Abstract
Leucine aminopeptidase A (LapA) is a late wound-response gene of tomato (Solanum lycopersicum). To elucidate the role of LapA, transgenic plants that overexpressed or abolished LapA gene expression were used. The early wound-response gene RNA levels were similar in wild-type and Lap-silenced (LapA-SI), -antisense (LapA-AS), and -overexpressing (LapA-OX) plants. By contrast, late wound-response gene RNA levels and protection against Manduca sexta damage were influenced by LapA RNA and protein levels. While LapA-OX plants had elevated levels of LapA RNAs and protein, ectopic expression of LapA was not sufficient to induce Pin (Ser proteinase inhibitor) or PPO (polyphenol oxidase) transcripts in nonwounded leaves. M. sexta larvae damaged less foliage and displayed delays in growth and development when feeding on LapA-OX plants. By contrast, LapA-SI and LapA-AS lines had lower levels of Pin and PPO RNAs than wild-type controls. Furthermore, larvae consumed more foliage and attained larger masses when feeding on LapA-SI plants. Jasmonic acid (JA) did not complement the wound-signaling phenotype of LapA-SI plants. Based on root elongation in the presence of JA, JA perception appeared to be intact in LapA-SI lines. Collectively, these data suggested that LAP-A has a role in modulating essential defenses against herbivores by promoting late wound responses and acting downstream of JA biosynthesis and perception.
INTRODUCTION
Animals and plants use oxygenated lipids in defense and immune responses. Similar to the animal prostaglandin pathway, which regulates pain and inflammation responses, plants produce an array of oxygenated lipids via the octadecanoid pathway to regulate plant defense and wound responses (Walling, 2000; Rojo et al., 2003; Howe, 2004; Wasternack et al., 2006). These oxylipins include the potent signaling molecules jasmonic acid (JA) and 12-oxo-phytodienoic acid, which are essential for mounting the direct and indirect defenses that deter insect infestations and necrotrophic pathogen attack and also mediate crosstalk with salicylic acid–dependent defenses.
JA activates a cascade of wound responses that are temporally (early and late) and spatially (local and systemic) regulated in tomato (Solanum lycopersicum) (Ryan, 2000). The early wound-response genes are activated within 30 min to 2 h after damage and encode gene products that aid to amplify wound signaling (Ryan, 2000). These gene products include the precursor of the wound peptide systemin (Prosystemin), lipoxygenases, mitogen-activated protein kinases, and an array of enzymes important in the biosynthesis of JA.
At later times (4 to 24 h), late wound-response genes that have a direct role in defense are activated. Late wound-response genes include the Ser proteinase inhibitors (Pin1 and Pin2), polyphenol oxidases (PPOs), arginase, Thr deaminase, and Leu aminopeptidase (LapA) (Howe, 2004). Pins and PPOs have well-established roles in deterring insect herbivory and reducing insect growth (Felton et al., 1989; Johnson et al., 1989). The inactivation of digestive proteases by Pins causes a compensatory induction of peptidases by the herbivore, which is thought to contribute to reduced larval growth (Broadway and Duffey, 1986). The wound-induced PPOs oxidize foliar chlorogenic acid to form chlorogenoquinone (Felton et al., 1989). Chlorogenoquinone, a highly reactive electrophilic molecule, covalently modifies the nucleophilic groups of amino acids and proteins, thereby reducing the ability of the herbivore to use these essential N sources. Recently, arginase and Thr deaminase were shown to have important antinutritive roles in defense by reducing the amounts of the essential amino acids Arg and Thr in the insect gut (Chen et al., 2005; Kang and Baldwin, 2006). To date, LAP-A's role in tomato defense has yet to be determined.
In tomato, the early and late wound-response genes are regulated by distinct mechanisms. Reactive oxygen species influence late wound-response gene transcript levels but do not affect the expression of early wound-response genes. Hydrogen peroxide (H2O2) accumulates rapidly (within 1 h) in response to wounding and systemin treatments (Orozco-Cárdenas and Ryan, 1999), and NADPH oxidase inhibitors prevent the accumulation of late wound-response RNAs (Pins and PPO-F) but do not diminish the accumulation of early wound-response gene transcripts (Prosystemin, AOS, and LOX-D) (Orozco-Cárdenas et al., 2001). Furthermore, antisense NADPH tomato plants have reduced levels of reactive oxygen species and have an impaired systemic induction of late wound-response genes (Sagi et al., 2004). Nitric oxide (NO) is the second reactive oxygen species important in regulating late wound responses. NO antagonizes the production of H2O2 and blocks activation of late wound-response genes in tomato, while NO does not interfere with the expression of early wound-response genes (Orozco-Cárdenas and Ryan, 2002). The mechanisms of H2O2 and NO action are currently unknown.
