAs, Membranes, and Cuticular Lipids
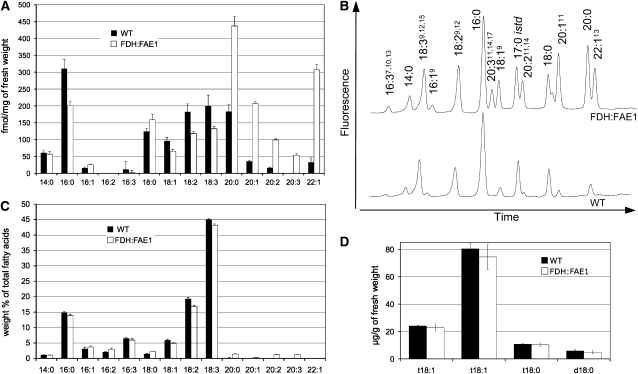
To further prove functional expression of FDH:FAE1, we analyzed acyl-CoA esters, the key substrates for elongation by the microsomal system and central metabolic intermediates of lipid metabolism. The fatty acyl-CoA esters were extracted from leaf samples, derivatized to fluorescent acyl-etheno-CoAs (Larson and Graham, 2001), and fractionated by HPLC. As expected, in FDH:FAE1 leaves, the longer fatty acids were overrepresented in the acyl-CoA pool (Figures 2A and 2C). In particular, the level of C20 acyl-CoA esters is significantly increased in FDH:FAE1 plants: to 239% of the wild-type level for C20:0, 580% for C20:1, and 596% for C20:2 (Figure 2A). Leaves of the FDH:FAE1 plants contained approximately ninefold higher levels of C22:1 acyl-CoA esters than leaves of the nontransformed control plants. The transgenic plants were able to synthesize C20:3, which were not found in wild-type leaves and accumulated fewer C18:1, C18:2, and C18:3 acyl-CoA esters (68, 65, and 66% of the wild-type levels, respectively) (Figure 2A). Acyl-CoA esters longer than C22 were not present in detectable levels in the FDH:FAE1 or wild-type plants, though this may reflect the poor resolution of VLCFA-CoAs with >22 carbons under the chromatographic conditions used. This experiment was repeated twice using plants from different cultivation setups, and similar results were obtained. These results prove that FAE1 is functional in the epidermis and are consistent with the previously proposed function of FAE1 (James et al., 1995; Millar and Kunst, 1997; Blacklock and Jaworski, 2006).
Figure 2.
Analysis of the Fatty Acid Composition of the Acyl-CoA and Total Lipid Pools and of LCBs in FDH:FAE1 and Wild-Type Plants.
(A) HPLC analysis of VLCFAs in acyl-CoA esters (mean ± se; n = 5). Acyl-CoAs were extracted from young rosette leaves of 6-week-old plants (0.5 to 1.0 cm), derivatized to fluorescent acyl-etheno-CoAs, and analyzed by HPLC.
(B) Representative HPLC profiles of acyl-CoAs in FDH:FAE1 and wild-type plants obtained as summarized in (A).
(C) Gas chromatography (GC) analysis of total fatty acids isolated from the same tissues as in (A) (mean ± se; n = 5). Fatty acids were transesterified and analyzed as methyl esters (FAMEs).
(D) LCBs were extracted and analyzed as dinitrophenyl derivatives by reverse-phase HPLC/MS (mean ± se; n = 4). In the figure, “d” denotes dihydroxy LCBs, and “t” denotes trihydroxy LCBs; the numbers designate the length and degree of desaturation of the acyl chain, respectively; t18:1(Z), 4-hydroxy-8-(cis)-sphingenine; t18:1(E), 4-hydroxy-8-(trans)-sphingenine; t18:0, 4-hydroxysphinganine; d18:0, dihydrosphinganine.
In agreement with the results of our acyl-CoA analysis, atypical C20 fatty acids also appeared in total leaf lipids extracted from the same batches of the FDH:FAE1 plants (Figure 2B), albeit in lower relative amounts (∼7- to 10-fold lower than that in the acyl-CoA fraction), suggesting that incorporation of atypically long FA chains into membrane lipids in the epidermis is a controlled process. Given that the epidermis constitutes a minor proportion of the total volume of the leaf, the expression of FDH:FAE1 is likely to cause a more pronounced increase in VLCFA levels in the epidermis while depleting shorter-chain fatty acids.
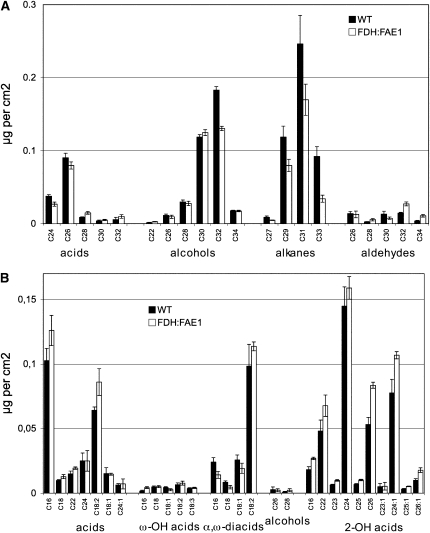
Elongation of fatty acids up to C34 is required for biosynthesis of the epi- and intracuticular wax in Arabidopsis. Therefore, based on the results of acyl-CoA profiling, one would anticipate that the chain lengths of fatty acids in wax, or the amount of wax synthesized, should be increased in the FDH:FAE1 plants if the level of KCS expression plays a significant role in controlling wax deposition (as it has been shown for CER6, another epidermis specific KCS; Hooker et al., 2002). However, contrary to this expectation, biochemical analysis of soluble lipids from epidermis revealed that the FDH:FAE1 plants had significantly (P < 0.05) reduced amounts of total epicuticular wax (∼80% of that in the wild type; Figure 3A). Among the functional classes of compounds in wax, the alkane fraction dropped to ∼57% of that in the wild type (Figure 3A). These wax analysis data are in good agreement with earlier results, as expression of FAE1 under the control of the constitutive CaMV35S promoter reduced wax load by up to 50% (Millar and Kunst, 1997; Millar et al., 1998). In many eceriferum mutants of Arabidopsis, a severe reduction of wax load correlates with a glossy green appearance and reduced fertility (Hannoufa et al., 1993; Hülskamp et al., 1995; Jenks et al., 1995), but the FDH:FAE1 plants displayed neither of these phenotypes.
Figure 3.
Composition Analysis of Epicuticular Waxes and Residual-Bound Lipids in Leaves.
(A) Wax was extracted by rapid dipping in chloroform and BSTFA derivatized and analyzed by GC and GC-MS (mean ± se; n = 5). Leaves of 6-week-old plants were harvested for analysis in (A) and (B).
(B) For cell wall–bound lipid analysis, leaves were first extensively defatted with chloroform-methanol. Bound lipids were analyzed using GC and GC-MS as described previously (Franke et al., 2005; Kurdyukov et al., 2006a) (mean ± se; n = 5).
The cuticular polyester cutin is the other major constituent responsible for the protective functions of the epidermis. It is difficult to purify cutin from Arabidopsis leaves and stems, but it has been demonstrated that the lipids left in leaves after extended exposure to organic solvents reflect the distribution seen in pure cutin (Bonaventure et al., 2004; Franke et al., 2005). To study whether the expression of FAE1 might play a role in the biosynthesis of cutin, we analyzed the residual lipids that remain bound after exhaustive extraction with chloroform/methanol in the FDH:FAE1 plants.
Misexpression of FAE1 elicits essentially no change in the typical cutin constituents, ω-hydroxylated fatty acids and α,ω-dicarboxy fatty acids, which are only 16 to 18 carbons in length. Compared with the wild type, no atypical fatty acids appeared in the fraction of residual bound lipids in FDH:FAE1 plants (Figure 3B). Supporting these results, a toluidine blue (TB) staining test did not reveal any overall defects in the cuticle of the FDH:FAE1 transgenic plants compared with wild-type plants (Figure 4U). Furthermore, the calcofluor white assay (Bessire et al., 2007) also implied that cuticle permeability was unchanged in FDH:FAE1 leaves (Figure 4V). A third method used to distinguish cuticular mutants, based on measuring the chlorophyll extraction rate (Lolle et al., 1997), also failed to uncover any significant changes in the FDH:FAE1 plants (Figure 4T), indicating that their cuticle is generally normal.
Figure 4.
Characterization of the Trichome Cell Death Phenotype in FDH:FAE1 Plants.
(A) and (B) Scanning electron micrographs showing the adaxial epidermis of rosette leaves. Note the fully developed trichomes on the wild-type leaf (A) and the arrested trichomes characteristic of FDH:FAE1 plants (B). Bars = 500 μm.
(C) Close-up view of FDH:FAE1 trichomes, including some that arrested or underwent collapse and degeneration at various developmental stages. Bar = 100 μm.
(D) and (E) Micrographs of young leaves (left panels) stained with PI and viewed using the DsRed filter set (right panels). Note that PI labels trichomes in the FDH:FAE1 leaf (E) but not the wild type (D). Bars = 500 μm.
(F) and (G) Micrographs of young leaves (left panels) stained with DCFH-DA and viewed using the GFP filter set (right panels). The DCFH-DA fluorescence reveals the presence of ROS in FDH:FAE1 trichomes (G) but not in wild-type trichomes (F). Bars = 500 μm.
(H) to (J) Trypan blue staining detects cell death in differentiating TEs in wild-type (H) and FDH:FAE1 (I) leaves and trichome death in FDH:FAE1 ([I] and [J]). Note that some mature trichomes in (I) are viable and are not stained. (J) shows detail of a FDH-FAE1 trichome. Bars = 250 μm in (H) and (I) and 50 μm in (J).
(K) to (M) Wild-type (K) and FDH:FAE1 ([L] and [M]) leaves stained with DAB to detect hydrogen peroxide. A brown reaction product indicates the presence of H2O2 in trichomes in FDH:FAE1 and in the vasculature of both wild-type and FDH:FAE1 leaves. (M) shows detail of a FDH-FAE1 trichome. Bars = 250 μm in (K) and (L) and 50 μm in (M).
(N) to (S) Aniline blue fluorescence test to detect callose. Bright blue-white fluorescent spots indicate the presence of callose in FDH:FAE1 trichomes ([O] and [P]) but not in wild-type leaves ([R] and [S]). (N) and (Q) are visible-light images, and (O) and (R) are UV-light images. (P) and (S) show enlarged details from (O) and (R), respectively. That the callose depositions are primarily localized to branch tips could be seen also in Supplemental Figure 2 online.
Bars = 500 μm in (N), (O), (Q), and (R) and 50 μm in (P) and (S).
(T) Kinetics of chlorophyll leaching from leaves of the indicated genotypes into ethanol solution. Leaves were incubated in 70% ethanol for the times indicated. Note that FDH:FAE1 leaves behave like the wild type, whereas fdh shows abnormal cuticle permeability.
