Abstract
The specification of cell fates during development requires precise regulatory mechanisms to ensure robust cell type patterns. Theoretical models of pattern formation suggest that a combination of negative and positive feedback mechanisms are necessary for efficient specification of distinct fates in a field of differentiating cells. Here, we examine the role of the R2R3-MYB transcription factor gene, AtMYB23 (MYB23), in the establishment of the root epidermal cell type pattern in Arabidopsis thaliana. MYB23 is closely related to, and is positively regulated by, the WEREWOLF (WER) MYB gene during root epidermis development. Furthermore, MYB23 is able to substitute for the function of WER and to induce its own expression when controlled by WER regulatory sequences. We also show that the MYB23 protein binds to its own promoter, suggesting a MYB23 positive feedback loop. The localization of MYB23 transcripts and MYB23-green fluorescent protein (GFP) fusion protein, as well as the effect of a chimeric MYB23-SRDX repressor construct, links MYB23 function to the developing non-hair cell type. Using mutational analyses, we find that MYB23 is necessary for precise establishment of the root epidermal pattern, particularly under conditions that compromise the cell specification process. These results suggest that MYB23 participates in a positive feedback loop to reinforce cell fate decisions and ensure robust establishment of the cell type pattern in the Arabidopsis root epidermis.
INTRODUCTION
Transcription factor networks orchestrate the temporal and spatial expression of genes to precisely direct developmental and metabolic events. The gene network controlling root epidermis cell fate in Arabidopsis thaliana represents a particularly well-studied example of a transcription factor network in plant development (Schiefelbein and Lee, 2006; Schellmann et al., 2007; Ishida et al., 2008). This network includes sets of transcription factors that promote the specification of the root hair or the non-hair cell types via a complex array of transcriptional regulatory events.
At the core of the root epidermis regulatory network is a putative transcription factor complex including a MYB-type transcription factor (WEREWOLF [WER]), basic helix-loop-helix (bHLH)-type transcription factors (GLABRA3 [GL3] and ENHANCER OF GLABRA3 [EGL3]), and a WD-repeat protein (TRANSPARENT TESTA GLABRA1 [TTG1]) (Galway et al., 1994; Lee and Schiefelbein, 1999; Bernhardt et al., 2003). This complex directs the specification of the non-hair cell fate, in part, by promoting expression of the homeodomain-leucine zipper transcription factor gene, GLABRA2 (GL2) (Masucci et al., 1996; Lee and Schiefelbein, 1999). In addition, this complex indirectly specifies the hair cell type by promoting the expression of one-repeat MYB genes, including CAPRICE (CPC) (Wada et al., 1997; Lee and Schiefelbein, 2002; Koshino-Kimura et al., 2005; Ryu et al., 2005) and two CPC-related genes, TRY (TRIPTYCHON) (Schellmann et al., 2002) and ETC1 (ENHANCER OF TRY AND CPC1) (Kirik et al., 2004; Simon et al., 2007). The one-repeat MYB proteins inhibit the formation and/or activity of the WER-GL3/EGL3-TTG1 complex, likely due to competition between the one-repeat MYBs and the WER protein (an R2R3 MYB) for binding to the GL3/EGL3 bHLH proteins (Wada et al., 2002; Bernhardt et al., 2003; Esch et al., 2003; Tominaga et al., 2007). This inhibition preferentially occurs in cells adjacent to the WER-GL3/EGL3-TTG1–bearing cells, due to cell-to-cell movement of the one-repeat MYBs (Wada et al., 2002; Kurata et al., 2005; Simon et al., 2007). This has led to a lateral inhibition model to explain cell fate specification, whereby the WER-GL3/EGL3-TTG1 complex specifies the non-hair cell fate through local induction of GL2 and induces the hair cell fate through the production and long-range action of CPC/TRY/ETC1 (Lee and Schiefelbein, 2002; Schellmann et al., 2007; Ishida et al., 2008).
The cell types in the Arabidopsis root epidermis are arranged in a position-dependent pattern. Root hair cells are formed over the anticlinal wall of underlying cortical cells (H position) and non-hair cells over the periclinal wall of underlying cortical cells (N position) (Dolan et al., 1993, 1994; Galway et al., 1994). This pattern is dependent on SCRAMBLED (SCM), a leucine-rich repeat receptor-like kinase, which appears to detect positional cues and preferentially inhibit WER expression in the H cell position (Kwak et al., 2005; Kwak and Schiefelbein, 2007). As a result, the WER-GL3/EGL3-TTG1 complex is preferentially active in the N cell position, which causes these cells to adopt the non-hair fate and, via lateral inhibition mediated by the one-repeat MYBs, prevents cells in the H cell position from adopting this fate (Schiefelbein and Lee, 2006; Ishida et al., 2008).
Theoretical studies of pattern formation suggest that, in addition to lateral inhibition, a local autoregulatory positive feedback loop is required to reinforce initial cell fate decisions and to provide robustness to resist alterations from external influences (Gierer and Meinhardt, 1972; Freeman, 2000; Meinhardt and Gierer, 2000; Benitez et al., 2007). However, to date, experimental evidence for a such a positive feedback loop has not been identified in the gene network governing root epidermal cell specification.
To further our understanding of the root epidermal regulatory network, we investigated the potential role of the MYB23 gene, which encodes a MYB domain transcription factor protein closely related to WER. Together with the trichome-specific GLABROUS1 (GL1), WER and MYB23 constitute a subgroup of R2R3-MYB proteins that share ∼95% amino acid sequence identity in the MYB motifs (which function in DNA binding and bHLH partner protein interaction [Williams and Grotewold, 1997; Zimmermann et al., 2004]) and ∼65% in a C-terminal domain responsible for transcriptional activation (Lee and Schiefelbein, 2001; Stracke et al., 2001). MYB23, but not GL1, is expressed in Arabidopsis roots within the cell division and differentiation zone (Kirik et al., 2001), and expression of a chimeric MYB23-EAR repressor protein induces ectopic root hair cells and reduced root length (Matsui et al., 2005). These findings imply a possible role for MYB23 in root development. Here, we show that MYB23 is preferentially expressed in epidermal cells in the N position and is positively regulated by WER. Furthermore, we demonstrate that the MYB23 protein is functionally equivalent to the WER protein, binds in a sequence-specific manner to the MYB23 promoter, induces expression of the MYB23 gene, and is required for complete epidermal pattern formation. These results indicate that MYB23 provides a positive feedback loop in the cell fate specification network in the root epidermis.
RESULTS
Expression of MYB23 in the Root Epidermis
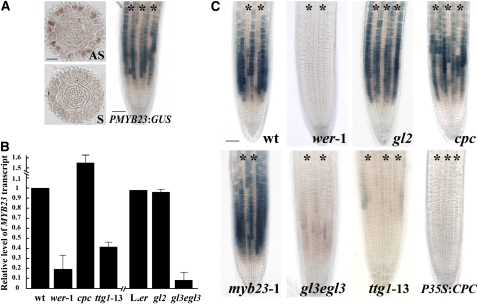
To begin to understand the role of MYB23 in root epidermis development, we examined its expression by performing RNA in situ hybridization and reporter gene fusion experiments. The in situ RNA hybridization was performed using a digoxigenin (DIG)-labeled MYB23 gene-specific antisense probe applied to transverse sections from the developing root. We detected the strongest signal in the N-position cells in the root epidermis (Figure 1A). This indicates that MYB23 RNA preferentially accumulates in the differentiating non-hair cells, a pattern similar to that seen for the transcript accumulation of epidermal patterning genes such as WER, CPC, and GL2 (Masucci et al., 1996; Lee and Schiefelbein, 1999, 2002; Wada et al., 2002).
Figure 1.
Expression of MYB23 in the Arabidopsis Root.
(A) Localization of MYB23 mRNA and PMYB23:GUS reporter expression. The left panels show in situ RNA hybridization of transverse sections from the developing root tip using antisense RNA (AS) and sense RNA (S) probes to the MYB23 gene sequence. Bar = 20 μm. The right panel shows a whole-mount view of the root tip of the PMYB23:GUS line stained for GUS activity using the 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) substrate. Bar = 50 μm.
(B) Relative levels of the MYB23 transcript in wild-type and mutant roots determined by quantitative real-time PCR analysis. Error bars represent sd values from three to six biological replicates each consisting of three technical replicates.