LapA1 and LapA2 are late wound-response genes encoding the abundant acidic subunits of the homohexameric metallopeptidase LAP-A (EC 3.4.11.1) (Gu et al., 1996b). The LAP-A1 and LAP-A2 mature proteins are 99% identical and reside within the chloroplast (Matsui et al., 2006; Narváez-Vásquez et al., 2007). LAP-A1 and LAP-A2 are 77% identical with LAP-N, which is expressed at low levels constitutively (Tu et al., 2003). LAPs are aminopeptidases that cleave the N-terminal amino acid from peptides and proteins. In addition to their distinctive programs of expression, LAP-A and LAP-N enzymes have distinct substrate specificities (Tu et al., 2003). LAP-A1 preferentially hydrolyzes substrates with N-terminal Leu, Met, and Arg, while LAP-N also hydrolyzes substrates with N-terminal Phe (Gu et al., 1999; Gu and Walling, 2000; Tu et al., 2003). LAPs are highly conserved, multifunctional proteins in eukaryotes and prokaryotes (Walling, 2004; Matsui et al., 2006). For example, the Escherchia coli XerB (a LAP homolog) is an aminopeptidase, site-specific recombinase and transcription factor (Colloms, 2004). In mammals, LAPs are capable of generating antigenic peptides for major histocompatibility complex I (MHCI) presentation and facilitate protein turnover in the lens and muscle tissue (Taylor, 1996; Beninga et al., 1998).
While the exact role of the tomato LAP-A in defense is not known, its expression programs and biochemical specificity are well characterized (Walling, 2004; Matsui et al., 2006). LapA RNAs are at low or undetectable levels in healthy leaves but are expressed in all floral organs and in fruit (Chao et al., 2000). Consistent with its regulation by the wound octadecanoid-signaling pathway, LapA RNAs and proteins increase in response to exogenous JA and systemin and are suppressed by salicylic acid (Chao et al., 1999). Furthermore, LapA RNAs, proteins, and activities increase in response to abscisic acid, mechanical wounding, tissue-damaging herbivores (Spodoptera littoralis and Manduca sexta), and some bacterial and oomycete pathogens (Pseudomonas syringae pv tomato and Phytophthora parasitica, respectively) (Pautot et al., 1993; Jwa and Walling, 2001; Pautot et al., 2001).
The hypothesis that LAP-A has a role in tomato defense is intriguing given the direct and indirect roles of endoproteases in plant defense (Pechan et al., 2002; Schaller, 2004; van der Hoorn and Jones, 2004) and persistence of LAP-A in the insect gut and frass (Chen et al., 2005, 2007). Furthermore, the role of aminopeptidases in the genesis of bioactive peptides in animals (Shrimpton et al., 2002) and, more recently, in plants (Srivastava et al., 2008) and the role of the 18-residue peptide systemin in activating wound responses in tomato (Ryan, 2000) provided motivation for examining the role of LAP-A in wound signaling and defense against tissue-damaging herbivores.
We previously investigated the susceptibility of LapA-antisense (LapA-AS) lines to herbivory and P. s. tomato infection (Pautot et al., 2001). These LapA-AS lines did not show an impact on defense using an excised leaf assay. However, LapA-AS lines express low levels of LapA RNAs and proteins, which might provide small amounts of LAP-A sufficient to fulfill LAP's enzymatic role in the tomato wound-defense response. Therefore, it was critical to isolate tomato plants with more stringent downregulation of LapA to clearly evaluate LAP's role in wound signaling and defense in intact plants.
This article describes the isolation of transgenic P35S:LapA tomato lines that have silenced (LapA-SI) and overexpressed (LapA-OX) LapA. These LapA-SI and LapA-OX lines, in conjunction with LapA-AS lines, allowed the role of LAP-A in regulating the early and late branches of the tomato wound-signaling pathway to be evaluated. The results indicate that LAP-A is a qualitative regulator of the late branch of wound signaling acting downstream or independently of JA biosynthesis and perception. Consistent with impaired late responses to wounding, LapA-SI lines were more susceptible to herbivory, and M. sexta larvae growth was enhanced on these lines. Reciprocally, LapA-OX lines were more resistant to herbivory and displayed reduced rates of M. sexta larvae growth and development.
RESULTS
Characterization of LapA-SI and LapA-OX Lines
To address the role of LAPs in defense, transgenic tomato lines harboring a P35S:LapA1 construct were characterized. Several transgenic lines that silenced Lap genes (LapA-SI) and/or ectopically expressed LapA (LapA-OX) were identified by monitoring LapA RNA and protein levels at 24 h after wounding. P35S:LapA1 plants displayed variation in the levels of LapA RNAs in healthy and wounded leaves; five characteristic T2 transgenic plants are shown (Figure 1A). Relative to wild-type plants, LapA RNAs were elevated in healthy leaves in P35S:LapA homozygous plants 1, 11, and 5. However, only lines 11 and 5 expressed LapA RNAs at sufficiently high levels in healthy and wounded leaves to be considered overexpressing lines (LapA-OX). Line 29 was in the process of silencing the transgene and endogenous LapA genes in the T2 generation, since RNA levels after wounding were lower in the silenced line 29 than in wild-type plants. In the T3 generation, this line had completely silenced LapA gene expression. Line 28 had completely silenced the transgene and endogenous LapA genes in the T2 generation.
Figure 1.
LapA RNA and Protein Levels.