(U) Staining of leaves of wild-type, FDH:FAE1, and fdh plants with TB to reveal cuticular defects. Adaxial (top panel) and abaxial (bottom panel) sides of leaves are shown. Note that only fdh tissues were positively stained. Bar = 5 mm.
(V) Calcofluor white test for cuticle permeability. Leaves of wild-type, FDH:FAE1, and fdh plants (top panel) were stained with calcofluor white and examined under UV light (bottom panel). The permeable fdh leaf was used as positive control; wild-type and FDH:FAE1 leaves were not stained; however, FDH:FAE1 dead trichomes could be visualized following an a more extended TB staining (see Supplemental Figure 3 online). Bar = 5 mm.
Unlike its lack of effect on typical cutin constituents, FDH:FAE1 expression induced an increase in the levels of saturated and monounsaturated 2-hydroxylated fatty acids in residual bound lipids (Figure 3B). The total amount of these compounds increased to 132% of that in the wild type. Increases in the individual length classes ranged from 110% for C24 to 180% for C26:1 (Figure 3B). Given that enzymatically isolated cuticles contain 5 to 6 times less 2-hydroxylated VLCFAs than do residual bound lipids of chloroform/methanol extracted leaves (Franke et al., 2005), only a minor fraction of them seems to be derived from apoplastic polyesters; however, these results do show that accumulation of 2-hydroxy-VLCFAs is elevated in FDH:FAE1 leaves. Because 2-hydroxylated VLCFA are known to be present in the sphingolipids, these results also suggest that the fatty acyl composition of sphingolipid molecular species is affected in FDH:FAE1 plants. Although this was not analyzed in further detail, we found no significant difference between transgenic and wild-type samples either in the absolute amounts of sphingolipids present (121.0 ± 4.6 μg/g in the wild type versus 112.7 ± 13.5 μg/g in FDH:FAE1 plants) or in their qualitative composition, as determined by analysis of sphingolipid-specific long-chain bases (LCBs). Specifically, the ratios of the predominant LCBs (t18:1, t18:0, and d18:0) in wild-type and FDH:FAE1 plants are very similar (Figure 2D). These LCBs (and the absence of d18:1) are diagnostic for glycosylinositol-phosphoceramides (GIPCs), rather than the glucosylceramides (GluCers). Thus, our data indicate that there is no alteration in the GIPC:GluCers ratio in the FDH:FAE1 plants nor perturbation to the overall levels of sphingolipids. Collectively, the data presented here show that FDH:FAE1 misexpression increases carbon chain lengths of acyl-CoAs but differently affects carbon chain lengths in various fatty acid classes.
The FDH:FAE1 Phenotype Is Independent of FDH, and FAE1 Is Not Able to Complement fdh
Mutations in FDH, the epidermis-specific promoter of which drives expression of FAE1 in our transgenic lines, result in epidermal organ fusions in rosette leaves and inflorescences, a feature which is easy to recognize (Lolle et al., 1992). The fdh mutation also causes a partially glabrous phenotype (Yephremov et al., 1999). The FDH gene encodes a putative KCS and is required for the formation of cuticle (Yephremov et al., 1999; Pruitt et al., 2000), but its precise metabolic function is not known. In fdh plants, we found relatively low levels of C18 and C20 fatty acids in acyl-CoAs, compared with wild-type plants, suggesting a deficiency in a C16-elongase, but the changes observed were not particularly pronounced. The direct biochemical effect of the fdh mutation is difficult to analyze (due in part to the restricted expression of FDH in leaf tissue) and also because it induces a strong response in the mutant plants (A. Yephremov and D. Voisin, unpublished data). FAE1 is active both with saturated and unsaturated FAs; in particular, the highest activity has been demonstrated in a condensation assay with 16:0, 16:1, and 18:0-CoAs (Blacklock and Jaworski, 2006). Since the enzymatic function of FAE1 as a KCS is well established, we decided to determine whether FAE1 could fully or partially complement the fdh mutation upon expression in the epidermis. If successful, this experiment would provide good biochemical support for the contention that FDH is a bona fide KCS with similar substrate specificity to FAE1. In addition, we sought to determine whether the fdh mutation might rescue the trichome growth.
We therefore created transgenic plants that express FAE1 under the control of the FDH promoter in the fdh mutant background. Because the recessive fdh mutant is sterile, the transgenic FDH:FAE1 plants were crossed with FDH/fdh heterozygotes, and five heterozygous FDH:FAE1 FDH/fdh F1 plants were selected by progeny testing. In total, 196 F2 plants were classified on the basis of phenotype, without prior knowledge of their individual genotypes. Based on subsequent PCR and DNA polymorphism analyses, we then defined six genetic classes in this segregating population: three nontransgenic and three transgenic classes (Table 1). In the case of FAE1 providing complementation of fdh, the transgenic fdh plants should not exhibit organ fusions or any other features of the fdh mutant. However, as summarized in the Table 1, all 25 fdh/fdh FDH:FAE1 transgenic plants showed a typical fdh phenotype, as did all nine nontransgenic fdh/fdh plants. These results show that FAE1 is not able to compensate for the loss of FDH.
Table 1.
Complementation Analysis of fdh/fdh FDH:FAE1 Transgenic Plants
| FDH Alleles | Transgene Allele | Organ Fusion Phenotype | Number of Plants | Average Rank of Trichome Phenotypea |
|---|---|---|---|---|
| FDH/FDH | – | No | 15 | 3,00 |
| FDH/FDH | FDH:FAE1 | No | 91 | 0.71 |
| FDH/fdh | – | No | 10 | 3.00 |
| FDH/fdh | FDH:FAE1 | No | 46 | 0.80 |
| fdh/fdh | – | Yes | 9 | 1.75 |
| fdh/fdh | FDH:FAE1 | Yes | 25 | 0.56 |
Scoring was based on a blind evaluation of trichome phenotypes, which were ranked according to overall trichome appearance from 3 (completely pubescent leaves of the wild type) to 0 (completely glabrous mutant).
Furthermore, the trichomeless phenotype was not rescued in the fdh/fdh FDH:FAE1 transgenic plants. On the contrary, because fdh mutants are partially glabrous, trichome differentiation appeared to be further inhibited in the fdh/fdh FDH:FAE1 double mutant (P < 0.001 by nonparametric Mann-Whitney test), suggesting that fdh and FDH:FAE1 separately contribute to suppressing the development of trichomes.
Cell Type–Specific Cell Death Accounts for the Glabrous Appearance of FDH:FAE1 Plants
Closer examination of the FDH:FAE1 plants using scanning electron microscopy revealed that many arrested trichomes are shrunken and ultimately degenerate, suggesting that cell death can explain the general lack of mature trichomes and the loss of GL2 expression (Figures 4A to 4C). To characterize the process in detail, wild-type and FDH:FAE1 leaves were exposed to trypan blue (Figures 4H to 4J) and the fluorescent DNA stain propidium iodine (PI), to which intact cells are impermeable (Figures 4D and 4E). FDH:FAE1 trichomes, unlike other epidermal and nonepidermal cells, were clearly stained, indicating that their eventual loss is due to cell-autonomously triggered death. As well as selectively staining trichome cells in FDH:FAE1 leaves, trypan blue also, but to a lesser extent, reacted with leaf veins in FDH:FAE1 and wild-type plants (Figures 4H to 4J). Staining was more often observed in the basal region of developing leaves where trichomes normally continue to develop (Szymanski et al., 1998), indicating that the trichome cell membrane becomes abnormally permeable during the early postinitiation stage. As in the case of pGL2:ICK1/KRP1 plants, which misexpress the cyclin-dependent kinase inhibitor ICK1/KRP1 (Schnittger et al., 2003), the distal portions of young leaves, which lacked living trichomes, were hardly stained at all by either dye. Using PI staining, we also observed that the number of trichome cells showing irregular chromatin condensation patterns tended to increase from the base toward the distal end of the FDH:FAE1 leaf. Since cell death does not contribute to the trichome phenotype of fdh (Yephremov et al., 1999), fdh trichomes were indistinguishable by PI staining from those in wild-type plants (Figure 4D).
Cell death is often preceded by a burst of reactive oxygen species (ROS) or reactive oxygen intermediates (Kroemer et al., 1995), such as hydrogen peroxide, oxyradicals, or organic hydroperoxides. Therefore, we assessed intracellular ROS levels in the FDH:FAE1 plants by staining them with 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA). This nonfluorescent compound is able to cross the cell membrane freely and can be hydrolyzed by cellular esterases to yield the membrane-impermeable dichlorodihydrofluorescein, which is then oxidized by ROS in the cell to generate the fluorescent dichlorofluorescein. Staining with DCFH-DA (Figures 4F and 4G) gives rise to intense fluorescent signals in developing trichomes in FDH:FAE1 but not in the wild-type plants (unless the latter were damaged). Nontrichome cells, including adjacent subsidiary cells, were not stained by DCFH-DA. As with PI staining, living trichomes in the basal portion of the leaf displayed an intense fluorescent signal, whereas the more distal regions did not. Diaminobenzidine (DAB) staining revealed that hydrogen peroxide accumulated in developing or mature FDH:FAE1 trichomes but not in wild-type trichomes or other cells except for differentiating tracheids (Figures 4K to 4M), further supporting the idea that ROS may be involved in the induction of cell death in FDH:FAE1 leaves. These findings show that developing FDH:FAE1 trichomes specifically accumulate increased amounts of intracellular ROS. The role of ROS has not been studied further, but treatment with the antioxidant N-acetyl cysteine (sprayed on leaves at 50 mM or added to the medium at 2 to 10 mM) did not rescue trichomes in FDH:FAE1 plants (see Supplemental Table 1 online).
To determine the levels of callose deposition, young leaves from FDH:FAE1 and wild-type plants were stained with the callose binding dye aniline blue (Figures 4N to 4S). Fluorescent staining indicative of callose was observed exclusively in the trichomes of FDH:FAE1 leaves. This result agrees with previous observations that the appearance of callose is correlated with the formation of papillae, preventing invasion by pathogens, in response to abiotic stresses and during cell death (Hardham et al., 2007). Callose synthesis and deposition appears to be cell-specifically induced in FDH:FAE1 trichomes.
It should be noted that, while the FDH:FAE1 transgenic plants have overall the same quality of cuticle as wild types, the TB staining test reveals a cuticular deficiency in their trichomes as they collapse (see Supplemental Figure 3 online). We noticed that living trichomes on emerging rosette leaves and some arrested trichomes at the basal part of bigger rosette leaves remain unstained, suggesting that it is not a defect of cuticle barrier that initiates the cell death program. Supporting this view, we have also observed that cultivation of FDH:FAE1 plants in vitro under high relative humidity did not rescue trichomes (data not shown).