(C) Wild-type and mutant roots bearing the PMYB23:GUS transgene and stained for GUS activity. Bar = 50 μm.
Asterisks in (A) and (C) indicate the H-position cells. [See online article for color version of this figure.]
To define the spatial pattern of MYB23 promoter activity, plants harboring a MYB23 promoter:β-glucuronidase reporter (PMYB23:GUS) transgene (Kirik et al., 2001) were examined. This line was previously reported to exhibit GUS expression in the cell division and differentiation zones of the root, but it was not analyzed in detail (Kirik et al., 2001). We discovered that PMYB23:GUS is preferentially expressed in the N-position cells in the developing root epidermis, resulting in a non-hair cell file pattern (Figure 1A). This expression profile, which displays maximal GUS activity in epidermal cells in the late meristematic zone, is most similar to that of GL2 and CPC (Masucci et al., 1996; Lee and Schiefelbein, 2002; Wada et al., 2002). This pattern is distinct from the WER expression pattern (Lee and Schiefelbein, 1999) because it initiates slightly later in development than WER, it is limited to the epidermis, and it is not detectable in the H cell position. Together, these results show that the MYB23 promoter activity and MYB23 transcript accumulation patterns are similar and occur preferentially in cells destined to adopt the non-hair cell fate in the root epidermis.
WER and Other Known Cell Fate Regulators Control MYB23 Expression
The WER MYB protein is known to be a positive regulator of several genes expressed in the developing non-hair cells, including GL2, CPC, and ETC1 (Lee and Schiefelbein, 1999; Simon et al., 2007). To determine whether WER also influences MYB23 gene expression, we examined MYB23 RNA accumulation and PMYB23:GUS activity in roots of the wer mutant. Quantitative real-time PCR analysis showed that the steady state level of MYB23 RNA in the wer-1 mutant root is reduced to ∼19% of the wild-type root level (Figure 1B). Expression of the PMYB23:GUS transgene was nearly undetectable in the wer-1 mutant roots (Figure 1C). These results indicate that WER induces MYB23 expression in the developing root epidermis.
To examine MYB23 regulation more broadly, we analyzed MYB23 transcript accumulation and PMYB23:GUS expression in various mutants defective in root epidermal cell specification. We discovered that, like wer, the ttg1 and gl3 egl3 mutant roots accumulate a significantly lower level of MYB23 RNA (42 and 10% of the wild-type level is present in the ttg1-13 and gl3-1 egl3-1 roots, respectively) (Figure 1B). Consistent with this, the ttg1-13 and gl3-1 egl3-1 mutant roots display a reduced amount of PMYB23:GUS expression, although the N cell expression pattern was retained (Figure 1C). By contrast, the cpc-1 mutant exhibited an increased level of MYB23 transcripts (∼56% greater than the wild type) (Figure 1B). This higher level of MYB23 RNA seems to be due to MYB23 promoter activity in some of the H-position epidermal cells of cpc-1 (Figure 1C).
To further assess the involvement of CPC, a line overexpressing CPC (P35S:CPC) was analyzed for its effect on PMYB23:GUS, and it was found to lack MYB23 promoter activity (Figure 1C). In the roots of the gl2-1 mutant, the MYB23 transcript level and PMYB23:GUS expression were indistinguishable from those in the wild-type root (Figures 1B and 1C). These results suggest that the MYB23 gene is positively regulated by the non-hair cell–promoting WER-GL3/EGL3-TTG1 complex, negatively regulated by the hair cell–promoting CPC protein, and unaffected by the downstream GL2 homeodomain protein.
The WER Protein Binds to the MYB23 Promoter
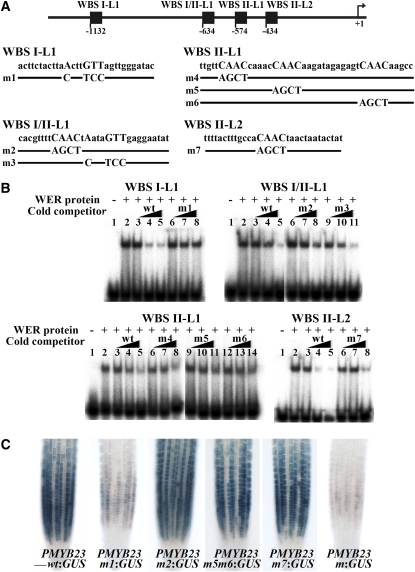
Recently, we and others have shown that the WER protein directly regulates CPC transcription through its binding to three sites in the CPC promoter, designated WBSI, WBSII/CPCMBSI, and CPCMBSII (Koshino-Kimura et al., 2005; Ryu et al., 2005). The core sequence of WBSI is ANNNGTT, and the core sequence of WBSII/CPCMBSI and CPCMBSII is CAAC. To explore the possibility that WER regulates MYB23 by binding directly to its promoter, we searched for WBSI-like (WBSI-L) or WBSII-like (WBSII-L) sequences in the MYB23 promoter. A total of nine WBSI-Ls and seven WBSII-Ls were identified within the 2-kb 5′ flanking region of MYB23 (which is the region sufficient to induce the normal expression of MYB23 based on the PMYB23wt:GUS reporter gene expression and myb23 mutant rescue experiments [Figure 2C; see Supplemental Figure 1 online]), and these were tested for WER binding in an electrophoretic mobility shift assay (EMSA). Using DNA fragments (29 to 45 nucleotides long) generated from each of these 16 sites, we detected binding of the WER protein to four sites. These are designated WBSI-L1, WBSI/II-L1, WBSII-L1, and WBSII-L2 and are located ∼1130, 630, 570, and 430 bp upstream of the MYB23 translation start site, respectively (Figure 2A).
Figure 2.
WER Binds to Specific Sequences in the MYB23 Promoter.
(A) Location of WER binding fragments in the MYB23 promoter. Fragments are shown relative to the start of translation (+1) (top), and the DNA sequence in wild-type and mutant versions of these fragments are shown at bottom.
(B) EMSA. Assays used WER protein exposed to radiolabeled wild-type DNA probes with or without various cold competitors. Competitions were performed using increasing amounts of wild-type DNA fragments or mutated derivatives as shown in A. Lane 1 of each gel contains the DNA probes without the WER protein, and lane 2 contains the radiolabeled wild-type DNA probes and the WER protein without a competitor. Increasing amounts (1×, 10×, and 50×) of the unlabeled wild-type DNA fragments (wt; lanes 3, 4, and 5, respectively) or the unlabeled mutated versions (m; lanes 6, 7, and 8, lanes 9, 10, and 11, and lanes 12, 13, and 14) were added as a cold competitor.
(C) Importance of the WER binding sites for the proper expression of MYB23 in the Arabidopsis root epidermis. For reporter constructs, several versions of 2-kb-long MYB23 promoters that have a mutation at the indicated sites (PMYB23m1, PMYB23m2, PMYB23m5m6, PMYB23m7, and PMYB23m for mutation at WBSI-L1, WBSI/II-L1, WBSII-L1, WBSII-L2, and at all four sites, respectively, as shown in A) were fused to a GUS reporter gene. Bar = 50 μm.
[See online article for color version of this figure.]
EMSA competition experiments were conducted to assess the importance of the core sequences for WER binding to these fragments. For the WBSI-L1, WER binding was not greatly reduced in the presence of competitor DNA fragments with a mutated core sequence (m1) as compared with the effect of wild-type competitor fragments (Figure 2B), which indicates sequence-specific binding of WER to this site. For WBSII-L2, the mutated fragment (m7) was able to compete with the probe DNA, but much less efficiently than the wild-type competitor (Figure 2B), perhaps indicating that flanking sequences may also function in efficient binding of the WER protein. The WBSI/II-L1 site contains the core sequence of WBSI (ANNNGTT) and the core sequence of WBSII (CAAC) juxtaposed to each other (Figure 2A). Thus, we tested two mutated versions of this site (m2, mutated in the WBSII-like sequence; m3, mutated in the WBSI-like sequence) and found that m2 did not compete with the wild-type fragment while m3 could compete with the wild-type fragment (Figure 2B). This suggests that only the WBSII-like sequence in this region is important for WER binding. The WBSII-L1 site contains three closely linked CAAC core sequences (Figure 2A). To define the core sequence(s) important for WER binding, we mutated each of these (m4, mutated in the first CAAC; m5, mutated in the second CAAC; m6, mutated in the third CAAC) and used them in EMSA. WER binding to the wild-type WBSII-L1 site was not affected by the m5 and m6 fragments, but the m4 fragment significantly reduced the WER-WBSII-L1 binding (Figure 2B), indicating that the second and third core sequences are required for effective WER protein binding to this site, while the first core sequence is less important.