(A) LapA RNA levels in healthy and wounded leaves. RNAs were extracted from healthy (H) and wounded (W) (24 h) leaves from wild-type UC82b and P35S:LapA plants. Five independent, homozygous P35S:LapA lines are displayed. Each lane represents RNAs from a pool of five plants.
(B) Protein blots. Total proteins were extracted from healthy and wounded (24 h) leaves from wild-type UC82b, LapA-SI (line 28), and LapA-OX (line 11) lines. Proteins (80 μg) were fractionated by two-dimensional PAGE. Gels were electroblotted and incubated with LAP-A antiserum. LAP-A (gray arrowhead), LAP-N (white arrowhead), and the 66-kD LAP-like proteins were detected. In some panels, the 77-kD LAP-like proteins are also displayed; the levels of the 66- and 77-kD proteins did not change in any of the genotypes or after treatments. The pH range of 8 to 4 is indicated. Masses of protein markers in kilodaltons are shown.
LAP protein levels in wild-type and LapA-SI and LapA-OX lines were evaluated. Two-dimensional PAGE immunoblots incubated with a polyclonal LAP-A antiserum detected low levels of the 55-kD LAP-N and LAP-A, as well as the 66- and 75-kD LAP-like proteins in healthy leaves of control plants (Figure 1B) (Gu et al., 1996b). After wounding, LAP-A levels increased in wild-type leaves. By contrast, LAP-A was not detected in healthy or wounded leaves from LapA-SI plants. In addition, the closely related LapN was also silenced, as evidenced by the lack of LAP-N protein accumulation in healthy and wounded leaves of LapA-SI lines. LapN silencing was presumed to be mediated by sense suppression due to its >86% nucleotide identity with LapA1 (Tu et al., 2003). The low levels of LAP-A protein in LapA-SI lines were correlated with reduced levels of LapA RNAs after wounding in LapA-SI lines relative to control plants (Figures 1A and 2A). LapA-SI lines had lower levels of LapA RNAs than the previously characterized transgenic tomato lines expressing a LapA antisense construct (LapA-AS) (Pautot et al., 2001) (Figure 2A).
Figure 2.
Wound-response Gene RNAs in Wild-Type, LapA-AS, and LapA-SI Plants.
Leaves from 3-week-old wild-type, LapA-AS (line 4) (Pautot et al., 2001), and LapA-SI (line 28) plants were wounded. The damaged (local) and apical nonwounded (systemic) leaves were collected at 0, 1, 2, 4, 8, 12, and 24 h after wounding. RNAs were extracted and RNA gel blots were hybridized to 32P-labeled gene probes. All blots were stripped and subsequently hybridized with an EF1-α probe to control for loading; one representative control blot is shown. These experiments were performed twice, and one representative experiment is shown. Each lane represents RNAs from a pool of five plants.
(A) Local and systemic accumulation of late wound-response gene (LapA, Pin1, Pin2, and PPO-F) RNAs.
(B) Local and systemic accumulation of early wound-response gene (AOS and LOX-D) RNAs.
Transgenic lines that ectopically expressed LapA (LapA-OX) RNAs and proteins were also characterized further. LAP-N and the 66- and 77-kD LAP-like protein levels were at similar levels in LapA-OX and wild-type plants. In healthy (nonwounded) LapA-OX leaves, LAP-A proteins were at levels similar to wounded wild-type leaves (Figure 1B). In addition, levels of LAP-A increased markedly after wounding of LapA-OX plants. The increased LAP-A levels were correlated with increased amounts of LapA RNA in healthy LapA-OX and wounded wild-type and LapA-OX lines (Figures 1B and 3A).
Figure 3.
Late Wound-response Gene RNAs in Wild-Type, LapA-OX, and LapA-SI Plants.
Leaves from 2-week-old wild-type, LapA-OX (line 11), and LapA-SI (line 28) were wounded, and the damaged (local) and apical nonwounded (systemic) leaves were collected at 0, 2, 4, 8, 12, 24, and 48 h after wounding. Total RNA was extracted, and the levels of LapA (A), Pin1 (B), Pin2 (C), and PPO-F (D) RNAs were determined by qPCR. Data for local and systemic leaves are shown in the left and right panels, respectively. Ubi3, HKG1, and EF1-α served as internal reference genes. The data are expressed as fold-change relative to RNAs present in the LapA-SI plant at 0 h. Data from two biological replicate experiments are displayed, and standard errors are shown.
LAP Is Necessary for the Wound Response
To determine the role of LAP-A in the tomato wound response, changes in four late wound-response gene RNAs after wounding were examined in 3-week-old control (wild-type), LapA-AS, and LapA-SI plants (Figure 2A). In wild-type plants, LapA, Pin1, Pin2, and PPO-F RNAs were first detected 2 to 4 h after wounding. These RNAs persisted for >24 h and were detected both locally and systemically. In LapA-AS and LapA-SI plants, LapA RNAs were at low levels or were undetectable, respectively, during the first 24 h after wounding. The timing and levels of Pin1, Pin2, and PPO-F RNAs were distinct in wounded leaves from wild-type, LapA-AS, and LapA-SI plants. The differences in Pin and PPO-F RNA levels in wild-type and transgenic lines were most obvious at 24 h in the wounded (local) and 12 to 24 h in apical, nonwounded (systemic) leaves. While the differences in Pin1 and Pin2 RNA levels in the LapA-AS and LapA-SI lines were most distinct, similar trends were seen with PPO-F RNAs. In general, wound response RNAs did not reach wild-type levels and did not persist as long in LapA-SI and LapA-AS plants. This phenotype was strongest in the LapA-SI lines and was detected, although less dramatically, in LapA-AS lines. Therefore, the impaired expression of late wound responses was correlated with the degree of Lap silencing in these lines.