Accumulation of Unrepaired DNA Single-Strand Breaks and Double-Strand Breaks in FDH:FAE1 Trichomes
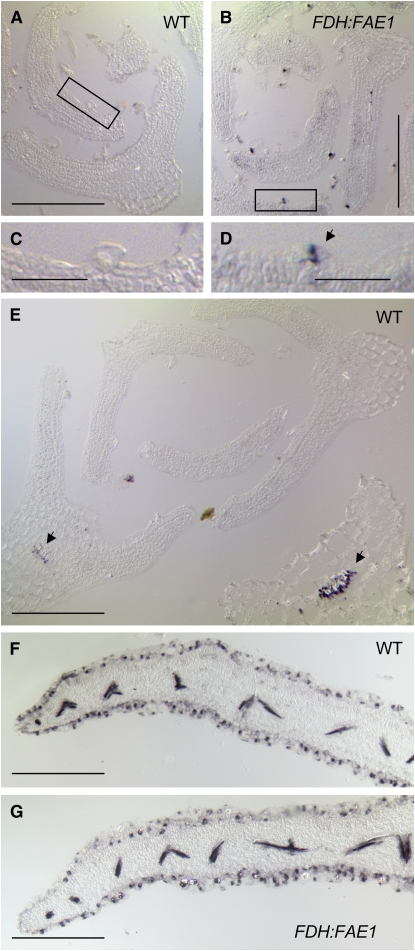
In many cell death models, the hallmark of the active, genetically controlled cell death process is fragmentation of DNA that occurs prior to death of the cell owing to the activation of specific nucleases (Walker et al., 2002). To study this issue, we employed a method that detects both single- and double-strand breaks using 3′-hydroxyl ends as primers for terminal deoxynucleotidyl transferase (TdT). This approach is known as terminal deoxynucleotide transferase dUTP nick end labeling (TUNEL) (Gavrieli, 1992), and it involves the direct labeling of the fragmented DNA with a nucleotide to which a fluorochrome, biotin, or digoxigenin has been conjugated (Walker et al., 2002). We developed a modification of this method in which unlabeled poly-dT tails produced by TdT were detected by in situ hybridization with a digoxigenin-labeled poly-dA oligonucleotide probe modified to contain locked nucleic acid (LNA) residues (LNA-A32-Dig). One advantage of this method is that the incubation time with TdT may be greatly extended to increase sensitivity. The other advantage is that, in principle, various fluorophore-labeled LNA gene probes may be combined to relate the appearance of DNA strand breaks to gene expression. We named this method TUNEL-LID (TUNEL-LNA in situ detection). Generally speaking, TUNEL-LID labeled cell nuclei as expected and showed little unspecific background staining. In sections of young leaves, FDH:FAE1 trichomes could be labeled with this technique, in contrast with wild-type trichomes and all other cell types (Figures 5A to 5C), suggesting the abnormal accumulation of unrepaired DNA strand breaks in these cells. In older rosette leaves, staining was also detected in the immature tracheal elements (TEs), which are known to undergo programmed cell death (PCD) (Turner et al., 2007) (Figures 5D and 5E). The specific labeling of FDH:FAE1 trichomes became obvious after typical incubation periods with alkaline phosphatase (30 to 60 min).
Figure 5.
Detection of DNA Fragmentation in Leaves by in Situ TUNEL-LID.
Relatively thick sections (14 μm) were cut to enhance the visibility of trichomes. DNA strand breaks were detected by enzymatically labeling the free 3′-OH, followed by hybridization of a specific digoxigenin-labeled probe to the added tag. Hybridization signals were visualized as blue-purple intranuclear precipitates. Bars = 200 μm in (A), (B), and (E) to (G) and 50 μm in (C) and (D).
(A) and (B) Cross sections of shoot apexes at a nonflowering stage, showing leaf primordia in the wild type (A) and FDH:FAE1 (B). Note specific TUNEL labeling in FDH:FAE1 trichomes (B).
(C) and (D) High-resolution views of the areas boxed in (A) and (B), respectively. In (B), the labeling of the FDH:FAE1 trichome (arrow) indicates the presence of DNA strand breaks.
(E) Differentiating TEs in older wild-type leaves (arrows) labeled under the same conditions as used in (A) and (B). Nuclear fragmentation is a normal part of TE formation, which involves PCD.
(F) and (G) Longer staining periods (up to overnight) result in a notable increase in sensitivity and reveal DNA strand breaks in the epidermis and developing vasculature of both wild-type (F) and FDH:FAE1 (G) leaves.
In contrast with single-strand breaks, spontaneously arising double-strand breaks (DSBs) cannot be repaired efficiently and could result in the cell cycle arrest and death. Presence of DSBs is considered to be a more specific marker of apoptosis (Didenko et al., 2003).
To detect DSBs specifically, we used in situ ligation, which relies on T4 DNA ligase-mediated attachment of labeled hairpin-forming oligonucleotide probes with specific ends to the ends of DNA in tissue sections (Didenko, 2002). We also developed this method further, taking advantage of a new fluorescent label (ATTO 590) with an emission maximum at 624 nm. This compound was used instead of a nonfluorescent biotin tag to internally label two probes: the LINS probe for blunt ends and the 3LINS probe for single 3′ dN overhangs. Both types of DNA ends are characteristic of the cleavage products produced by the two major DNases activated by apoptotic signaling pathways (Didenko et al., 2003). In addition to allowing the application of stringent washing conditions at 70°C (which was necessary after DSB labeling), the method has the advantage that red fluorescent labeling of apoptotic DNA can be combined with blue or green fluorescent labeling of nuclear DNA. For dual or triple labeling experiments, the nucleic acid dye EvaGreen, because of its high fluorescence yield and stability, and 4',6-diamidino-2-phenylindole (DAPI) were used. Both nucleic acid dyes gave similar fluorescent patterns, although the DAPI staining sometimes showed higher contrast.
In sections of young rosette leaves, intense ATTO 590 labeling was observed in most FDH:FAE1 trichome nuclei (Figures 6B and 6C, arrows), whereas no labeling was seen in wild-type trichome nuclei (Figure 6A, arrows). Omitting ligase from the reaction resulted in the absence of ATTO 590 labeling. As evidenced also by staining with EvaGreen (Figure 6B), some FDH:FAE1 trichomes displayed chromatin condensation at the periphery of the nucleus; however, most apparently possessed highly deformed or collapsed nuclei (Figure 6C). EvaGreen and DAPI also stained cytoplasmic RNA, although less efficiently, and this contributed to the background signal in very young leaf primordia, thereby interfering with chromatin visualization. However, nuclei in other cell types that were stained with EvaGreen or DAPI were not labeled with ATTO 590 in FDH:FAE1 or wild-type plants. In particular, cells in the leaf vasculature did not show nuclear labeling with ATTO 590 (Figure 6D), confirming that nuclear fragmentation does not occur in the course of differentiation of procambium into TEs, although TUNEL-positive nuclei are observed there (Fukuda, 2000).
Figure 6.
Detection of Blunt-End and Single 3′ dN Overhang DNA Fragments by in Situ Ligation.
Confocal fluorescence images of ATTO 590 labeling (red) and EvaGreen staining (green) and transmission images (gray) of cross sections were obtained from FDH:FAE1 ([B] and [C]) and wild-type plants ([A] and [D]) at a nonflowering stage. EvaGreen preferentially stained the nuclear DNA and, to a lesser extent, cytoplasm due to indiscriminate staining of RNA. Bars = 20 μm.
(A) Wild-type leaf primordium. Trichome nuclei are not labeled by the ATTO 590 oligonucleotide probe (arrow).
(B) FDH:FAE1 leaf primordium showing bright ATTO 590 labeling of fragmented chromatin (arrow), which has undergone condensation at the periphery of the trichome nucleus. Note the absence of ATTO 590 labeling in other epidermal cells in (A) and (B).
(C) FDH:FAE1 leaf showing disorganized chromatin labeled with the ATTO 590 probe in two trichomes (arrows).
(D) ATTO 590 fluorescence and transmission images of vascular bundles in a wild-type leaf. The thickened cell walls of TEs display background fluorescence signals. No evidence for labeling of double-stranded DSBs is seen in this or any other sections of younger or older leaves from wild-type or FDH:FAE1 plants.
We also attempted to detect the presence of nucleosomal fragmentation, leading to the appearance of a characteristic fragment ladder starting at ∼200 bp upon gel electrophoresis of nuclear DNA. Because only a small proportion of cells (i.e., the trichomes) seemed to be affected in FDH:FAE1 plants, we used linker-mediated PCR (LM-PCR) amplification (Staley et al., 1997), which allows the detection of low levels (<2%) of apoptotic cells (McLachlan et al., 2000), to screen leaf samples. After amplification, the expected fragment ladders were observed in our positive controls (allowing detection of as little as 1.2% fragmented DNA), but not in FDH:FAE1 or wild-type control leaf samples. It is worth noting that nucleosomal fragmentation of DNA does not appear to be indispensable for PCD, as some cell types produce only high molecular weight fragments (Walker et al., 2002). Therefore, such nucleosomal DNA fragments must be essentially absent in FDH:FAE1 plants, or their frequency must be so low that it is below the detection threshold of the technique. We concluded that DNA fragmentation that occurs in the course of cell death in FDH:FAE1 trichomes is detectable by both TUNEL-LID and the in situ ligation assay.
Endogenous Nuclear DNA Fragmentation in the Epidermis
Using TUNEL-LID, we observed that longer staining periods (e.g., overnight instead of 30 to 60 min) with alkaline phosphatase resulted in visualization of DNA strand breaks associated with differentiating TEs (beginning at the time of leaf primordium formation) and, surprisingly, with nuclei in the epidermis; other cell types in leaves and apical meristems were not labeled (Figures 5D and 5E). Although trichomes are not present on the abaxial surfaces of rosette leaves, the nucleus-specific labeling was observed on both adaxial and abaxial sides of rosette leaves (Figures 5F and 5G), and it disappeared nearly totally when the probe concentration used in the hybridization was reduced by 50%, clearly indicating that the signal is specific to the LNA-A32-Dig probe. Because many (if not all) nuclei in wild-type epidermal cells showed the DNA strand break-specific labeling, the breaks appear to arise during the normal differentiation program (i.e., in the absence of cell death). When a lower hybridization temperature (61°C) and longer staining periods (up to overnight) were used, TUNEL-LID signals were observed in essentially all nuclei, as expected.
Chemical Genetic Screens of FDH:FAE1 Identify Cell Death Inhibitors in the PLA2 Pathway
Screening for secondary mutations or chemical compounds that suppress a mutant phenotype are genetic and chemical genetic approaches, respectively, toward a pathway-based analysis.