To further validate these WER binding sites in the MYB23 promoter, we made stable transgenic lines containing the GUS reporter gene fused to the wild-type version or mutated versions of the 2-kb-long MYB23 promoter (Figure 2C). While the wild-type version (PMYB23wt:GUS) showed strong GUS activity in the N-position epidermal cells, the reporter gene mutated at the WBSI-L1 site (PMYB23m1:GUS) showed very low activity. The reporter genes mutated at the other three sites individually (PMYB23m2:GUS, PMYB23m5m6:GUS, and PMYB23m7:GUS), by contrast, did not show a significant difference in their GUS activities compared with the wild-type version. However, when all four mutations were combined (PMYB23m:GUS), the reporter gene activity was lower than the activity of the PMYB23m1:GUS (Figure 2C). These results show that the WBSI-L1 plays an important role in the proper expression of MYB23 and that WBSI/II-L1, WBSII-L1, and WBSII-L2 exert a minor combined role.
MYB23 Localizes to the Nucleus of N-Position Epidermal Cells
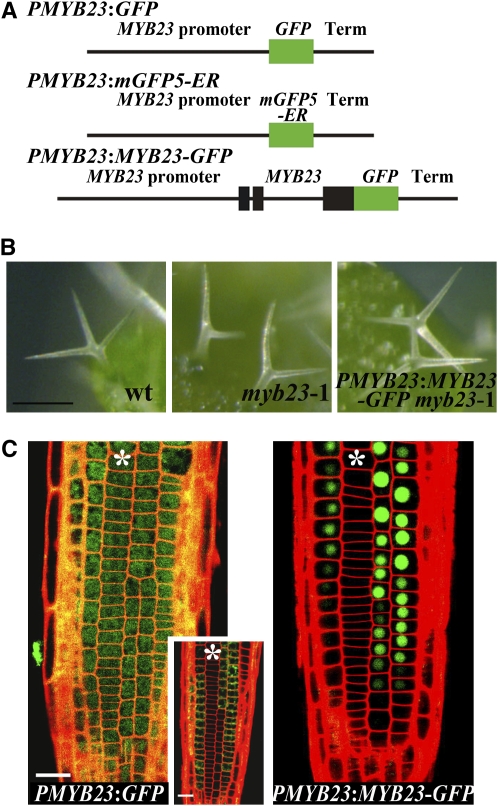
As a predicted transcription factor protein produced from N cell–specific transcripts, MYB23 may be expected to localize and act within the nucleus of cells in the N position of the root epidermis. On the other hand, some transcription factors participating in root epidermal cell specification (e.g., CPC and GL3) are able to move from cell to cell (Bernhardt et al., 2005; Kurata et al., 2005). To define the location of the MYB23 protein, we generated and analyzed a transgenic line bearing an in-frame MYB23-GFP (for green fluorescent protein) reporter translational fusion (PMYB23:MYB23-GFP) (Figure 3A). This transgene was expressed under the control of MYB23 regulatory sequences (3.1-kb 5′ flanking DNA fragment and 1.0-kb 3′ flanking DNA fragment) previously determined to be sufficient in genomic complementation experiments to rescue the trichome-branching defect in the myb23 mutant (Kirik et al., 2005).
Figure 3.
Subcellular Localization of the MYB23 Protein.
(A) Structures of the MYB23 translational and transcriptional GFP fusion constructs.
(B) Complementation of the trichome-branching defect of myb23-1 using the MYB23 translational fusion construct. Bar = 100 μm.
(C) GFP expression in the developing root epidermis of plants bearing a PMYB23:GFP transcriptional fusion (inset, PMYB23:mGFP5-ER transcriptional fusion) or PMYB23:MYB23-GFP translational fusion. Asterisks indicate the H-position cells. Bar = 20 μm.
[See online article for color version of this figure.]
We found that this translational fusion was able to rescue the branching defect in the myb23 mutant trichomes (Figure 3B), indicating that the MYB23-GFP protein functions like the native MYB23 in Arabidopsis. Analysis of the PMYB23:MYB23-GFP plants showed that GFP fluorescence was present in the nucleus of the developing root epidermal cells located in the N position (Figure 3C). As a negative control, we generated a GFP transcriptional fusion driven by the same regulatory sequences that were used for the PMYB23:MYB23-GFP construction (PMYB23:GFP), which exhibited fluorescence generally distributed throughout the cells (Figure 3C). This overall distribution seems to be caused by the nonspecific diffusion of the GFP, which is expressed in the N-position cells, based on the fact that the fluorescent signal from the endoplasmic reticulum (ER)–targeted GFP driven by the same regulatory sequences is found only in the N-position cells (Figure 3C, inset). These results indicate that the MYB23 protein accumulates in the nuclei of the developing non-hair cells and does not move from cell to cell.
MYB23 Can Substitute for WER
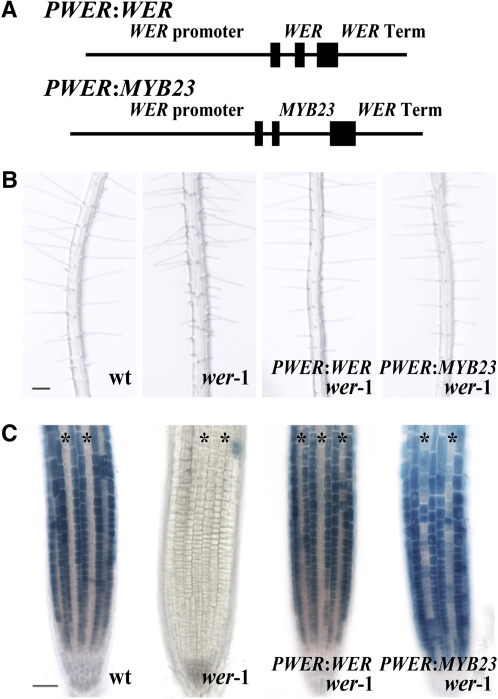
The MYB23 protein is structurally similar to WER, exhibiting 57% amino acid sequence identity overall as well as 95% identity in their N-terminal R2R3 MYB domains and 68% identity in their C-terminal transcriptional activation domains (Lee and Schiefelbein, 1999). To determine whether the MYB23 protein is functionally similar to WER in root epidermal cell specification, we tested the ability of the MYB23 coding region, driven by the WER regulatory sequences, to complement the wer mutant. Based on our prior study of WER regulation (Lee and Schiefelbein, 2001), we knew that a 4.0-kb fragment 5′ to the start codon and a 1.1-kb fragment 3′ to the stop codon together represented the cis-regulatory sequences sufficient to direct normal WER gene function (Table 1). These same 5′ and 3′ WER regulatory sequences were fused to the MYB23 coding sequences to generate the PWER:MYB23 construct (Figure 4A). When this construct was introduced into a wer-1 mutant background, the resulting PWER:MYB23 wer-1 plants showed normal position-dependent cell type patterning in the root epidermis, in contrast with the excessive hair cell specification in the wer mutant (Figure 4B; Table 1).
Table 1.
Specification of Cell Types in the Root Epidermis
| Genotype | H Position Hair Cell (%) | Non-hair Cell (%) | N Position Hair Cell (%) | Non-hair Cell (%) |
|---|---|---|---|---|
| Wild type | 98.1 ± 0.8 | 1.9 ± 0.8 | 1.7 ± 1.0 | 98.3 ± 1.0 |
| wer-1 | 99.8 ± 0.4 | 0.2 ± 0.4 | 97.8 ± 0.6 | 2.2 ± 0.6 |
| PWER:WER wer-1 | 95.2 ± 4.9 | 4.8 ± 4.9 | 1.4 ± 4.2 | 98.6 ± 4.2 |
| PWER:MYB23 wer-1 | 99.0 ± 0.5 | 1.0 ± 0.5 | 3.5 ± 1.5 | 96.5 ± 1.5 |
| myb23-1 | 97.4 ± 2.0 | 2.6 ± 2.0 | 6.0 ± 0.2 | 94.0 ± 0.2 |
| myb23-3 | 99.2 ± 0.5 | 0.8 ± 0.5 | 4.7 ± 1.0 | 95.3 ± 1.0 |
| cpc | 28.5 ± 0.9 | 71.5 ± 0.9 | 0.1 ± 0.2 | 99.9 ± 0.2 |
| cpc myb23-1 | 41.8 ± 3.6 | 58.2 ± 3.6 | 0.2 ± 0.2 | 99.8 ± 0.2 |
| MYB23-SRDX | 99.8 ± 0.3 | 0.2 ± 0.3 | 62.2 ± 14.3 | 37.8 ± 14.3 |
At least sixty 4-d-old seedlings were examined for each plant line. Values indicate means ± sd.