These data indicated that a LAP deficiency altered the quantity and timing of late wound-response transcript accumulation. To quantitate these changes, 2-week-old wild-type and LapA-SI plants were wounded. Damaged and apical (systemic) leaves were collected from 0 to 48 h after wounding, and RNAs were subjected to quantitative PCR (qPCR) analyses; relative RNA levels are displayed in Figure 3. All late wound-response gene RNAs increased locally and systemically by 4 h and reached peak levels by 8 (Pin2) or 12 h (LapA, Pin1, and PPO-F). It should be noted that when compared with the wounding time course in Figure 2A, the trends in RNA accumulation patterns were similar but there was a temporal shift in the responses. This is likely due to the age of the plants at the time of wounding (2-week-old versus 3-week-old plants) and/or the different wounding regimens that were used in these experiments. In wild-type plants, LapA, Pin1, and Pin2 RNAs increased 52-, 47-, and 22-fold, respectively, and PPO-F RNAs increased 10-fold in wounded leaves. The systemic changes in LapA and Pin2 RNAs in wild-type plants were similar in magnitude to the damaged leaves. By contrast, the systemic increases in Pin1 and PPO-F RNAs were less pronounced.
When late wound-response gene RNA levels in LapA-SI plants were examined by qPCR, LapA RNAs were barely detected in the LapA-SI line (Figure 3A). Similar to RNA gel blot analyses, qPCR showed that Pin2 RNAs (8 h) and Pin1 and PPO-F RNAs (12 h) were 38 to 40% of the levels attained in the wild-type plants at these times (Figures 3B to 3D). The reduced levels of late wound-response RNAs in LapA-SI plants relative to wild-type plants was also seen in the apical, nonwounded leaves at these times.
To examine if LAP modulated the levels of early wound-response gene RNAs, the levels of Allene Oxide Synthase (AOS) and Lipoxygenase-D (LOX-D) RNAs in 3-week-old wild-type, LapA-AS, and LapA-SI plants was visualized by RNA gel blot analyses (Figure 2B). AOS and LOX-D RNAs increased locally and systemically by 1 h and reached peak levels by 1 to 2 h after damage. AOS and LOX-D RNA levels were similar in wild-type and LapA-SI lines and were slightly decreased in the wounded LapA-AS line. These data showed that the early wound-response RNAs were not correlated with LapA RNA levels. AOS and LOX-D RNA levels were also quantitated by qPCR in healthy and wounded 2-week-old wild-type and LapA-SI plants (Figures 4A and 4B). These data showed a marked variation in the levels of LOX-D and AOS RNA levels in the replicate experiments but showed that the maximum LOX-D and AOS RNA levels attained in the wild-type or LapA-SI plants were similar. Collectively, these data indicated that Lap deficiency did not alter early wound-response gene expression but altered the accumulation of RNAs of genes (Pin1, Pin2, and PPO-F) regulated by the late branch of wound signaling.
Figure 4.
Early Wound-response Gene RNAs in Wild-Type, LapA-OX, and LapA-SI Plants.
Leaves from 2-week-old wild-type, LapA-OX (line 11), and LapA-SI (line 28) were wounded, and the damaged (local) and apical nonwounded (systemic) leaves were collected at 0, 1, 2, 4, 8, and 12 h after wounding. Total RNA was extracted, and the levels of LapA, LOX-D, and AOS RNAs were determined by qPCR. Ubi3, HKG1, and EF1-α served as internal reference genes. The data are expressed as fold-change relative to RNAs present in the LapA-SI plant at 0 h. Data from two biological replicate experiments are displayed, and standard errors are shown.
LAP-A Is Not Sufficient for the Wound Response
The spatial and temporal accumulation of wound response RNAs in the LapA-OX and wild-type plants was also analyzed. qPCR analysis of LapA transcript levels in 2-week-old plants showed that LapA transcripts were present at 26-fold higher levels in healthy (0 h) leaves in LapA-OX plants than wild-type plants (Figure 3A). However, no increases in Pin1, Pin2, or PP0-F RNA transcripts were detected in healthy LapA-OX leaves, indicating that LAP-A was not sufficient to induce wound-response gene transcript accumulation.