Successful rescue of trichomes after spraying with KCS inhibitors (Figure 1) shows that the suppression of cell-specific cell death in the FDH:FAE1 epidermis offers a powerful system for the latter approach. To initiate the candidate-based chemical genetic screening, we analyzed the literature regarding potential targets to be screened and inhibitors of cell death, including lipoapoptosis. It appears that two pathways in ceramide and lysophosphatidylcholine (LPC) biosynthesis, both involving acyl-CoA, converge to determine critical cell fate decisions (Figure 7). Although the precise mechanisms remain to be determined, ceramide metabolites and LPC were shown to be death effectors in several animal systems, while their respective biosynthesis inhibitors suppressed PCD (Chalfant and Spiegel, 2005; Han et al., 2007). In plants, the strong evidence for involvement of ceramide metabolites in PCD has been provided by the molecular identification of the ACCELERATED CELL DEATH5 gene as one that encodes a ceramide kinase (Liang et al., 2003) and FUMONISIN B1 RESISTANT11, coding for an LCB1 subunit of serine palmitoyltransferase (SPT) (Shi et al., 2007). LPC is produced by the breakdown of phosphatidylcholine (PC), catalyzed by PC-specific PLA2. The effective appearance of cell death symptoms following Botrytis cinerea infection or herbicide paraquat treatment in transgenic Arabidopsis plants that overexpress PLA2 (coded by the patatin-like PLP2 gene) (La Camera et al., 2005) suggests the pro-PCD role of the PLA2 in plants as well.
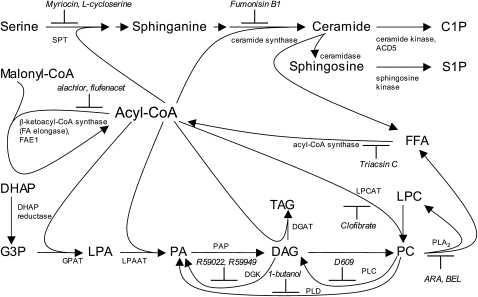
Figure 7.
Simplified Scheme of Cell Death–Related Acyl-CoA Metabolism and Chemical Inhibitors.
Biosynthesis of ceramides requires two acylation steps catalyzed by SPT and ceramide synthase, which are membrane-bound enzymes active at the cytosolic face of the endoplasmic reticulum. The eukaryotic glycerolipid pathway in the endoplasmic reticulum involves three acylation steps from G3P to TAG. PC is the main constituent of cellular membranes. For simplicity, PE and PS are not shown here. C1P, ceramide 1-phosphate; DAG, diacylglycerol; DGAT, DAG acyltransferase; DGK, diacylglycerol kinase; DHAP, dihydroxyacetone phosphate; FFA, free fatty acid; G3P, glycerol 3-phosphate; GPAT, glycerol-3-phosphate acyltransferase; LPA, lysophosphatidic acid; LPCAT, acyl-CoA:lysophosphatidylcholine acyltransferase; LPAAT, lysophosphatidic acid acyltransferase; PAP, phosphatidic acid phosphohydrolase; PLD, phospholipase D; S1P, sphingosine 1-phosphate; SPT, serine palmitoyltransferase; TAG, triacylglycerol.
To study whether foliar application of bioactive chemicals can suppress the PCD phenotype, we sprayed FDH:FAE1 transgenic plants at various concentrations (mostly in the low micromolar range) and monitored the trichomes daily for rescue of the lethality. The list of test chemicals included enzyme inhibitors of phospholipid and sphingolipid synthesis, protein kinase, and phosphatase inhibitors, lipids, hormones, etc. (>30 compounds in total; see Supplemental Table 1 online). Although all chemicals caused visible symptoms at higher concentrations (growth retardation, formation of local lesions, hyponasty, etc.), their application did not generally prevent cell death (see Supplemental Table 1 online).
Out of all compounds tested against the ceramide and LPC pathways, only a few compounds, including aristolochic acid (ARA) and bromoenol lactone (BEL; also referred to as haloenol lactone suicide substrate) were found to definitely possess anti-PCD activity on FDH:FAE1 trichomes (Figure 8). The effect was concentration dependent in the range of 10 to 40 μM. Both compounds are known PLA2 inhibitors (Figure 7); however, they differ with regard to their mode of action and chemical structure. While ARA is a noncompetitive, reversible inhibitor of PLA2, BEL is a potent, irreversible, mechanism-based inhibitor of PLA2. The identification of two structurally and mechanistically distinct inhibitors of PLA2 blocking cell death (ARA and BEL) strongly suggests that PLA2 is indispensable for inducing and/or executing PCD in FDH:FAE1. To verify that PLA2 inhibitors do not inhibit elongation of fatty acids, we examined the acyl-CoA composition in control and ARA-treated FDH:FAE1 plants (see Supplemental Figure 4 online). The VLCFA levels were not suppressed by ARA but elevated significantly (by 1.5-fold) compared with mock-treated controls, showing that ARA does not inhibit enzymes involved in the VLCFA biosynthesis.
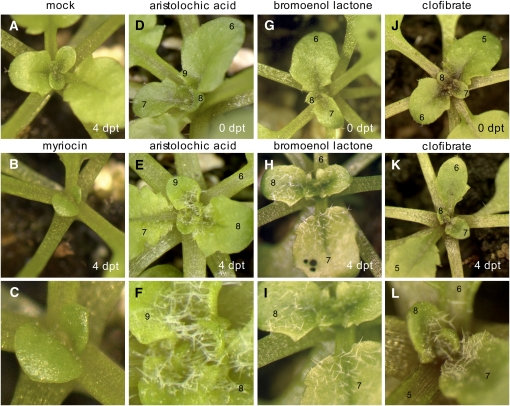
Figure 8.
Results of the Chemical Genetic Screen Designed to Identify Chemicals That Rescue Trichomes from Lipid-Induced PCD.
Images were taken within 4 days post-treatment (dpt) as indicated. The images in (C), (F), (I), and (L) show blown-up portions of the images in (B), (E), (H), and (K), respectively. Leaves are numbered from the base to the apex of the plant. Plants shown here are representative of multiple samples (∼100 plants).
(A) Mock-treated FDH:FAE1 plant.
(B) and (C) The FDH:FAE1 plant exposed to a high concentration of myriocin (125 μM), an inhibitor of the de novo ceramide synthesis pathway. Note that myriocin suppresses the growth of leaf primordia but cannot block trichome death.
(D) to (F) Chemical rescue of trichome development in FDH:FAE1 with a PLA2 inhibitor, 30 μM ARA. Trichomes that were dead at the time of spraying were not recovered.
(G) to (I) Chemical rescue of trichome development in FDH:FAE1 with a mechanistically different PLA2 inhibitor, 30 μM BEL. Note rescue of trichome death after spraying.
(J) to (L) Chemical rescue of trichome development in FDH:FAE1 with 0.02% clofibrate, a lipid-lowering drug showing hepatoprotective effect in animals. Clofibrate is a putative LPLAT inhibitor, but this function has not been confirmed in vitro and in plants.
On the other hand, blocking de novo sphingolipid synthesis with SPT inhibitors l-cycloserine and myriocin or the sphingosine N-acyltransferase (ceramide synthase) inhibitor Fumonisin B1 (Figure 7) was not effective in preventing the death of trichomes at the concentrations tested (10 to 40 μM; see Supplemental Table 1 online), suggesting that the canonical sphingolipid biosynthesis pathway is not a major regulator of PCD in FDH:FAE1.
Profiling Membrane Lipids Using Mass Spectrometry
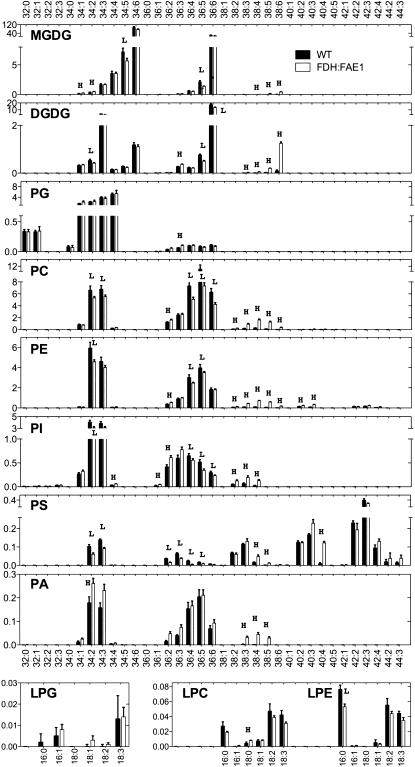
Since the above results suggested the role of membrane lipids and PLA2s in trichome death in FDH:FAE1, we performed electrospray ionization (ESI) and tandem mass spectrometry (MS/MS) analysis to determine whether the levels of membrane lipids are altered in plant tissues. ESI-MS/MS allowed identification of alterations in >140 diverse polar glycerolipids, including six head-group classes of phospholipids (PC, phosphatidylethanolamine [PE], phosphatidylinositol, phosphatidylserine [PS], phosphatidic acid [PA], and phosphatidylglycerol) and two head-group classes of galactolipids (monogalactosyldiacylglycerol and digalactosyldiacylglycerol). In each class, molecular species varying in the lengths and desaturation levels of acyl groups were identified according to their mass spectra and with previous references (Welti et al., 2002; Devaiah et al., 2006).
Major phospholipids, in which the proportions of VLCFAs are usually low, accumulated higher amounts of VLCFAs in FDH:FAE1 plants in a qualitatively similar fashion (Figure 9). For example, VLCFAs are present in a small proportion in PC (1.9%) in wild-type plants, but FDH:FAE1 plants accumulate VLCFAs in 12.6% of all PC species. Similar proportions are 4.3 and 15.0% for PE, and 1.3% and 4.7% for phosphatidylinositol, respectively. The greatest magnitude of increase was found in PA (0.3 to 8.7%), while the lowest increase (76.2 to 86.6%) was detected in PS species, which already contain a significant fraction of VLCFAs. Thus, these results are in agreement with our results above and the results of previous studies (Millar and Kunst, 1997; Millar et al., 1998), consistent with the interpretation that overexpression of FAE1 leads to an increase in the VLCFAs levels in phosholipids. Higher levels of VLCFAs in galactolipids, such as digalactosyldiacylglycerol in FDH:FAE1 plants, are also in a good agreement with the previous results (Millar et al., 1998).
Figure 9.
Lipid Species Profiling to Detect Changes in FDH:FAE1 Leaf Tissues.
Lipids were extracted from young leaves and analyzed by ESI-MS/MS as described in Methods. The y axis of each plot indicates the amount of lipids (nmol/mg dry weight); y axis values are the means ± se (n = 6). The x axes indicate lipid molecular species (total acyl carbons: double bonds). Acyl chain composition of major lipid molecular species could be found in Welti et al. (2002). L and H indicate that the value in FDH:FAE1 is lower or higher than that of wild-type plants; P < 0.01, Mann-Whitney test. MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; PI, phosphatidylinositol; PG, phosphatidylglycerol; LPG, lysophosphatidylglycerol; LPE, lysophosphatidylethanolamine.
More remarkably, however, the results indicate that total levels of lysophospholipids, which are minor phospholipid species in Arabidopsis, in the three classes analyzed (LPC, LPE, and LPG) are not essentially changed in FDH:FAE1 plants. Since lysophospholipids are obtained when a FA is enzymatically released from phospholipids by phospholipase A, this suggests that no change in overall activity of PLA occurs in plant tissues in FDH:FAE1.