Figure 4.
MYB23 Can Substitute for WER Function.
(A) Schematic representation of constructs containing the WER or MYB23 coding sequences and introns fused to the same WER 5′ and 3′ regulatory sequences.
(B) Four-day-old seedlings illustrating root hair production. Bar = 100 μm.
(C) Four-day-old seedlings harboring the PGL2:GUS transgene were stained for GUS activity. Asterisks indicate the H-position cells. Bar = 50 μm.
[See online article for color version of this figure.]
To further define the molecular basis of this complementation, we examined the expression of the PGL2:GUS reporter gene in these lines (Figure 4C). WER is well known to be required for the position-dependent expression of GL2 (Lee and Schiefelbein, 1999, 2002), so that GUS activity was hardly detected in the wer mutant roots harboring PGL2:GUS. The expression pattern of PGL2:GUS was completely restored in the PWER:MYB23 wer-1 line as well as in the PWER:WER wer-1 line, which is consistent with the normal cell type patterning in these two lines (Figure 4B; Table 1). These data indicate that the MYB23 protein is biochemically similar to WER for regulating cell fate in the root epidermis.
A Dominant Negative Form of MYB23 Affects MYB23, GL2, and CPC Gene Expression
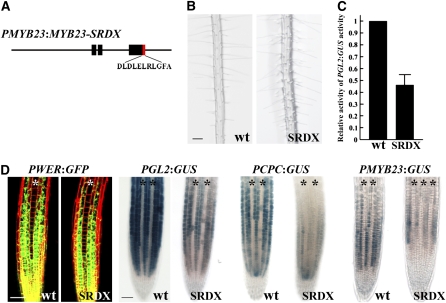
The ability of the MYB23 protein to substitute for the WER protein in root epidermal cell specification implies that MYB23 is able to regulate the same set of genes as WER regulates, such as the GL2, CPC, and MYB23 genes. To examine this in a different manner, we converted the MYB23 protein into a transcriptional repressor by fusing the modified repressor domain of SUPERMAN (SRDX; LDLDLELRLGFA [Hiratsu et al., 2003]) in frame to the 3′ end of the MYB23 coding region (Figure 5A). This MYB23-SRDX fusion was placed under the control of the MYB23 regulatory sequences, as defined above (3.1 kb 5′ to the start codon and 1.0 kb 3′ to the stop codon), to generate the PMYB23:MYB23-SRDX construct. Plants harboring this construct exhibited excessive root hair cell specification, due to root hair cells produced in the N position (Figure 5B; Table 1). This result is consistent with the previously reported effect of a P35S:MYB23-SRDX construct, which causes essentially all root epidermal cells to adopt the hair cell fate and also affects root elongation (Matsui et al., 2005).
Figure 5.
Effect of a Dominant Negative Form of MYB23.
(A) Structure of the MYB23 translational fusion constructs with the SRDX repressor domain.
(B) Root hair phenotype for the wild-type and PMYB23:MYB23-SRDX lines. Bar = 100 μm.
(C) Relative activity of the GL2 promoter in the root tips of wild-type and PMYB23:MYB23-SRDX lines. GUS activity in the root tips of the wild type or the PMYB23:MYB23-SRDX line harboring PGL2:GUS was quantitatively analyzed from 4-d-old seedlings. At least 20 seedlings were used for each. The error bar indicates sd from four biological replicates (each biological replicate consists of three technical replicates).
(D) Expression of transcriptional reporters in the developing root epidermis of the wild-type and PMYB23:MYB23-SRDX lines. Asterisks indicate the H position. Bar = 50 μm.
[See online article for color version of this figure.]
To determine whether this phenotypic effect of MYB23-SRDX is caused by a change in GL2 expression, we analyzed GL2 promoter activity by incorporating the PGL2:GUS reporter into this line. We found a significant reduction in the level of PGL2:GUS expression in the PMYB23:MYB23-SRDX roots as compared with the wild type (Figure 5C). To further determine the effect of the MYB23-SRDX on specific gene expression in root epidermis development, we examined expression of the transcriptional reporters PWER:GFP, PGL2:GUS, PCPC:GUS, and PMYB23:GUS in the PMYB23:MYB23-SRDX line as compared with the wild type. Although PWER:GFP reporter gene expression was not altered in the PMYB23:MYB23-SRDX line, the PGL2:GUS, PCPC:GUS, and PMYB23:GUS lines exhibited a decrease in reporter expression in the developing non-hair cells, even though the basic expression patterns were not changed (Figure 5D). Together, these results suggest that the MYB23 promoter is active early enough to be involved in cell fate specification in the Arabidopsis root epidermis, even though its expression starts somewhat later than WER expression, and that MYB23 normally participates in the regulation of its own gene expression as well as the regulation of the GL2 and CPC genes.
WER Does Not Regulate Its Own Gene Expression
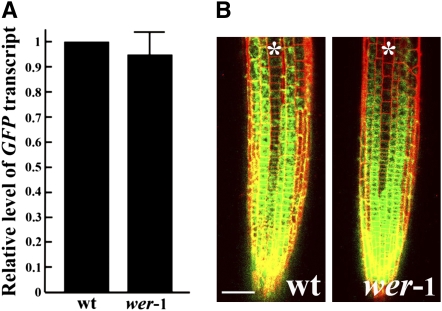
Our results here show that MYB23 expression is promoted by WER and that MYB23 is able to function in a similar manner as WER in root epidermal cell specification, which implies that MYB23 participates in a positive feedback loop in the root epidermis regulatory network. To determine whether WER might also participate in a positive feedback loop, whereby it promotes the transcription of its own gene, we analyzed WER transcript accumulation and PWER:GFP expression in the wer mutant background. Real-time PCR analysis showed that the steady state level of GFP transcripts in the wer mutant root tip was not significantly different from that in the wild type (Figure 6A). Also, the PWER:GFP expression pattern in the root tip was not altered in the wer-1 mutant (Figure 6B). This suggests that WER does not regulate its own gene expression.
Figure 6.
WER Does Not Regulate Its Own Gene Expression.
(A) Quantitative real-time PCR analysis of GFP transcript accumulation in roots of the wild type and the wer-1 mutant harboring PWER:GFP. Error bar represents sd from three biological replicates each consisting of three technical replicates.
(B) Expression pattern of the PWER:GFP transcriptional reporter fusion in the developing root tips of the wild type and the wer-1 mutant. Asterisks indicate the H position. Bar = 50 μm.
[See online article for color version of this figure.]