LapA-OX plants responded to the signals generated after wounding by increasing LapA RNA levels at a rate similar to that observed in 2-week-old wild-type plants. The elevation in LapA RNAs occurred in both local, wounded, and apical nonwounded leaves (Figure 3A). Although there was some variation the levels of Pin1 and Pin2 RNAs detected by qPCR in the replicate experiments, one consistent distinction between wild-type and LapA-OX plants was observed (Figures 3B to 3D). Pin1 and Pin2 RNAs persisted for longer periods of time in wounded LapA-OX plants than wild-type plants, as seen at 48 h after wounding; a similar trend is seen for PPO-F RNAs. RNA gel blot analysis of a longer time-course experiment, which spanned 24 to 96 h after wounding, showed that Pin1 RNAs persisted for longer periods of time in the LapA-OX relative to wild-type plants (see Supplemental Figure 1 online). By contrast, qPCR analyses showed that early wound-response AOS and LOX-D RNA accumulation patterns were similar in LapA-OX and wild-type leaves (Figures 4A and 4B).
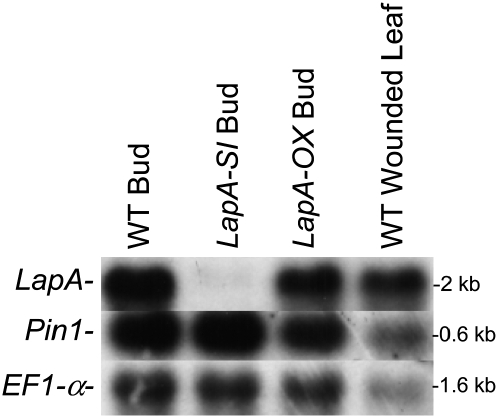
LAP-A Does Not Influence Pin RNA Levels in Floral Buds
Wound-response RNAs, such as LapA and Pin1, also accumulate during flower development in wild-type tomato plants (Peña-Cortes et al., 1991; Pautot et al., 2001; Li et al., 2004). Therefore, the impact of LAP deficiency and ectopic expression of LAP-A on wound-response gene expression in developing wild-type, LapA-SI, and LapA-OX floral buds was examined (Figure 5). LapA and Pin1 transcripts were abundant in wild-type floral buds, which was similar to previous findings (Peña-Cortes et al., 1991; Pautot et al., 2001). Consistent with the LAP-silencing phenotype observed in foliage, LapA transcripts were undetectable in LapA-SI floral buds. Surprisingly, Pin1 RNAs accumulated to similar levels in LapA-SI, LapA-OX, and wild-type buds. These data indicated that unlike wounding, the accumulation of Pin RNAs during development was not influenced by LAP-A.
Figure 5.
LapA and Pin1 RNAs in Wild-Type, LapA-SI, and LapA-OX Floral Buds.
Unopened floral buds (8 mm) were harvested and RNAs extracted from wild-type, LapA-SI (line 28), and LapA-OX (line 11) plants. RNA gel blots with floral bud and wounded wild-type leaf (positive control) RNAs were hybridized to 32P-labeled LapA, Pin1, and EF1-α gene probes. EF1-α served as a control for loading as described in Figure 2. Each lane represents RNAs from a pool of five plants.
LAP-A Acts Downstream of JA Biosynthesis
JA is a key regulator of wound responses and activates both early and late genes (Howe, 2004). To determine if LAP acted prior to or after JA biosynthesis, the ability of exogenous JA to restore the late wound response in LapA-SI and LapA-AS plants was tested. Excised shoots from wild-type, LapA-SI, and LapA-AS plants were treated with 10 μM JA for 16 h (Gu et al., 1996a). Figure 6A shows that wild-type plants responded to exogenous JA by accumulating LapA and Pin1 transcripts to high levels. By contrast, Pin1 transcripts were barely detected in LapA-SI and LapA-AS plants after JA treatments. The inability of JA to restore late wound responses was consistent with LAP acting downstream of JA biosynthesis.
Figure 6.
Exogenous JA Treatment of Wild-Type, LapA-SI, and LapA-OX Shoots.
Shoots were excised from wild-type, LapA-SI, LapA-AS, and LapA-OX plants and placed in 0.005% ethanol or 10 μM JA in 0.005% ethanol for 0 to 24 h. Leaves were collected, RNAs isolated, and RNA gel blots performed as described in Figure 2. All blots were stripped and subsequently hybridized with an EF1-α probe to control for loading. These experiments were replicated, and one representative experiment is shown.
(A) Levels of LapA, Pin1, and EF1-α RNAs in the wild type, LapA-SI line 28 (SI 28), LapA-SI line 29 (SI 29), and the LapA-AS line 4 (AS) 16 h after JA treatment.
(B) Levels of LapA, Pin1, and EF1-α RNAs in the wild type and LapA-SI (line 28) at 0, 2, 4, 6, 18, and 24 h after JA treatment.
(C) Levels of LapA, Pin1, and EF1-α RNAs in the wild type and LapA-OX (line 11) leaves after 16 h of JA treatment.
To determine if Lap silencing shifted the timing of wound-response gene RNA accumulation in response to exogenous JA, LapA and Pin1 RNAs levels were determined in wild-type and LapA-SI lines at 0, 2, 4, 6, 18, and 24 h after 10 μM JA treatment (Figure 6B). In wild-type plants, LapA RNAs were abundant at 6 h and increased until 24 h. Similarly, Pin1 RNAs were at high levels from 6 to 24 h. By contrast, LapA RNAs were undetectable in JA-treated LapA-SI plants. Unlike responses in wild-type plants, Pin1 RNAs were at lower levels and were more transient in LapA-SI plants. For example, while Pin1 RNAs were abundant after 24 h in wild-type plants, Pin1 RNAs had declined by 24 h in LapA-SI plants. By contrast, JA-treated LapA-OX plants showed no difference in the accumulation of either LapA or Pin1 transcripts compared with the wild type (Figure 6C). The fact that JA did not restore wound response gene expression in LapA-SI plants to wild-type levels supported the hypothesis that LAP acts downstream of JA biosynthesis.