Also interesting is that while levels of VLCFA phospholipids were increased at the expense of non-VLCFA species in most classes, C34 PA species (i.e., phosphatidic acid containing 18:0 and 16:0), which do not contain VLCFAs, also accumulated to higher levels in FDH:FAE1 (150%) relative to the wild type. PA has been shown to induce cell death in leaves through the ROP-mediated ROS generation pathway (Park et al., 2004), supporting the interpretation that the PA formation, increased in response to FAE1 expression, potentiates the PCD. However, inhibitors of two enzymes that produce PA (Figure 7), phospholipase D antagonist 1-butanol, and diacylglycerol kinase inhibitors (R59022 and R59949), were not effective in preventing the death of trichomes (see Supplemental Table 1 online), warranting further investigation.
DISCUSSION
Cell Type–Specific Death in FDH:FAE1: A Case of a Lipoapoptosis-Like Process in Plants
This study presents the unexpected finding that epidermal misexpression of FAE1 in Arabidopsis leads to the death of a specific cell type, trichomes, via a lipid metabolic pathway. Loss of viability and membrane integrity, generation of ROS, and DNA fragmentation lend support to the conclusion that trichomes undergo PCD. Using the new TUNEL-LID method, we detected DNA strand breaks in the nuclei of epidermal cells and in differentiating vascular bundle cells (which can thus serve as a positive internal control) in wild-type and FDH:FAE1 plants. Under certain conditions, FDH:FAE1 trichomes could clearly be distinguished from wild-type trichomes by specific labeling, suggesting that DNA strand breaks could not be efficiently repaired, leading to chromosomal breaks. It is known that if DNA strand breaks are not repaired prior to replication, they are generally harmful, prevent cell cycle progression, and appear to be sufficient to initiate the events leading to PCD (Nelson and Kastan, 1994). However, in addition to being primarily responsible for cell death, DNA strand breaks may be a consequence of a PCD-activated nuclease action or result from ROS attack on DNA in FDH:FAE1 trichomes.
In situ ligation revealed that the DNA strand breaks in FDH:FAE1 trichome nuclei give rise to blunt-end and single 3′ dN overhang DNA fragments. It is remarkable that we did not observe DSB-specific labeling in the vasculature in the same experiment. It was previously reported that nucleosomal DNA fragmentation, nuclear shrinkage, and noticeable chromatin condensation do not occur during TE differentiation when collapse of the vacuole triggers cell death, whereas these characteristic features are displayed during apoptosis-like PCD in plants when nuclei appear to be the first target of degradation (Fukuda, 2000). It is noteworthy that, in FDH:FAE1 trichomes, we detected intracellular ROS, including H2O2, which are also characteristic of the HR, while oxidative bursts have not been detected during TE formation (Jones and Dangl, 1996) pointing to a marked difference between these PCDs.
We have shown that levels of long-chain acyl-CoAs esters are increased in the FDH:FAE1-expressing plants, suggesting a possible link with lipoapotosis, a process that is induced by accumulation of excessive amounts of long-chain saturated fatty acids, via an acyl-CoA–dependent pathway, in a variety of cell types in animals (Listenberger and Schaffer, 2002). It is important to mention that lipoapoptosis was not defined purely on morphological grounds but rather as a fatty acid–induced PCD in animals. Bearing in mind that the course of PCD in plants is different from that of apoptosis (e.g., cell fragmentation and phagocytosis of apoptotic bodies do not occur) (Danon et al., 2000; van Doorn and Woltering, 2005), the term “lipoapoptosis-like process” to describe a VLCFA-induced PCD in plants is used here to emphasize the fact that both cell death programs are possibly triggered by the presence of a critical threshold of certain fatty acids or lipids.
Although long-chain acyl-CoAs can themselves act as signaling molecules, they are classically viewed as precursors to numerous other metabolites containing very long aliphatic chains, including sphingolipids, some of which are known to be potent proapoptotic molecules in animals and plants (Chalfant and Spiegel, 2005). However, the identification of the responsible death effectors in lipoapoptosis remains controversial with evidence both for and against the involvement of sphingolipids. One explanation may be that cell type–specific processes for channeling FAs to particular metabolic fates may define the mechanism by which PCD is activated (Listenberger et al., 2001). Since the FDH promoter is active in all epidermal cells (Yephremov et al., 1999; Efremova et al., 2004), differences in key molecular and cellular processes in diverse cell types are likely to be the causes for the observed trichome death in FDH:FAE1 plants, in turn mediated by differences in the metabolic configuration of acyl-exchange pathways in different cells. It is reasonable to hypothesize that GL2 or other trichome-specific genes play some role in eventual manifestation of the death process.
There is an interesting parallel between our findings and data on palmitate- and stearate-induced lipoapoptosis in Chinese hamster ovary cells and in human coronary artery endothelial (HCAE) cells that was also found to be independent of ceramide synthesis (Listenberger et al., 2001; Staiger et al., 2006). In these studies, inhibition of ceramide synthase and specific inhibition of SPT by fumonisin B1 and l-cycloserine, respectively, did not rescue cells from lipoapoptosis (see Supplemental Table 1 online). Our data are also in a good agreement with the results of Han et al. (2007), who reported a critical role for the PLA2-catalyzed formation of LPC in hepatocite lipoapoptosis. In common with these studies, we found that myriocin did not prevent PCD (Figure 8), while selective PLA2 inhibitors were quite effective. The fungal metabolite myriocin is a potent inhibitor of SPT, which catalyzes the first rate-limiting step in the de novo sphingolipid biosynthesis (Figure 7) (Kolter and Sandhoff, 1996). When this step is chemically or genetically blocked in plants, the sphingolipid-mediated pathway of cell death is interrupted (Spassieva et al., 2002; Shi et al., 2007). Treatment with a major metabolite of ceramide, ceramide-1-phosphate, which is known to partially block PCD induction by C2 ceramide (Liang et al., 2003), also did not promote cell survival in FDH:FAE1 (see Supplemental Table 1 online). Collectively, these experiments further support a link between PLA2 and lipoapoptotis-like PCD and suggest that the initiation of cell death in FDH:FAE1 may not require de novo sphingolipid synthesis or can occur through a ceramide-independent pathway.
Fatty Acid–Mediated Cell Death Pathway and PLA2 Operate in Hypersensitive and Stress Response
Also unexpected was the finding that selective PLA2 inibitors prevent PCD in FDH:FAE1 plants. While PCD could be a component of normal development in plants, it occurs during plant–pathogen interactions and can play a dual role in the pathogenesis, both promoting and counteracting the spread of disease through a HR around the site of infection (Rogers, 2005; Van Breusegem and Dat, 2006; Hofius et al., 2007). The involvement of lipid-mediated signaling in the HR cell death is supported by the identification of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN DEFICIENT4 (PAD4), SUPPRESSOR OF SA INSENSITIVITY2 (SSI2), SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1, and MYB30 genes in Arabidopsis. EDS1 and its interacting partner, PAD4, encode lipase-like proteins, important activators of salicylic acid signaling that is involved in cell death control, but their putative enzymic functions have not been proven (Wiermer et al., 2005). SSI2/FAB2, which encodes a plastidial stearoyl-acyl carrier protein-desaturase, preferentially converts stearoyl-ACP (18:0-ACP) to oleoyl-ACP (18:1Δ9-ACP). The recessive ssi2 mutation results in spontaneous lesion formation but confers constitutive PATHOGENESIS-RELATED gene expression and renders plants more resistant against bacterial and oomycete pathogens (Shah et al., 2001). The misregulation of cell death control in ssi2 and fad7 fad8 FA desaturation mutants is reminiscent of the pro-(lipo)apoptotic effect of saturated FAs and the anti-(lipo)apoptotic effect of unsaturated FAs in animal cells (Busch et al., 2005; Shah, 2005). The sfd1 mutant is a ssi2 suppressor, defective in dihydroxyacetone phosphate reductase/G3P dehydrogenase, which catalyzes the interconversion of dihydroxyacetone phosphate to G3P (Nandi et al., 2004).
Additionally, MYB30 is worthy of further consideration: MYB30 is a transcription factor, positive regulator of PCD, and a key transcriptional activator of the HR (Vailleau et al., 2002). Most interestingly, MYB30 putative target genes, identified by DNA microarrays, encode four enzymes involved in VLCFA biosynthesis: KCS, hydroxy-CoA dehydratase, trans-2,3-enoyl-CoA reductase, β-ketoacyl-CoA reductase, and other lipid-related proteins (Raffaele et al., 2008). These genes are significantly induced at early time points after infection of Arabidopsis with the avirulent strains 147 of Xanthomonas campestris pv campestris and DC3000 (avrRpm1) of Pseudomonas syringae pv tomato. The expression of three KCS genes, KCS1, KCS2, and FDH, is controlled by MYB30. Based on these findings, a model was proposed in which MYB30 acts as a strong, positive regulator of biosynthesis of VLCFAs and that, as yet unidentified, VLCFA-derived signaling molecules are capable of regulating the HR cell death and defense responses (Raffaele et al., 2008). Our results are indicative of possible involvement of PLA2 in this VLCFA-triggered PCD process.
Plants have two groups of PLA2s: secreted PLA2s and patatin-like PLA2s (PAT-PLA), which are related in amino acid sequence to calcium-independent PLA2s in animals. Studies with animal PLA2 inhibitors, such as ARA, BEL, ETYA (5,8,11,14-eicosatetraynoic acid), PACOCF3, and AACOCF3, showed that they too can inhibit plant PLA2s in vitro and in planta (Holk et al., 2002; Ryu, 2004). BEL is deemed to be not capable of inhibiting secreted PLA2, which uses a different catalytic mechanism; therefore, it is most probable that a PAT-PLA enzyme is responsible for executing the PCD process in FDH:FAE1 plants. However, this remains to be established in further studies. Arabidopsis possesses a PAT-PLA family containing nine (La Camera et al., 2005) or 10 proteins (Holk et al., 2002), which are localized to the cytoplasm or associated with the plasma membrane. PAT-PLA PLP2 is induced in response to biotic and abiotic stresses (Ryu, 2004), and its knockout is probably embryo lethal (La Camera et al., 2005). Because soluble PAT-PLA activity was sharply induced in tobacco (Nicotiana tabacum) infected with tobacco mosaic virus, it was proposed that PAT-PLAs reaction products contribute to HR cell death against pathogens (Dhondt et al., 2000). PLP2 expression in transgenic plants was found to potentiate the appearance of PCD symptoms imposed by the superoxide radical-generating herbicide paraquat and B. cinerea and also promoted higher multiplication of avirulent P. syringae (avrRpt2) (La Camera et al., 2005). Of interest is also the presence of members of the PAT-PLA superfamily in genomes as diverse as bacteria, fungi, and animals. Furthermore, Pseudomonas aeruginosa type III secretory toxin ExoU, which is a factor directly responsible for cell death resulting in acute lung injury, is a potent PAT-PLA (Tamura et al., 2004).