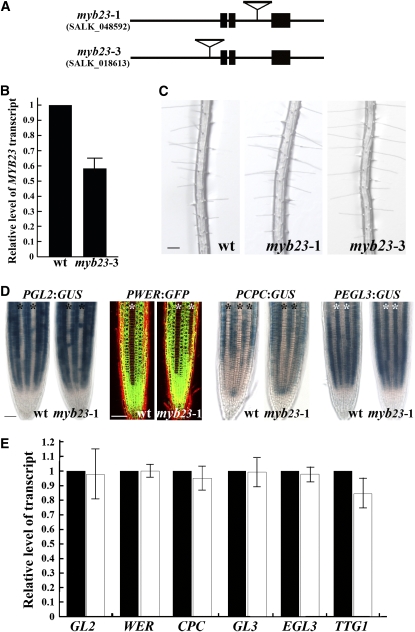
MYB23 Is Required for Complete Cell Specification in the Root Epidermis
To test the biological significance of MYB23 and its positive feedback loop in root epidermis development, we analyzed two myb23 mutant lines from the SALK T-DNA insertion collection (myb23-1 and myb23-3). The myb23-1 allele has a T-DNA insertion in its second intron and has been previously reported to have no detectable MYB23 transcripts (Figure 7A) (Kirik et al., 2005). The myb23-3 allele has an insertion in its 5′ promoter region, ∼260 bp upstream to the start codon, and we found that it has reduced accumulation of MYB23 RNA in its roots (58% of the wild type) (Figures 7A and 7B). These myb23 mutant lines appear to have a normal root hair density (Figure 7C), but a quantitative analysis shows that both of the mutant alleles, myb23-1 and myb23-3, have a small but statistically significant increase in hair cell production in the N position (ectopic hair cells; Table 1). To analyze myb23 further, we introduced the PGL2:GUS (Masucci et al., 1996), PWER:GFP (Lee and Schiefelbein, 1999), PEGL3:GUS (Zhang et al., 2003; Bernhardt et al., 2005), and PCPC:GUS (Wada et al., 2002) transcriptional reporters into the myb23-1 mutant by genetic crosses. We did not observe any significant difference in the pattern of expression for any of these transcriptional reporters in the myb23-1 mutant, as compared with the wild type (Figure 7D). Furthermore, the steady state RNA levels of the known transcriptional regulators were not significantly changed in the myb23-1 mutant roots, except for a slight reduction in TTG1 transcripts (Figure 7E). Lastly, we tested the expression pattern of the MYB23 gene in the myb23-1 mutant, using the PMYB23:GUS reporter, and discovered no difference from the wild-type pattern (Figure 1B). Together, these results suggest that MYB23 gene function plays a minor role in the normal establishment of the cell pattern and gene regulatory activities in the developing root epidermis.
Figure 7.
Phenotypic and Molecular Analysis of myb23 Mutants.
(A) Structure of the myb23-1 and myb23-3 insertion mutants.
(B) Quantitative real-time PCR analysis of MYB23 transcript levels in the wild type and the myb23-3 mutant. The error bar represents sd from three biological replicates each consisting of three technical replicates.
(C) Root hair phenotype of the wild type and myb23 mutants. Bar = 100 μm.
(D) Transcriptional reporter expression in the wild-type and the myb23-1 mutant. Asterisks indicate the H-position cells. Bar = 50 μm.
(E) Quantitative real-time PCR analysis of gene-specific RNA levels from the roots of the wild type (black bars) and the myb23-1 mutant (white bars). Error bars represent sd from four to six biological replicates each consisting of three technical replicates.
[See online article for color version of this figure.]
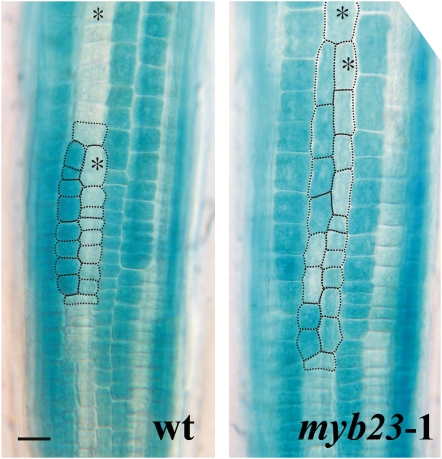
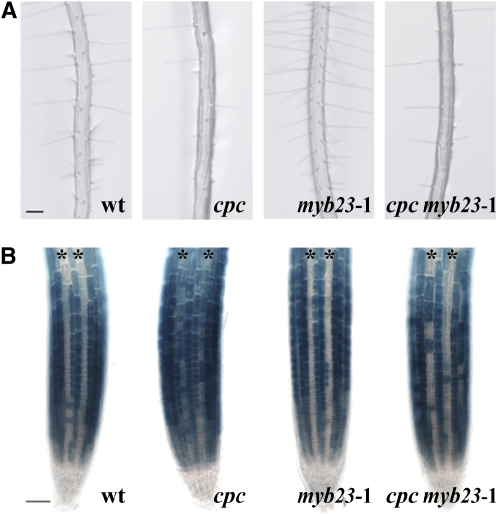
Because the myb23 mutant has relatively minor defects, we next considered the possibility that MYB23 and its positive feedback loop might help reinforce the existing cell fate determination mechanisms, so that a greater effect of myb23 might be observed under conditions where the cell specification process is compromised. We tested this possibility in two ways. First, we examined cell specification in epidermal clones in the myb23-1 mutant and the wild type. Epidermal clones result from rare longitudinal divisions usually in the H-position cells that effectively cause one of the epidermal daughter cells (and its descendants) to change their position relative to the underlying cortical cells at a relatively late stage in epidermis development (Berger et al., 1998). In the wild type, the cells in the epidermal clones are able to accurately respecify their fates in a normal position-dependent manner (Table 2). However, we discovered that epidermal clones in the myb23-1 mutant are less able to respecify their fates properly, because approximately five times more myb23 clones possess misspecified cells than do wild-type clones (Figure 8; Table 2). These misspecified cells from myb23 clones preferentially occur in the N cell files (Table 2), implying that the myb23 mutant is less able to direct the non-hair fate in cells that have switched their position from the H to the N cell position. Unlike fate specification in the early stage, which involves a gradual establishment of epidermal cell pattern through several cell divisions, fate specification in these epidermal clones requires rapid establishment of the cell fate. This could explain why the positive feedback regulation via MYB23 is more important in these epidermal clones.
Table 2.
Cell Patterning within Epidermal Clones from the Wild Type and myb23-1
| Cells Showing Abnormal PGL2:GUS Expression
|
|||
|---|---|---|---|
| Genotype | Correctly Patterned Clones (%)a | PGL2:GUSExpressing Cells in the N Position (%)b | PGL2:GUSNon-expressing Cells in the H Position (%)c |
| Wild-type PGL2:GUS | 94.4 ± 2.9 | 99.3 ± 0.6 | 99.1 ± 0.7 |
| myb23-1PGL2:GUS | 71.6 ± 5.2 | 91.6 ± 1.2 | 96.2 ± 1.1 |
Values indicate means ± sd.
Percentage of clones containing PGL2:GUS–expressing cells exclusively in the N position and PGL2:GUS non-expressing cells exclusively in the H position.
Percentage of PGL2:GUS–expressing cells among all cells in the N position in epidermal clones.
Percentage of PGL2:GUS non-expressing cells among all cells in the H position in epidermal clones.
Figure 8.
The myb23 Mutant Alters Cell Patterning in Epidermal Clones.
PGL2:GUS expression is shown in epidermal clones from the wild type (left) with normal patterning and from the myb23-1 mutant (right) with abnormal patterning. Asterisks indicate the H-position cells. Bar = 20 μm. [See online article for color version of this figure.]
As a second method to impose suboptimal cell specification conditions, we analyzed homozygous double mutant combinations that included myb23-1 and a second mutation in one of the cell specification genes. We found that the wer-1 myb23-1, ttg1-13 myb23-1, and gl3-1 egl3-1 myb23-1 mutants showed a similar root epidermis phenotype as each mutant in the absence of the myb23-1 mutation (data not shown), which was expected given these strong single mutant phenotypes and the presumed role of MYB23 in the same pathway. However, the root epidermis phenotype of the cpc-1 myb23-1 double mutant was intermediate between the cpc-1 single mutant and the wild-type pattern, showing that the myb23-1 defect partially rescued the cpc-1 mutant (Figure 9A; Table 1). Specifically, in the cpc-1 myb23-1 double mutant, 41.8% ± 3.6% of its H-position cells adopted the hair cell fate, while only 28.5% ± 0.9% of the H-position cells adopted the hair cell fate in the cpc-1 mutant (Table 1). To test whether this change is associated with a change in GL2 expression, we analyzed PGL2:GUS expression in the cpc-1 myb23-1 line (Figure 9B). The PGL2:GUS expression can be detected in the H-position cells (ectopic expression) as well as in the N-position cells in the cpc-1 mutant root (Figure 9B) (Lee and Schiefelbein, 2002), but in the cpc-1 myb23-1 double mutant, many of the H-position cells do not express PGL2:GUS. This implies that MYB23 is required for non-hair cell specification in the H cell position of the cpc-1 mutant root.
Figure 9.
The myb23 Mutation Partially Suppresses the cpc Mutant Phenotype.
(A) Root hair phenotypes of the wild type, cpc, myb23-1, and cpc myb23-1 mutants. Bar = 100 μm.