JAI1 Signaling Is Intact in LapA-SI Plants
Tomato JASMONATE INSENSITIVE1 (JAI1), an F-box protein similar to Arabidopsis thaliana CORONATINE INSENSITIVE1 (Li et al., 2004), mediates recognition of the JA-Ile conjugate (Chini et al., 2007; Thines et al., 2007). Consistent with previous studies (Li et al., 2004), the jai1/jai1 mutant, which cannot perceive JA-Ile, elongates its roots in the presence of 1 mM JA (Figure 7). To determine if JA perception was altered in LapA-SI lines, wild-type and LapA-SI root elongation was measured in the presence and absence of 1 mM JA. Root elongation of wild-type (UC82b) and LapA-SI seedlings was inhibited with 1 mM JA treatments (Figure 7). Similar to Castlemart and the jai1 heterozygous and homozygous plants, intermediate JA concentrations (1 to 100 μM JA) did not reveal discernable changes in root elongation between the genotypes. These data indicated that LapA-SI lines could perceive JA and had intact JAI1 signaling. Collectively, these data and the data presented in Figures 5 and 6 suggested that LAP acts downstream of JA biosynthesis and JAI-dependent perception of JA-Ile.
Figure 7.
Root Elongation Phenotypes of Wild-Type, LapA-SI, and jai1 Mutants.
Wild-type, LapA-SI (line 28), homozygous (jai1), or heterozygous (+/jai1) seeds were germinated in the dark in water (control) or 1 mM JA for 4 d.
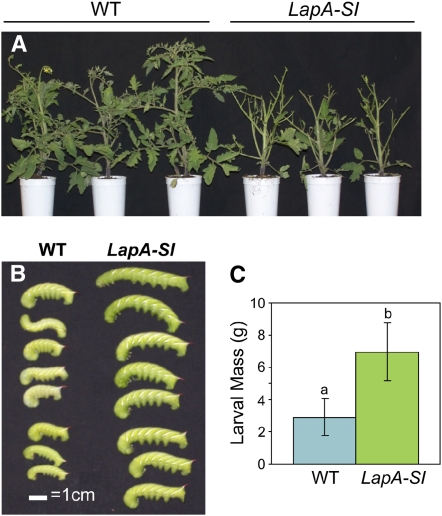
LapA-SI Lines Have a Compromised Defense to M. sexta
The impaired wound response exhibited in LapA-SI lines (Figures 2A and 3A to 3D) may alter the relative susceptibility/resistance to herbivore attack. This hypothesis is based on the fact that the reduced levels of Pin and PPO transcripts observed in LapA-SI lines may result in lower levels of these proteins, which act directly to deter herbivore feeding and growth (Felton et al., 1989; Johnson et al., 1989). To assess the relative resistance/susceptibility to herbivore feeding, wild-type and LapA-SI lines were infested with M. sexta 1st-instar larvae. After 11 d of feeding, M. sexta larvae consumed ∼65% of the foliage from wild-type (control) plants and >95% of the foliage from LapA-SI lines (Figure 8A).
Figure 8.
Wild-Type and LapA-SI Plant Responses to Herbivory.
Wild-type (WT) and LapA-SI plants (line 28) were infested with M. sexta larvae for 11 d.
(A) The foliage from wild-type (left) and LapA-SI (right) plants at day 11.
(B) M. sexta larva raised on wild-type (left) or LapA-SI (right) plants.
(C) Masses of M. sexta larvae reared on wild-type (n = 8) and LapA-SI (n = 9) plants. Masses were significantly different (P < 0.05; Student's t test), and standard errors are shown.
Consistent with increases in plant material consumed, M. sexta larvae that fed upon the LapA-SI plants attained masses (6.97 ± 1.80 g) that were twofold greater (P < 0.05) than the masses of larvae raised on control plants (2.90 ± 1.13 g) (Figures 8B and 8C). The head capsule sizes of the M. sexta larvae, which are an accurate indicator of instar stage (Nijhout and Williams, 1974), were measured to determine if the rate of larval development was altered on plants that consumed LapA-SI or wild-type foliage. Larvae raised on LapA-SI and control plants had head capsule diameters of 5.8 ± 0.6 mm and 5.2 ± 0.9 mm, respectively. These data indicated that the larvae raised on wild-type and LapA-SI plants were all in their fifth instar and the absence of LAP-regulated defenses influenced the growth but not the rate of larvae development.