PAT-PLAs, which appeared to combine phospholipase A1 (PLA1) and PLA2 activities, potentially generate free fatty acids and a range of lysoglycerolipids, such as LPC, LPE, LPI, LPS, monogalactosyl monoacylglycerol, and digalactosyl monoacylglycerol. Although LPC has been identified as a death effector in an animal model (Han et al., 2007), it is unclear at this stage whether the release of free fatty acids or a lysoglycerolipid, or both, could give rise to a pro-PCD signal (or an anti-PCD signal depleted in FDH:FAE1 plants) and whether there is a role for perturbation and destabilization of membrane structure that reduces cell viability. It is also plausible that PAT-PLA–induced reorganization of plasma membrane triggers relocalization of raft proteins and stimulates a signal transduction cascade that eventually culminates in cell death. Although the focus of this report is on the unexpected, lipoapoptosis-like death of trichomes, it is interesting to note that the FDH:FAE1 plants display altered disease susceptibility when infected with bacteria and fungi, suggesting that all epidermal cell types may be competent in responding to the putative PLA2-generated lipid mediators. Further studies are currently underway to elucidate the mechanism of this phenomenon, but it would be beyond the scope of this article to address them.
A Possible Role for Fatty Acid Remodeling of Phospholipids in Fatty Acid–Induced Cell Death
Another, yet not excluding, possibility is that PLA2 inhibitors block a remodeling pathway (Lands' cycle) involved in the dynamic turnover of acyl groups in glycerophospholipids. The pathway is thought to involve the PLA2-catalyzed hydrolysis of the sn-2 acyl residue, followed by reacylation of the resulting lysophospholipid mediated by acyl-CoA:lysophospholipid acyltransferases (LPLATs), such as acyl-CoA:lysophosphatidylcholine acyltransferase (Yamashita et al., 1997). This metabolic process has been studied in a variety of tissues in animals, plants, and fungi (Ståhl et al., 2007); however, its role in cell death has not been determined. It is interesting that one of a few compounds capable of interfering with trichome cell death appeared to be the antihyperlipidaemic and hepatoprotective drug clofibrate (Figure 8). Clofibrate [ethyl 2-(4-chlorophenoxy)-2-methylpropanoate], mainly known as an agonist of peroxisome proliferator-activated receptors, which are absent in the genome of plants, was also reported to act as an LPLAT inhibitor (Riley and Pfeiffer, 1986). Given that plant LPLAT genes were recently identified (Ståhl et al., 2007; Hishikawa et al., 2008), a cellular site of action for clofibrate in plants would be interesting to study.
Although details of the acyl remodeling pathway remain to be determined, the model predicts that both PLA2 and LPLAT chemical inhibitors should be effective in controlling acyl composition of membranes and acyl-CoAs. In such a scenario, their anti-PCD action in FDH:FAE1 plants may be related to the protective effects on cellular membranes, which maintain the fatty acid composition in the presence of high levels of VLCFAs in the acyl-CoA pool. Supportive data for this hypothesis have been obtained from quantitative analysis of acyl-CoA esters extracted from control and ARA-treated plants. As the model predicted, the treatment with a PLA2 inhibitor increased VLCFA levels in the acyl-CoA fraction (see Supplemental Figure 4 online). However, a full confirmation of this hypothesis would require an additional analysis of membrane lipid composition and the use of other inhibitors. Moreover, the analysis of mutants and transgenic lines with reduced activity of PLA2 or LPLAT in the future should give us insight into the mechanisms underlying acyl remodeling and its potential link to PCD.
METHODS
Arabidopsis thaliana Plant Material and Growth Conditions
Arabidopsis, Columbia (Col-0) ecotype, was used as a wild type in all experiments. The GL2:GUS expression line was obtained from the ABRC at Ohio State University (Columbus, OH). The fdh-3940S1 allele used for complementation analysis (referred here as fdh) was described previously (Yephremov et al., 1999). Arabidopsis plants were grown for analyses in a greenhouse at 22 to 23°C and 50 to 60% humidity and kept on an 8-h photoperiod (short day) for the first 6 to 7 weeks after sowing and on a 16-h photoperiod (long day) thereafter. Some plants were grown for propagation or renewal of seeds in a controlled environment chamber.
Generation of Transgenic Plants and Transgenic Complementation
To construct the binary vector pBPF:FAE1, the FAE1 cDNA from pT7T318U (Millar and Kunst, 1997) was cloned as a SalI-XbaI fragment into the XhoI/XbaI sites of the binary vector pBPF-SX (Efremova et al., 2004), placing it under the control of the FDH promoter. Wild-type Arabidopsis plants (Col-0) were transformed with pBPF:FAE1 by vacuum infiltration as described previously (Bechtold et al., 1993), and BASTA was employed for selection of transgenic plants. More than 30 independent transformants, all showing a similar trichome phenotype, were obtained. Homozygous lines were established in four families, and plants were backcrossed to the Col ecotype to test segregation and identify lines carrying a single copy of the transgene. Expression of FAE1 driven by the FDH promoter was confirmed by RT-PCR using pAnos (5′-TATTACATGCTTAACGTAATTCAACAG-3′) and FAE-T (5′-AAAACGGTCGGTCCTAATTTGATG-3′) as primers.
To determine whether FDH:FAE1 can complement the fdh mutant, fdh heterozygotes were crossed with FDH:FAE1 transgenic plants representing five different families. The progeny of double heterozygotes was then examined for organ fusions and trichome phenotypes. In an independent, blind experiment, progeny plants were tested for the presence of the transgene by PCR with the pAnos and FAE-T primers.
To construct the GL2-GFP protein fusion, a 5.6-kb genomic portion of the GL2 gene was PCR-amplified using GL2XP (5′-GATAAAGCCCGGGAATTGTAGATAAATCATCTGCTA-3′) and GL2XE (5′-TGTGACTTGCCCGGGGCAATCTTCGATTTGTAGACTTCTCTTAATG-3′) primers appended with XmaI recognition sequences (underlined). This fragment comprised 2124-bp sequence upstream the initiation codon and GL2 open reading frame excluding the termination codon. It has been cloned in frame to GFP into the unique XmaI site in the polylinker of the pBctGFP binary vector (A. Yephremov, unpublished data). Wild-type and gl2 plants were transformed with the GL2-GFP construct, and transgenic plants were selected with BASTA as described above. Expression the GL2-GFP protein was confirmed by complementation of the gl2 mutant, and GFP fluorescence. The GL2-GFP plants were crossed to FDH:FAE1 plants, and double transgenic plants were selected by epifluorescence microscopy and PCR.
Genotyping
Genotyping of the FDH locus was conducted in a blind experiment with an automated DHPLC instrument equipped with a DNASep column (WAVE; Transgenomic) as described previously (Efremova et al., 2004). This approach relies on different melting behavior of PCR amplicons derived from FDH, fdh-3940S1, which carries a GT insertion, and their heteroduplex.
Chemical Treatments
Three-week-old FDH:FAE1 transgenic and wild-type plants with seven to eight rosette leaves were sprayed three times daily with mock solutions (controls) and the solutions to be tested. Plants were examined using a Leica MZFL-III microscope prior to the first treatment and on days 4, 5, and 7 after treatment. The following concentrations of chemicals were used if not otherwise indicated: 10, 20, 30, and 40 μM (see Supplemental Table 1 online for the list of compounds, concentrations, solvents, cellular targets, and effects).
Microscopy and Staining Techniques
Cryoscanning electron microscopy was performed as described previously (Yephremov et al., 1999).
Young rosette leaves (∼1 cm) or rosettes of 5- to 6-week-old plants were stained with PI and DCFH-DA (both from Sigma-Aldrich) by incubation for 15 to 30 min and then washed with water for 20 min. PI was used at 0.1 mg/mL in PBS. The 50 mM stock solution of DCFH-DA was prepared in dimethylformamide and diluted to 5 μM in water before staining. Uptake of DCFH-DA and PI by living cells was examined using a Leica MZFL-III fluorescence microscope equipped with GFP3 (excitation 470/40 nm; emission 525/50 nm) and DsRED (excitation 546/10 nm; emission 600/40 nm) filters (AHF-Analysentechnik), respectively.
Staining with trypan blue lactophenol, aniline blue, and DAB was performed as described by Koch and Slusarenko (1990), Adam and Somerville (1996), and Schraudner et al. (1998), respectively.
TUNEL-LID
Tissues from 6-week-old plants grown on an 8-h/16-h day/night cycle were fixed in methanol:acetic acid (3:1) at −20°C overnight, and chlorophyll was then removed by extraction with 70% ethanol at −20°C overnight (longer incubation times and several changes of the ethanol solution may be necessary to ensure extraction). The fixed tissues were dehydrated in an ethanol series (70, 85, 95, and 2× 100% for 5 min each), transferred from 100% ethanol through an ethanol-Histoclear series to 100% Histoclear (Histoclear: ethanol 1:2, Histoclear: ethanol 2:1, 100% Histoclear for 5 min each), and embedded into Paraplast plus (Tyco Healthcare) at 60°C. Sections (14 μm) were cut with a microtome and mounted on SuperFrost Plus adhesion microscope slides (Menzel-Gläser). Before the TUNEL treatment, sections were deparaffinized (3× 100% Histoclear, Histoclear:ethanol 2:1, Histoclear:ethanol 1:2, 2× 100% ethanol), rehydrated in a graded series of ethanol (95, 85, 70, 50, and 30% for 2 min each), washed in a 2× SSPE buffer at 70°C for 20 min, incubated with 1 mg/mL Proteinase K solution for 20 min at 37°C, and carefully washed again in 2× SSPE at 70°C and in 0.85% NaCl at room temperature. A 200-μL aliquot of tailing reaction mixture, comprising 160 units of TdT (Fermentas) and 1 mM dTTP in the supplied TdT buffer was applied to each slide. Slides were incubated at 37°C for 1 h, washed in 2× SSPE and 0.85% NaCl, and dehydrated using an ethanol series (70, 85, 95, and 100% for 5 min each). The probe LNA-A32-Dig used to detect poly-dT tails was a digoxigenin-labeled 32-nucleotide poly-dA oligonucleotide containing 11 LNA residues (aaAaaAaaAaaAaaAaaAaaAaaAaaAaaAaaAaa; LNA residues are shown in upper case). The probe was synthesized, labeled, and purified by anion-exchange HPLC by the manufacturer (Exiqon). Its melting temperature is predicted to be 61°C (http://lna-tm.com). This temperature was optimal for dot blots with oligo(dT)-labeled λ phage DNA digests; however, we found that the best results were obtained when in situ hybridization assays were conducted at 64°C using 5 nM probe. The hybridization buffer contained 50% deionized formamide, 20 mM Tris-HCl, pH 7.0, 0.3 M NaCl, 0.2 mM EDTA, 0.5 mg/mL yeast tRNA, 10% dextran sulfate, and 1× Denhardt's solution (Sigma-Aldrich). A 100-μL aliquot of hybridization mix was applied to each slide, covered with a cover slip, and incubated overnight at 64°C in a humid chamber. Slides were then washed with 3× SSPE (twice for 5 min each at room temperature and twice for 20 min at 64°C) and NTE (0.5 M NaCl, 1× TE, pH 7.5; twice for 5 min at room temperature). Slides were treated with 40 μg/mL RNase A in NTE for 30 min, washed with NTE (twice for 5 min at room temperature), 1.5× SSPE (once for 30 min at 64°C), and 0.3×SSPE (twice for 30 min at 64°C). Digoxigenin-labeled hybrids were detected using antidigoxigenin antibody conjugated to alkaline phosphatase diluted 1:3000 in buffer 1 (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% BSA) for 1 h at room temperature. Slides were then washed in buffer 1 containing 0.1% Tween 20 (for 10 min, 15 min, and 2× 20 min) and equilibrated in the staining buffer (100 mM Tris-HCl, pH 9.5, 150 mM NaCl, 50 mM MgCl2, and 0.1% Tween 20). For staining, the slides were incubated in the staining buffer containing the color substrates 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt and nitro-blue tetrazolium chloride as recommended by the manufacturer (Roche). The signal appeared after 30 to 60 min. The reaction was stopped by washing the slides in deionized water.