(B) PGL2:GUS expression in the root epidermis of the wild type, cpc, myb23-1, and cpc myb23-1 mutants. Asterisks indicate the H position. Bar = 50 μm.
[See online article for color version of this figure.]
MYB23 Positively Regulates Its Own Expression via Direct Binding to Its Promoter
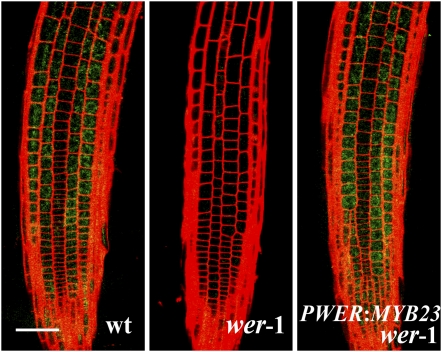
To further clarify the MYB23 positive feedback loop in cell fate specification, we examined the effect of overexpression of MYB23 on the MYB23 promoter activity. Since WER promoter activity is much greater than cauliflower mosaic virus (CaMV) 35S promoter activity in the root, we used the PWER:MYB23 construct and introduced it into the wer-1 PMYB23:mGFP5-ER line. As mentioned above, the PMYB23:mGFP5-ER expression is undetectable in the wer mutant root (Figure 10). However, when MYB23 was expressed in the wer mutant root epidermis by the WER promoter, the fluorescent signal from PMYB23:mGFP5-ER was much higher, indicating that the MYB23 promoter activity is positively regulated by the MYB23 protein.
Figure 10.
The MYB23 Protein Induces Its Expression in the Arabidopsis Root Epidermis.
The PWER:MYB23 construct was introduced into the wer-1 mutant harboring PMYB23:mGFP5-ER (see Figure 3A for this construct), and the GFP expression in the root epidermis was observed using a confocal microscope. Bar = 50 μm.
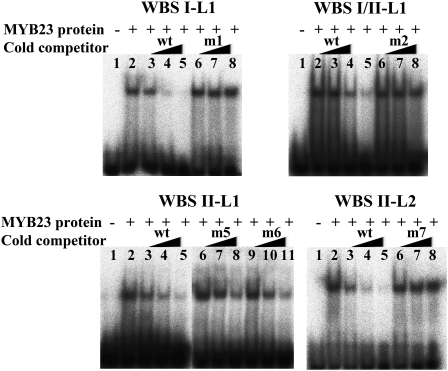
Next, we directly tested whether the MYB23 protein is able to bind to its own promoter. For this purpose, the MYB23 protein was expressed and purified from Escherichia coli and used in EMSA experiments. These competition experiments showed that MYB23 specifically binds to all four of the WER binding sites (WBSI-L1, WBSI/II-L1, WBSII-L1, and WBSII-L2) in the same manner as WER (Figure 11). Taken together, these results show that MYB23 is able to induce its own expression by directly binding to its promoter.
Figure 11.
MYB23 Binds to Specific Sequences in Its Own Promoter.
EMSA using MYB23 protein exposed to radiolabeled wild-type DNA probes with or without various cold competitors. Competitions were performed using increasing amounts of wild-type DNA fragments or mutated derivatives as shown in Figure 2A. Lane 1 of each gel contains the DNA probes without the MYB23 protein, and lane 2 contains the radiolabeled wild-type DNA probes and the MYB23 protein without a competitor. Increasing amounts (1×, 10×, and 50×) of the unlabeled wild-type DNA fragments (wt; lanes 3, 4, and 5, respectively) or the unlabeled mutated versions (m; lanes 6, 7, and 8 and lanes 9, 10, 11) were added as a cold competitor.
DISCUSSION
A MYB23 Positive Feedback Loop Enhances WER-Directed Cell Patterning
In this study, we analyzed the role of MYB23 in root epidermis development. We showed that it is expressed preferentially in the developing non-hair cells of the root epidermis, that its promoter activity and RNA accumulation are regulated by the WER, GL3/EGL3, TTG1, and CPC, and that the MYB23 protein localizes to the nucleus of the developing non-hair cells. We further showed that MYB23 protein activity is similar to that of WER, because MYB23 can functionally substitute for WER in root epidermal cell pattern formation. Also, we showed that MYB23 can bind to its own promoter in a sequence-specific manner and induce its own gene's expression. The ability of MYB23 to function like WER, to be positively regulated by WER in the developing non-hair cells, and to positively regulate its own expression by direct binding shows that MYB23 is part of a positive transcriptional feedback loop.
A positive feedback loop is a system in which the output signal returns to influence the level of the input. If the output signal increases, it increases the level of input, which results in a further increased level of the output. Theoretical work on pattern formation mechanisms have shown that positive feedback loops are an important feature, because they ensure that once a developmental decision has begun to be made, the positive feedback loop helps to lock in the decision and make it less sensitive to perturbation or noise (Gierer and Meinhardt, 1972; Brenner et al., 1981; Freeman, 2000; Page et al., 2003). In particular, positive feedback loops play important roles as binary switches between alternative cell fates to amplify a small fluctuation determining the choice. Positive feedback regulation may be indirect, with several components in a cascade feedback loop, or may be direct, with an autoactivation mechanism.
As an example of positive feedback regulation with direct autoactivation, Drosophila neuronal precursor cells express proneural genes, and self-stimulation of one of the proneural genes, scute (sc), is essential for the high accumulation of Sc, which is required for maintaining the fate of neuronal precursor cells and sense organ development (Culi and Modolell, 1998). Efforts to mathematically model the root epidermis patterning system have assumed such an autoregulatory feedback loop, even though this has not, until recently, been experimentally identified (Pesch and Hulskamp, 2004; Benitez et al., 2007). In these mathematical models, WER has been hypothesized to promote its own expression to provide the autoregulatory feedback. Here, we have shown that WER does not regulate its own transcription; rather, MYB23 participates in an autoregulatory positive feedback loop as a WER-equivalent protein to accomplish the same outcome.
The MYB23 positive feedback appears to be necessary for precise establishment of the epidermal cell pattern, based on three lines of evidence. First, myb23 mutants have a slight increase in ectopic hair cells (Table 1). Second, epidermal clones of the myb23-1 mutant exhibit a greater likelihood to possess pattern defects (Figure 8; Table 2), indicating that MYB23 is particularly important for the relatively late and rapid cell fate switching that occurs during pattern formation in these clones. Third, the myb23-1 mutation partially suppresses the cpc-1 mutant phenotype (Figure 9), suggesting that the lateral inhibition–impaired cpc mutant is dependent on MYB23 function for non-hair cell specification. This effect of myb23-1 is reminiscent of the effect of wer-1 on suppressing the cpc phenotype; the wer cpc double mutant has 81% of its H-position cells adopting the hair cell fate (Lee and Schiefelbein, 1999, 2002).
These results provide new insight into the Arabidopsis root epidermis regulatory network (Figure 12). In response to the SCM signaling pathway, WER appears to play a primary role in initiating the differential accumulation of the WER-GL3/EGL3-TTG1 complex in the N versus H cells. This complex, in turn, promotes the expression of GL2 (to specify the non-hair fate), CPC/TRY (to mediate lateral inhibition and direct adjacent cells to adopt the hair fate), and MYB23. The MYB23 protein, due to its functional equivalence to WER, is able to contribute to increased production of the WER/MYB23-GL3/EGL3-TTG complex and further promote the establishment of the cell fates. Because MYB23 is immobile (as shown by the accumulation of MYB23-GFP in the cells that express the MYB23 gene [Figure 3]), the MYB23 feedback loop ensures greater differential expression of the GL2, CPC/TRY, and MYB23 genes between the N and H cells. In this way, MYB23 is able to enhance and reinforce the initial cell fate decision.
Figure 12.
Model for the Role of MYB23 in Root Epidermal Cell Patterning.
The MYB23 is part of a positive feedback loop, because MYB23 gene transcription is promoted by a complex that includes the MYB23 protein. This WER/MYB23-GL3/EGL3-TTG1 complex determines cell pattern by specifying the non-hair fate (in cells accumulating this complex) and the hair fate (in adjacent cells). Solid lines with arrowheads indicate transcriptional regulation with positive effect, while blunt-ended lines indicate transcriptional regulation with negative effect. The dashed line for CPC/TRY indicates movement from the N-position cell to the H-position cell, and the dashed line for MYB23 indicates movement inside a cell to form a protein complex. [See online article for color version of this figure.]