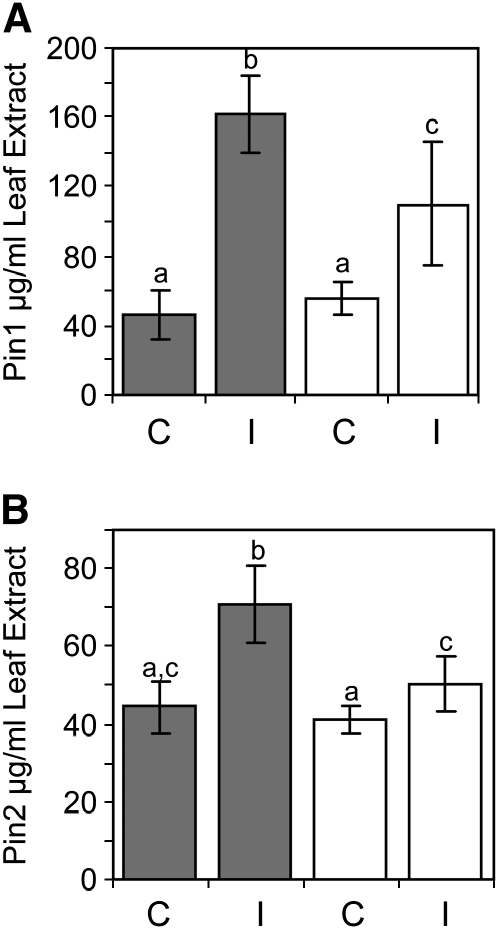
Measurement of the levels of Pin1 and Pin2 proteins in noninfested and M. sexta–infested leaves provided additional evidence that reduced defenses were a cause of the enhanced susceptibility of LapA-SI plants relative to wild-type plants. Basal levels of Pin1 or Pin2 proteins were similar in healthy wild-type and LapA-SI leaves (P > 0.05) (Figure 9). After 24 h of M. sexta infestation, Pin1 and Pin2 protein levels increased 3.5- and 2-fold in wild-type leaves, respectively (Figures 9A and 9B). By contrast, the increase in Pin1 protein was only 1.9-fold in LapA-SI leaves, and there was more plant-to-plant variation than in wild-type plants (Figure 9A); furthermore, significantly less Pin1 accumulated in LapA-SI than wild-type plants (109 and 161 μg/mL leaf extract, respectively). Overall, Pin2 levels did not change as dramatically as Pin1 levels in wild-type or LapA-SI leaves, but distinctions between wild-type and LapA-SI lines were clearly observed. While Pin2 levels increased 1.5-fold in wild-type plants after 24 h of M. sexta feeding, Pin2 levels in infested LapA-SI leaves were not significantly different from levels in noninfested wild-type or LapA-SI leaves (Figure 9B). These data indicated that the enhanced insect performance on LapA-SI lines was correlated with the reduced defenses reflected by reduced Pin RNA and protein levels.
Figure 9.
Pin Protein Levels in Wild-Type and LapA-SI Lines after M. sexta Feeding.
Leaves from healthy (control, C) or M. sexta–infested (I) (24 h) wild-type (gray bars) and LapA-SI (white bars) plants were collected. Leaf juice was extracted and Pin protein levels (μg per mL of leaf juice) were measured using a radial immunodiffusion assay (Ryan, 1967). Pin levels that were significantly different (P < 0.05; Student's t test) are indicated by different lowercase letters. Means are based on samples from five plants per treatment, and standard errors are shown. These experiments were performed twice, and one representative experiment is shown.
(A) Pin1 protein levels.
(B) Pin2 protein levels.
LapA-OX Lines Are More Resistant to M. sexta
To determine if elevated levels of LAP-A provide resistance to herbivory, wild-type and LapA-OX line plants were infested with 1st-instar M. sexta larvae. After 14 d of feeding, M. sexta larvae consumed approximately two-thirds of the foliage from wild-type plants and approximately one-third of the foliage from LapA-OX plants (Figure 10A). These data indicated that LapA-OX plants were less palatable to the insects. The mass of the insects that consumed wild-type leaves was 6.65 ± 1.26 g. By contrast, the masses of the insects that fed upon LapA-OX lines were variable ranging from 1.94 to 5.96 g. The mean mass (3.95 ± 1.79 g) of these insects was significantly different (P < 0.05) from insects grown on wild-type plants (Figures 10B and 10C).
Figure 10.
Wild-Type and LapA-SI Plant Responses to Herbivory.
Wild-type and LapA-OX plants (line 11) were infested with M. sexta larvae for 14 d.
(A) The foliage from LapA-OX (right) and wild-type (left) plants at day 14.
(B) M. sexta larvae raised on LapA-OX (right) or wild-type (left) plants.
(C) Masses of M. sexta larvae reared on wild-type (n = 8) and LapA-OX (n = 10) plants. Masses were significantly different (P < 0.05; Student's t test).
(D) Comparison of insect stature and head capsule sizes of 5th and 4th instar M. sexta larvae that consumed wild-type or LapA-OX foliage.
Head capsule diameters were measured to determine if the development of the larvae raised on LapA-OX was altered relative to insects that consumed wild-type foliage. All insects that fed on wild-type plants were in their fifth instar with capsule diameters of 6.2 ± 0.2 mm. By contrast, insects that consumed LapA-OX leaves experienced significant delays in development. While 67% were in their 5th instar (6.1 ± 0.1 mm), the remaining 33% of the larvae were in their 4th instar (4.2 ± 0.2 mm) (Figure 10D).