In Situ DNA Ligation Assay
The in situ ligation assay was performed using the same plant materials and preparation techniques as for the TUNEL-LID tailing reaction. Sections were first preincubated with 1× ligation buffer containing 40 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, and 5 mM ATP, pH 7.8 (Fermentas), for 15 min at room temperature and then with the ligation solution (180 μL/slide) containing 1× ligation buffer, 5% PEG 4000 (Fermentas), T4 DNA ligase (0.15 units/μL) (Fermentas) and the labeled hairpin-forming oligonucleotides LINS and 3LINS (5 pmol/μL of each). LINS (GAATTCCCGGGATCCtGGATCCCGGGAATTC) and 3LINS (NGAATTCCCGGGATCCtGGATCCCGGGAATTC) were labeled internally during synthesis with ATTO 590 (t denotes dT with the attached fluorescent label, N denotes any of the four nucleotides) and HPLC purified by the manufacturer (IBA). The slides were covered with cover slips, and the ligation was allowed to proceed overnight at room temperature in a moist, light-protected chamber. The slides were washed twice for 10 min with 2× SSPE at 70°C and stained with 0.1× EvaGreen fluorescent DNA stain (Biotium) in 2× SSPE at 70°C to reveal nuclei or/and RNA. Unbound EvaGreen was removed by washing twice with 2× SSPE at room temperature for 10 min, and the slides were examined on a Leica TCS SP2 AOBS spectral confocal microscope. Fluorescence was observed with excitation at 561 nm and emission at 570 to 651 nm for ATTO 590 and excitation at 488 nm and emission at 495 to 545 nm for EvaGreen. To reveal nucleic acids, some slides were also stained with DAPI (2.5 μL/mL in PBS) for 5 min and washed in PBS for 5 min at room temperature. The DAPI signal was excited with UV light and recorded at 415 to 470 nm.
DNA Fragmentation Analysis Using LM-PCR
For LM-PCR, genomic DNA, isolated from Arabidopsis using the DNeasy plant mini kit (Qiagen), was ligated to blunt-end linkers consisting of oligonucleotides LM24 (5′-AGCACTCTCGAGCCTCTCACCGCA-3′) and LM12 (5′-TGCGGTGAGAGG-3′). Ligation products were purified using a Microcon 30 device (50-bp cutoff for double-stranded DNA; Millipore) and amplified using the Advantage 2 PCR kit (Clontech Laboratories) with LM24 as a primer. The PCR program was 94°C for 1 min, followed by 15 cycles of 94°C for 20 s, 56°C for 30 s, 68°C for 1 min 30 s, and 20 cycles of 94°C for 20 s, 56°C for 30 s, and 68°C for 1 min 30 s with a 5-s increment per cycle, and concluded by an extension cycle of 68°C for 7 min. The LM-PCR products were analyzed by electrophoresis on 1.2% agarose gels. Positive control ladders were produced from λ phage DNA (Fermentas) digested with SspI (Biolabs) and ligated to the same linkers.
Sphingolipid Analysis
Sphingoid base analysis was conducted essentially as described previously using sphingosine (d20:1) as an internal standard (Borner et al., 2005; Sperling et al., 2005). Briefly, 500-mg mixtures (fresh weight) of 22 to 24 4-week-old seedlings of wild-type and transgenic plants were hydrolyzed in 10% (w/v) Ba(OH)2 for 20 h at 110°C, and LCBs were extracted with chloroform:dioxane:water (6:5:1; v/v/v). LCBs were then converted to their dinitrophenyl derivatives with methanolic 1-fluoro-2,4-dinitrobenzene, extracted with chloroform:methanol:water (8:4:3; v/v/v), and purified by thin layer chromatography on silica plates. The samples were analyzed by reverse-phase HPLC/MS, and individual compounds were quantified with respect to the internal standard.
HPLC Analysis of Acyl-etheno-CoA Derivatives and Analysis of the Total Fatty Acids
Twenty-milligram portions of leaf material were collected from 4-week-old plants grown under short-day conditions, frozen in liquid nitrogen, and extracted for subsequent quantitative analysis of fluorescent acyl-etheno-CoA derivatives by HPLC and GC analysis of the total fatty acids. HPLC [Agilent 1100 LC system; Phenomenex LUNA 150 × 2 mm C18(2) column] was performed using the methodology and gradient conditions described previously (Larson and Graham, 2001; Larson et al., 2002). HPLC analysis of acyl-etheno-CoA derivatives in ARA-treated plants was conducted similarly, using 4-week-old plants. Analysis of the total fatty acid composition of leaves was performed as described previously (Sayanova et al., 2007).
Profiling Membrane Lipids
Samples were prepared from young leaves (<1 cm) of 6-week-old Arabidopsis plants grown under short-day conditions. Each sample contained tissues from seven plants with a pooled dry weight of 5 to 10 mg, and six replicates for each genotype were analyzed. To inhibit lipolytic activities, tissues were transferred immediately into 3 mL of isopropanol with 0.01% butylated hydroxytoluene at 75°C and extracted several times with chloroform/methanol as a recommended by Kansas Lipidomics Research Center (http://www.k-state.edu/lipid/lipidomics/leaf-extraction.html). Automated ESI-MS/MS analysis was performed in the Kansas Lipidomics Research Center Analytical Laboratory essentially as described previously (Welti et al., 2002; Devaiah et al., 2006).
Characterization of Cuticle
The following experiments were employed to characterize the properties of the cuticular barrier and were performed as described previously: TB staining and evaluation of chlorophyll leaching from rosette leaves into ethanol (Kurdyukov et al., 2006b), fatty acid composition analysis of wax, and residual bound lipids in leaves (Franke et al., 2005; Kurdyukov et al., 2006a).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes characterized in this study are as follows: FDH (At2g26250), FAE1 (At4g34520), and GL2 (At1g79840).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Localization of the GL2-GFP Protein Fusion in Trichome Nuclei in FDH:FAE1 Plants.
Supplemental Figure 2. Aniline Blue Staining for Callose.
Supplemental Figure 3. Toluidine Blue Staining for Cuticular Defects in FDH:FAE1 Trichomes.
Supplemental Figure 4. Effect of Aristolochic Acid Treatment on VLCFAs in Acyl-CoA Esters in FDH:FAE1 Plants.
Supplemental Table 1. Summary Data Set for Compounds Tested in the Cell Death Assay.
Supplemental Movie 1. Localization of the GL2-GFP Protein Fusion in Trichome Nuclei in FDH:FAE1 Plants.
Supplementary Material
Acknowledgments
We thank Ljerka Kunst for the FAE1 cDNA clone, Martin Hülskamp for bringing the cell death phenotype of the FDH:FAE1 plants to our attention, Elmon Schmelzer for his help with fluorescence microscopy, Ruth Welti and Mary Roth for the ESI-MS/MS identification of lipid molecular species in our samples, Klaus Tietjen for a sample of flufenacet, and Paul Hardy for his helpful comments and critical reading of the manuscript. We would especially like to thank Isa Will for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alexander Yephremov (efremov@mpiz-koeln.mpg.de).
Online version contains Web-only data.