Role of MYB23 in Preventing CPC/TRY Inhibition in N Cells
A long-standing unresolved issue in root epidermal cell specification has centered around the ability of the N-position cells to be unaffected by the CPC/TRY inhibitors that they produce. One possible explanation is that an inhibitor of CPC/TRY may exist in the N-position cells or an activator of CPC/TRY may be present in the H-position cells. However, CPC expression throughout the epidermis in the P35S:CPC transgenic line causes all cells to adopt the hair fate (Wada et al., 1997; Lee and Schiefelbein, 2002). A second possibility is that the CPC/TRY proteins might be activated only during translocation from one cell to another. However, this is not consistent with the ability of a movement-defective version of CPC to induce hair cell fate in N-position cells (Kurata et al., 2005). A third possibility is efficient translocation of CPC/TRY proteins out of the cells where they are produced. In support of this view, a HA-tagged CPC protein was detected preferentially in the nuclei of the H-position cells in the PCPC:HA-CPC cpc-1 line, which produced a normal epidermal cell pattern, while the 2XGFP-tagged CPC protein accumulated in all epidermal cells in the PCPC:CPC-GFP cpc-1 line, which possesses root hair cells in both the N and H positions (Kurata et al., 2005).
Our results provide one more possible explanation: that the CPC/TRY and MYB23 genes are regulated in a similar manner, which ensures that CPC/TRY-producing cells will also produce MYB23. Thus, the WER-equivalent MYB23 protein can compete with the CPC/TRY for binding to the bHLH proteins and maintain a significant level of the WER/MYB23-GL3/EGL3-TTG1 active complex in the N cells. This explanation would predict that, in the absence of the MYB23 positive feedback loop, misspecification of the N-position cells would increase. Consistent with this, we have observed a relative increase in hair cell specification in the N-position cells in the myb23-1 mutant and the myb23-1 cpc double mutant.
Similarities and Differences in the MYB23, WER, and GL1 MYB Gene Subfamily
The MYB23 and WER proteins share ∼57% amino acid sequence identity, and together with GL1, they constitute a subgroup in the Arabidopsis MYB transcription factor family (Stracke et al., 2001). Each of these proteins contains a conserved MYB domain including the amino acid signature [D/E]Lx2[R/K]x3Lx6Lx3R (Zimmermann et al., 2004) that mediates interaction with R/B-like bHLH transcription factors as well as the amino acid signature WVx2DxFELSxLx2M in the C terminus responsible for transcriptional activation (Lee and Schiefelbein, 2001; Jiang et al., 2004). GL1, the first identified R2R3 MYB gene in Arabidopsis, is involved in trichome cell fate specification in leaves and stems (Oppenheimer et al., 1991). GL1 is expressed in the epidermal cells of shoots and induces GL2 gene expression, and these GL2-expressing cells differentiate into trichome cells (Larkin et al., 1993; Szymanski et al., 1998). Although WER and GL1 participate in different developmental processes and show nonoverlapping expression patterns, they encode functionally equivalent proteins (Lee and Schiefelbein, 2001). In addition, MYB23 and GL1 were shown to be functionally equivalent in trichome initiation (Kirik et al., 2005). Detailed studies revealed that MYB23 also influences trichome branching, which is not controlled by GL1 (Kirik et al., 2005). Diversification of these two genes was proposed to be due to the differences in the cis-regulatory sequences for the trichome initiation stage and in the coding sequences for trichome branching.
Based on the functional similarity between WER, GL1, and MYB23 described above, it was conceivable that WER and MYB23 might be functionally equivalent in root epidermis development. Indeed, the MYB23 and WER proteins are both preferentially localized to nuclei in N-position cells (Ryu et al., 2005) (Figure 3C), and MYB23 was able to completely rescue the phenotype of the wer mutant when it was expressed under the control of the WER promoter (Figure 4; Table 1). Because an unrelated MYB protein, At MYB2, is not able to substitute for WER in the root epidermis (Lee and Schiefelbein, 2001), functional equivalence among MYB proteins is apparently only possible between MYBs in this subgroup.
Interestingly, MYB genes related to the GL1/WER/MYB23 subgroup have been found only in eudicots (WER, MYB23, and GL1 from Arabidopsis; Cs WER from Cucumis sativus [GenBank accession number AAM23006]). This suggests that the ancestral gene for this subgroup originated after the relatively recent split of the monocots and eudicots, and this was followed by amplification and diversification. It may be possible to tell which one of the genes in this subgroup represents the ancestral form, the relationship between the genes, and the timing of the split, once enough sequence information on the MYB genes in this subgroup is obtained from many eudicots.
METHODS
Plant Materials and Growth Conditions
The following mutant lines of Arabidopsis thaliana have been described: wer-1 (Lee and Schiefelbein, 1999), cpc (Wada et al., 1997), gl3-1 (Koornneef et al., 1982), egl3-1 (Zhang et al., 2003), gl2-1 (Koornneef, 1981), myb23-1 (Kirik et al., 2005), and ttg1-13 (Larkin et al., 1999). The plants harboring P35S:CPC, PWER:GFP, PMYB23:GUS, PGL2:GUS, and PCPC:GUS were also described previously (Masucci et al., 1996; Wada et al., 1997, 2002; Lee and Schiefelbein, 1999; Kirik et al., 2001). The reporter genes or multiple mutations were introduced into each line by genetic crosses and subsequent verification of homozygous lines by phenotypic and/or molecular tests.
For plant growth, seeds were surface-sterilized, germinated, and grown vertically on agarose-solidified medium containing mineral nutrients at 22°C under continuous light conditions (Schiefelbein and Somerville, 1990).
In Situ RNA Hybridization
The in situ RNA hybridization procedure has been described (Lee and Schiefelbein, 1999). DIG-labeled RNA probes were designed to hybridize specifically to MYB23 RNA only, so they included the 5′ untranslated region (392 to 120 bp upstream from the translation start site), the variable C-terminal region (1310 to 1555 bp downstream from the translation start site), and the 3′ untranslated region (300-bp fragment from the translation stop codon).
Histochemical GUS Staining and Quantitative Assay
GUS activity was examined histochemically by staining 4-d-old seedlings as described (Lee and Schiefelbein, 2002). GUS activity was measured quantitatively also from 4-d-old seedling root tips as described previously (Hung et al., 1998).
Quantitative Real-Time RT-PCR
Total RNA was extracted from the root tips of 4-d-old seedlings using the RNeasy Plant Mini kit (Qiagen) and treated with RNase-free DNase I (Invitrogen) according to the manufacturer's instructions. Two micrograms of total RNA was used for the reverse transcription using the AccuScript High Fidelity 1st Strand cDNA Synthesis kit (Stratagene), and 100 ng of resulting first-strand cDNA was used as a PCR template for the quantitative real-time RT-PCR. Quantitative PCR analysis was performed using SYBR Premix EX Taq (Takara) with an Mx3000P real-time PCR machine (Stratagene). Samples were denatured for 1 min at 95°C; followed by 40 cycles of 15 s of denaturation at 95°C, 30 s of annealing at 60°C, and 30 s of elongation at 72°C; followed lastly by one cycle of 1 s of denaturation at 95°C, 30 s of annealing at 65°C, and 30 s of denaturation at 95°C. After the renaturation, the melting parameters were assessed. EF1 was used as an internal reference to normalize the relative level of each transcript. Each experiment was repeated three to six times, and each time the experiment included triplicate samples.
EMSA
The EMSA was performed essentially as described earlier (Ryu et al., 2005). DNA fragments were labeled by T4 polynucleotide kinase and [γ-32P]ATP (>3000 Ci/mmol, 10 mCi/mL) and purified on a gel. Labeled DNA fragments and the WER protein were mixed, incubated, and resolved on a 6% polyacrylamide gel in 0.5× tri-borate-EDTA (TBE) buffer. The gel was dried and the signal was visualized using BAS-2500 (Fuji Film).
Protein Expression and Purification
Expression of the WER and MYB23 protein in Escherichia coli BL21 (DE3) and purification of the protein using His·Bind Quick 900 cartridges (Novagen) were performed as described previously (Ryu et al., 2005).
Confocal Microscopy
GFP expression in seedlings was counterstained with 5 μg/mL propidium iodide solution for 5 min and examined using a Zeiss LSM510 Meta confocal microscope, with excitation at 488 nm and detection with a 500- to 530-nm band-path filter for GFP and excitation at 543 nm and detection with a 560-nm long-path filter for propidium iodide (Lee and Schiefelbein, 1999).
Gene Constructs and Plant Transformation
To examine the subcellular localization of the MYB23 protein, the genomic DNA fragments of the MYB23 coding region were PCR-amplified and fused to the N terminus of GFP (mGFP5 without the N-terminal signal peptide sequence and the C-terminal sequence for retention of GFP in the ER) (Haseloff et al., 1997) in-frame. This chimeric MYB23-GFP was inserted between a 3.1-kb 5′ flanking region DNA fragment and a 1-kb 3′ flanking region DNA fragment from the MYB23 gene (PMYB23:MYB23-GFP). For PMYB23:GFP or PMYB23:mGFP5-ER, the soluble GFP coding region or ER-targeted version of the GFP coding region (mGFP5-ER) was inserted between the same 5′ and 3′ flanking region DNA fragments, respectively (Haseloff et al., 1997).
To generate the MYB23-SRDX construct, genomic DNA fragments of the MYB23 coding region were PCR-amplified using a primer that contains the SRDX nucleotide sequence (Hiratsu et al., 2003) at the end, which resulted in the fusion of the SRDX to the C terminus of the MYB23 coding region. This MYB23-SRDX DNA fragment was inserted between a 3.1-kb 5′ flanking region DNA fragment and a 1-kb 3′ flanking region DNA fragment from the MYB23 gene (PMYB23:MYB23-SRDX).
PWER:WER has been described previously (Lee and Schiefelbein, 2001). For PWER:MYB23, the same regulatory sequences (a 4.0-kb 5′ flanking region to the start codon and a 1.1-kb 3′ flanking region to the stop codon) were used to direct the MYB23 expression. To express and purify the MYB23 protein from E. coli, the MYB23 coding region excluding the region coding for the last 24 amino acids was PCR-amplified and inserted in the vector pET28(a). All of the DNA fragments PCR-amplified using iProof High-Fidelity DNA polymerase (Bio-Rad) were sequenced and confirmed to be error-free.
We used pCB302 vector as a binary vector for plant transformation (Xiang et al., 1999). Plant transformation was achieved by electroporating constructs into the Agrobacterium tumefaciens strain GV3101 followed by introduction into Arabidopsis using the floral dip method as described previously (Clough and Bent, 1998). T1 seeds were harvested, and transgenic plants were selected by spraying commercially available basta three times per week for 2 weeks.
Primers
Primer sequences used are listed in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CPC (At2g46410), EGL3 (At1g63650), GL2 (At1g79840), GL3 (At5g41315), MYB23 (At5g40330), TTG1 (At5g24520), and WER (At5g14750). Germplasm identification numbers from this article are as follows: cpc-1 (CS6399), gl2-1 (CS65), gl3-1 (CS66), myb23-1 (SALK_048592), myb23-3 (SALK_018613), and wer-1 (CS6349).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of the trichome-branching defect of myb23-1 using the MYB23 gene including the 2-kb 5′ flanking DNA fragment and the 1-kb 3′ flanking DNA fragment (PMYB23-2kb:MYB23).
Supplemental Table 1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Su-Hwan Kwak, Christine Bernhardt, and M.M.L. laboratory members for helpful advice and suggestions. This research was supported by grants from the Crop Functional Genomics Center of the 21st Century Frontier Program (Code CG2152) and the Plant Signaling Network Research Center (SRC), the Ministry of Education, Science and Technology/Korea Science and Engineering Foundation (2009-0079304/R0602494), to M.M.L. and by the National Science Foundation (Grants IBN0316312 and IOS-0744599 to J.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Myeong Min Lee (mmlee@yonsei.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Benitez, M., Espinosa-Soto, C., Padilla-Longoria, P., Diaz, J., and Alvarez-Buylla, E.R. (2007). Equivalent genetic regulatory networks in different contexts recover contrasting spatial cell patterns that resemble those in Arabidopsis root and leaf epidermis: a dynamic model. Int. J. Dev. Biol. 51 139–155. [DOI] [PubMed] [Google Scholar]
- Berger, F., Hung, C.Y., Dolan, L., and Schiefelbein, J. (1998). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194 235–245. [DOI] [PubMed] [Google Scholar]
- Bernhardt, C., Lee, M.M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Murray, J.D., and Wolpert, L. (1981). Theories of Biological Pattern Formation. (London: Royal Society); 191 pages.
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Culi, J., and Modolell, J. (1998). Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relations and patterning in the root epidermis of Arabidopsis. Development 120 2465–2474. [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Esch, J.J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., Neizer, J., and Marks, M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894. [DOI] [PubMed] [Google Scholar]
- Freeman, M. (2000). Feedback control of intercellular signalling in development. Nature 408 313–319. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166 740–754. [DOI] [PubMed] [Google Scholar]
- Gierer, A., and Meinhardt, H. (1972). A theory of biological pattern formation. Kybernetik 12 30–39. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localisation of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Hung, C.Y., Lin, Y., Zhang, M., Pollock, S., Marks, M.D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, T., Kurata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59 365–386. [DOI] [PubMed] [Google Scholar]
- Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 5 R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., Lee, M.M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., Schiefelbein, J., and Hulskamp, M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Schnittger, A., Radchuk, V., Adler, K., Hulskamp, M., and Baumlein, H. (2001). Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 235 366–377. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1981). The complex syndrome of ttg mutants. Arabidopsis Inf. Serv. 18 45–51. [Google Scholar]
- Koornneef, M., Dellaert, L.W., and van der Veen, J.H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. 93 109–123. [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura, Y., Wada, T., Tachibana, T., Tsugeki, R., Ishiguro, S., and Okada, K. (2005). Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 46 817–826. [DOI] [PubMed] [Google Scholar]
- Kurata, T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132 5387–5398. [DOI] [PubMed] [Google Scholar]
- Kwak, S.H., and Schiefelbein, J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302 118–131. [DOI] [PubMed] [Google Scholar]
- Kwak, S.H., Shen, R., and Schiefelbein, J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307 1111–1113. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Pollock, S., and Marks, M.D. (1993). Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Walker, J.D., Bolognesi-Winfield, A.C., Gray, J.C., and Walker, A.R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128 1539–1546. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122 1253–1260. [DOI] [PubMed] [Google Scholar]
- Matsui, K., Hiratsu, K., Koyama, T., Tanaka, H., and Ohme-Takagi, M. (2005). A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol. 46 147–155. [DOI] [PubMed] [Google Scholar]
- Meinhardt, H., and Gierer, A. (2000). Pattern formation by local self-activation and lateral inhibition. Bioessays 22 753–760. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493. [DOI] [PubMed] [Google Scholar]
- Page, K., Maini, P.K., and Monk, N.A.M. (2003). Pattern formation in spatially heterogeneous Turing reaction-diffusion models. Physica D. 181 80–101. [Google Scholar]
- Pesch, M., and Hulskamp, M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14 422–427. [DOI] [PubMed] [Google Scholar]
- Ryu, K.H., Kang, Y.H., Park, Y.H., Hwang, I., Schiefelbein, J., and Lee, M.M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132 4765–4775. [DOI] [PubMed] [Google Scholar]
- Schellmann, S., Hulskamp, M., and Uhrig, J. (2007). Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem. Soc. Trans. 35 146–148. [DOI] [PubMed] [Google Scholar]
- Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., Thumfahrt, J., Jurgens, G., and Hulskamp, M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J., and Lee, M.M. (2006). A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis. Physiol. Plant. 126 503–510. [Google Scholar]
- Schiefelbein, J.W., and Somerville, C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M., Lee, M.M., Lin, Y., Gish, L., and Schiefelbein, J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311 566–578. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171. [DOI] [PubMed] [Google Scholar]
- Tominaga, R., Iwata, M., Okada, K., and Wada, T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M.D., Shimura, Y., and Okada, K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129 5409–5419. [DOI] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277 1113–1116. [DOI] [PubMed] [Google Scholar]
- Williams, C.E., and Grotewold, E. (1997). Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J. Biol. Chem. 272 563–571. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40 711–717. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Gonzalez, A., Zhao, M., Payne, C.T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.