References
- Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9 341–356. [DOI] [PubMed] [Google Scholar]
- Bach, L., Michaelson, L.V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., Da Costa, M., Boutin, J.P., Miquel, M., and Tellier, F. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 105 14727–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In-planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316 1194–1199. [Google Scholar]
- Bessire, M., Chassot, C., Jacquat, A.C., Humphry, M., Borel, S., Petétot, J., Métraux, J.P., and Nawrath, C. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26 2158–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklock, B.J., and Jaworski, J.G. (2006). Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 346 583–590. [DOI] [PubMed] [Google Scholar]
- Boger, P., Matthes, B., and Schmalfuss, J. (2000). Towards the primary target of chloroacetamides - New findings pave the way. Pest Manag. Sci. 56 497–508. [Google Scholar]
- Bonaventure, G., Beisson, F., Ohlrogge, J., and Pollard, M. (2004). Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 40 920–930. [DOI] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., MacAskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A.K., Gurisik, E., Cordery, D.V., Sudlow, M., Denyer, G.S., Laybutt, D.R., Hughes, W.E., and Biden, T.J. (2005). Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic β-cells from lipoapoptosis. Diabetes 54 2917–2924. [DOI] [PubMed] [Google Scholar]
- Chalfant, C.E., and Spiegel, S. (2005). Sphingosine 1-phosphate and ceramide 1-phosphate: Expanding roles in cell signaling. J. Cell Sci. 118 4605–4612. [DOI] [PubMed] [Google Scholar]
- Danon, A., Delorme, V., Mailhac, N., and Gallois, P. (2000). Plant programmed cell death: A common way to die. Plant Physiol. Biochem. 38 647–655. [Google Scholar]
- David, D., Sundarababu, S., Gerst, J.E., and Gerst, J.E. (1998). Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 143 1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, S.P., Roth, M.R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., Welti, R., and Wang, X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 67 1907–1924. [DOI] [PubMed] [Google Scholar]
- Dhondt, S., Geoffroy, P., Stelmach, B., Legrand, M., and Heitz, T. (2000). Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 23 431–440. [DOI] [PubMed] [Google Scholar]
- Didenko, V.V. (2002). Detection of specific double-strand DNA breaks and apoptosis in situ using T4 DNA ligase. Methods Mol. Biol. 203 143–151. [DOI] [PubMed] [Google Scholar]
- Didenko, V.V., Ngo, H., and Baskin, D.S. (2003). Early necrotic DNA degradation: Presence of blunt-ended DNA breaks, 3′ and 5′ overhangs in apoptosis, but only 5′ overhangs in early necrosis. Am. J. Pathol. 162 1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova, N., Schreiber, L., Bar, S., Heidmann, I., Huijser, P., Wellesen, K., Schwarz-Sommer, Z., Saedler, H., and Yephremov, A. (2004). Functional conservation and maintenance of expression pattern of FIDDLEHEAD-like genes in Arabidopsis and Antirrhinum. Plant Mol. Biol. 56 821–837. [DOI] [PubMed] [Google Scholar]
- Franke, R., Briesen, I., Wojciechowski, T., Faust, A., Yephremov, A., Nawrath, C., and Schreiber, L. (2005). Apoplastic polyesters in Arabidopsis surface tissues - A typical suberin and a particular cutin. Phytochemistry 66 2643–2658. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2000). Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 44 245–253. [DOI] [PubMed] [Google Scholar]
- Gavrieli, Y. (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanevati, M., and Jaworski, J.G. (2002). Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur. J. Biochem. 269 3531–3539. [DOI] [PubMed] [Google Scholar]
- Han, M.S., Park, S.Y., Shinzawa, K., Kim, S., Chung, K.W., Lee, J.H., Kwon, C.H., Lee, K.W., Park, C.K., and Chung, W.J. (2007). Lysophosphatidylcholine as a death effector in lipoapoptosis of hepatocytes. J. Lipid Res. 49 84–97. [DOI] [PubMed] [Google Scholar]
- Hannoufa, A., Mcnevin, J., and Lemieux, B. (1993). Epicuticular waxes of eceriferum mutants of Arabidopsis thaliana. Phytochemistry 33 851–855. [Google Scholar]
- Hardham, A.R., Jones, D.A., and Takemoto, D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10 342–348. [DOI] [PubMed] [Google Scholar]
- Hishikawa, D., Shindou, H., Kobayashi, S., Nakanishi, H., Taguchi, R., and Shimizu, T. (2008). Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA 105 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlousek-Radojcic, A., Evenson, K.J., Jaworski, J.G., and Post-Beittenmiller, D. (1998). Fatty acid elongation is independent of acyl-Coenzyme A synthetase activities in leek and Brassica napus. Plant Physiol. 116 251–258. [Google Scholar]
- Hofius, D., Tsitsigiannis, D.I., Jones, J.D.G., and Mundy, J. (2007). Inducible cell death in plant immunity. Semin. Cancer Biol. 17 166–187. [DOI] [PubMed] [Google Scholar]
- Holk, A., Rietz, S., Zahn, M., Quader, H., and Scherer, G.F.E. (2002). Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker, T.S., Millar, A.A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp, M., Kopczak, S.D., Horejsi, T.F., Kihl, B.K., and Pruitt, R.E. (1995). Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 8 703–714. [DOI] [PubMed] [Google Scholar]
- James, D.W., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H.K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks, M.A., Tuttle, H.A., Eigenbrode, S.D., and Feldmann, K.A. (1995). Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 108 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M., and Dangl, J.L. (1996). Logjam at the Styx: Programmed cell death in plants. Trends Plant Sci. 1 114–119. [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter, T., and Sandhoff, K. (1996). Inhibitors of glycosphingolipid biosythesis. Chem. Soc. Rev. 25 371–381. [Google Scholar]
- Kroemer, G., Petit, P., Zamzami, N., Vayssiere, J.L., and Mignotte, B. (1995). The biochemistry of programmed cell-death. FASEB J. 9 1277–1287. [DOI] [PubMed] [Google Scholar]
- Kunst, L., Taylor, D.C., and Underhill, E.W. (1992). Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 30 425–434. [Google Scholar]
- Kurdyukov, S., Faust, A., Nawrath, C., Bar, S., Voisin, D., Efremova, N., Franke, R., Schreiber, L., Saedler, H., Metraux, J.P., and Yephremov, A. (2006. b). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov, S., Faust, A., Trenkamp, S., Bar, S., Franke, R., Efremova, N., Tietjen, K., Schreiber, L., Saedler, H., and Yephremov, A. (2006. a). Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224 315–329. [DOI] [PubMed] [Google Scholar]
- La Camera, S., Geoffroy, P., Samaha, H., Ndiaye, A., Rahim, G., Legrand, M., and Heitz, T. (2005). A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J. 44 810–825. [DOI] [PubMed] [Google Scholar]
- Larson, T.R., Edgell, T., Byrne, J., Dehesh, K., and Graham, I.A. (2002). Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J. 32 519–527. [DOI] [PubMed] [Google Scholar]
- Larson, T.R., and Graham, I.A. (2001). A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 25 115–125. [DOI] [PubMed] [Google Scholar]
- Liang, H., Yao, N., Song, L.T., Luo, S., Lu, H., and Greenberg, L.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger, L.L., Ory, D.S., and Schaffer, J.E. (2001). Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 276 14890–14895. [DOI] [PubMed] [Google Scholar]
- Listenberger, L.L., and Schaffer, J.E. (2002). Mechanisms of lipoapoptosis: Implications for human heart disease. Trends Cardiovasc. Med. 12 134–138. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Berlyn, G.P., Engstrom, E.M., Krolikowski, K.A., Reiter, W.D., and Pruitt, R.E. (1997). Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 189 311–321. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Cheung, A.Y., and Sussex, I.M. (1992). Fiddlehead - An Arabidopsis mutant constitutively expressing an organ fusion program that involves interactions between epidermal cells. Dev. Biol. 152 383–392. [DOI] [PubMed] [Google Scholar]
- McLachlan, C.S., Yin, J.L., Driussi, C., Jusuf, P.R., Hambly, B., and McGuire, M.A. (2000). A semi-quantitative PCR method for the detection of low levels of apoptotic DNA fragmentation in a heart failure model. Jpn. J. Physiol. 50 281–284. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12 121–131. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Wrischer, M., and Kunst, L. (1998). Accumulation of very-long-chain fatty acids in membrane glycerolipids is associated with dramatic alterations in plant morphology. Plant Cell 10 1889–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, A., Welti, R., and Shah, J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W.G., and Kastan, M.B. (1994). DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol. Cell. Biol. 14 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Ruberti, I., Morelli, G., and Aoyama, T. (2002). Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 29 359–369. [DOI] [PubMed] [Google Scholar]
- O'Neill, C.M., Gill, S., Hobbs, D., Morgan, C., and Bancroft, I. (2003). Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64 1077–1090. [DOI] [PubMed] [Google Scholar]
- Park, J., Gu, Y., Lee, Y., Yang, Z., and Lee, Y. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related Small G Protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S., Gable, K., Beaudoin, F., Cahoon, E., Jaworski, J., Napier, J.A., and Dunn, T.M. (2006). Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281 9018–9029. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E., Vielle-Calzada, J.P., Ploense, S.E., Grossniklaus, U., and Lolle, S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S., Vailleau, F., Leger, A., Joubes, J., Miersch, O., Huard, C., Blee, E., Mongrand, S., Domergue, F., and Roby, D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, W.W., and Pfeiffer, D.R. (1986). The effect of Ca2+ and acyl coenzyme A: Lysophospholipid acyltransferase inhibitors on permeability properties of the liver mitochondrial inner membrane. J. Biol. Chem. 261 14018–14024. [PubMed] [Google Scholar]
- Rogers, H.J. (2005). Cell death and organ development in plants. Curr. Top. Dev. Biol. 71 225–261. [DOI] [PubMed] [Google Scholar]
- Ryu, S.B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9 229–235. [DOI] [PubMed] [Google Scholar]
- Sayanova, O., Haslam, R., Venegas Caleron, M., and Napier, J.A. (2007). Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: Identification of an acyl-Coenzyme A C20 Δ5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol. 144 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hülskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraudner, M., Moeder, W., Wiese, C., Camp, W.V., Inze, D., Langebartels, C., and Sandermann, H. (1998). Ozone-induced oxidative burst in the ozone biomonitor plant, tobacco Bel W 3. Plant J. 16 235–245. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2005). Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 43 229–260. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25 563–574. [DOI] [PubMed] [Google Scholar]
- Shi, L., Bielawski, J., Mu, J., Dong, H., Teng, C., Zhang, J., Yang, X., Tomishige, N., Hanada, K., and Hannun, Y.A. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17 1030–1040. [DOI] [PubMed] [Google Scholar]
- Spassieva, S.D., Markham, J.E., and Hille, J. (2002). The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32 561–572. [DOI] [PubMed] [Google Scholar]
- Sperling, P., Franke, S., Luthje, S., and Heinz, E. (2005). Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43 1031–1038. [DOI] [PubMed] [Google Scholar]
- Ståhl, U., Stålberg, K., Stymne, S., and Ronne, H. (2007). A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett. 582 305–309. [DOI] [PubMed] [Google Scholar]
- Staiger, K., Staiger, H., Weigert, C., Haas, C., Haring, H.-U., and Kellerer, M. (2006). Saturated, but not unsaturated, fatty acids induce apoptosis of human coronary artery endothelial cells via nuclear factor-κB activation. Diabetes 55 3121–3126. [DOI] [PubMed] [Google Scholar]
- Staley, K., Blaschke, A.J., and Chun, J. (1997). Apoptotic DNA fragmentation is detected by a semiquantitative ligation-mediated PCR of blunt DNA ends. Cell Death Differ. 4 66–75. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171. [DOI] [PubMed] [Google Scholar]
- Tamura, M., Ajayi, T., Allmond, L.R., Moriyama, K., Wiener-Kronish, J.P., and Sawa, T. (2004). Lysophospholipase A activity of Pseudomonas aeruginosa type III secretory toxin ExoU. Biochem. Biophys. Res. Commun. 316 323–331. [DOI] [PubMed] [Google Scholar]
- Trenkamp, S., Martin, W., and Tietjen, K. (2004). Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. USA 101 11903–11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S., Gallois, P., and Brown, D. (2007). Tracheary element differentiation. Annu. Rev. Plant Biol. 58 407–433. [DOI] [PubMed] [Google Scholar]
- Vailleau, F., Daniel, X., Tronchet, M., Montillet, J.-L., Triantaphylidès, C., and Roby, D. (2002). A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 99 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem, F., and Dat, J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn, W.G., and Woltering, E.J. (2005). Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10 117–122. [DOI] [PubMed] [Google Scholar]
- Walker, P.R., Carson, C., Leblanc, J., and Sikorska, M. (2002). Labeling DNA damage with terminal transferase: Applicability, specificity, and limitations. Methods Mol. Biol. 203 3–19. [DOI] [PubMed] [Google Scholar]
- Wang, W., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 20 3163–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H.-E., Rajashekar, C.B., Williams, T.D., and Wang, X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced changes in Arabidopsis. J. Biol. Chem. 277 31994–32002. [DOI] [PubMed] [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., Sugiura, T., and Waku, K. (1997). Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and mtabolism of bioactive lipids in mammalian cells. J. Biochem. 122 1–16. [DOI] [PubMed] [Google Scholar]
- Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.