DISCUSSION
The role of hexameric LAPs in tomato defense was elucidated in this study. In tomato, as in a subset of the Solanaceae, there are two major and distinctive forms of LAP: LAP-A and LAP-N (Chao et al., 2000). LAP-A and LAP-N are distinctively regulated in response to stress and development. LAP-A is an abundant protein after wounding (Gu et al., 1996a; Chao et al., 1999). While LAP-N is readily detected by LAP antisera in all organs, LAP-N is present in vanishingly small quantities, and LapN encodes a rare class RNA (Tu et al., 2003). LAP-A and LAP-N have different substrate specificities (Gu et al., 1999; Tu et al., 2003). For example, LAP-N has 50-fold lower activity on substrates with N-terminal Leu than LAP-A. Since LapA-SI and LapA-AS plants downregulated both LAP-A and LAP-N, it was possible that the herbivore-susceptibility phenotype could have been attributed to one or both of these enzymes. However, the fact that plants ectopically expressing only LAP-A confer resistance to herbivory suggests that LAP-A, and not LAP-N, can exert its effects in modulating insect defense.
LAP-A is important in deterring herbivory-induced damage and controlling insect growth in tomato. Similarly, when the Solanum nigrum (nightshade) LAPs were downregulated using a transient virus-induced gene-silencing strategy (Hartl et al., 2008), masses of M. sexta larvae were greater than insects that fed upon control plants. Unlike tomato, where LapA RNAs and proteins are usually absent or detected at low levels in healthy leaves, S. nigrum leaves accumulate substantial amounts LapA RNAs and proteins in nondamaged leaves (Chao et al., 2000). At this time, it is unclear if the mechanisms for LAP action are similar in S. nigrum and tomato and whether or not the wound-induced LAP-A or LAP-N is important in defense in S. nigrum.
The data presented here indicated that LAP-A modulates the wound-signaling pathway that it important for deterring herbivory in tomato. When LapA RNAs and proteins were reduced in tomato foliage via RNA silencing (LapA-SI) or antisense (LapA-AS) strategies, local and systemic wounding signaling was impaired. The diminished wound signaling observed in LapA-SI and LapA-AS lines was consistent with an early hypothesis that LAP-A is important in the genesis of the bioactive wound peptide systemin as initially proposed by Pautot et al. (1993). This hypothesis was based on the role of aminopeptidases in the activation of animal bioactive peptides (Shrimpton et al., 2002), the importance of systemin in tomato wound signaling (Ryan, 2000), and the fact that in vitro LAP-A can process a putative 19-residue precursor of systemin (Leu-systemin) to its 18–amino acid form (Gu and Walling, 2000).
The results of this study indicated that LAP-A may not play a role in wound signaling through the regulation of systemin biogenesis in tomato. First, systemin is known to increase JA biosynthesis and activate wound signaling by inducing the expression of both early and late wound-response genes in tomato (Ryan, 2000). Second, LAP-A only modulates the expression of late wound-response genes (Pin1, Pin2, and PPO-F) and not early wound-response genes (AOS and LOX-D). Third, high levels of LapA RNAs and proteins were not sufficient to induce Pin RNA accumulation in healthy LapA-OX leaves. These data indicated that LAP-A did not generate a primary wound signal, such as systemin. Rather, LAP-A acted after generation of a wound signal(s) to modulate the late branch of wound signaling. Finally, exogenous application of JA to LapA-SI and wild-type plants showed that JA did not restore wound signaling in LapA-SI lines, indicating that LAP-A acted downstream of JA biosynthesis. Collectively, these data and the facts that LAP-A and Prosystemin are localized to distinct cellular compartments indicated that LAP-A is unlikely to influence the biogenesis of systemin (Narváez-Vásquez and Ryan, 2004; Narváez-Vásquez et al., 2007).
Analysis of LapA-SI, LapA-AS, and LapA-OX lines showed that LAP-A was a modulator of the late branch of the tomato wound response. Neither the LapA-OX plants nor the LapA-SI or LapA-AS plants had altered timing or magnitude of early wound-response gene expression. By contrast, analysis of these transgenic plants showed that LAP-A regulated the late branch of wound signaling. This conclusion was based on the facts that LapA-SI lines accumulated late wound-response RNAs (Pin1, Pin2, and PPO-F) and proteins (Pin1 and Pin2) to lower levels, and these RNAs disappeared more rapidly in wounded leaves of LapA-SI and LapA-AS than in wild-type plants. In addition, LapA-OX plant Pin RNAs persisted for longer periods of time in LapA-OX plants.
These data strongly support the bifurcation of JA-regulated early and late wound signaling in tomato (Orozco-Cárdenas et al., 2001). While the site of LAP-A action is currently unknown, several hypotheses can be proposed. LAP-A could modulate the enzymes essential for covalent modifications to JA. LAP-A could enhance JAR1-mediated conjugation of Ile to JA, which is critical for JA activity (Staswick and Tiryaki, 2004) or deter JA modifications that drive JA into inactive storage forms (Gidda et al., 2003; Miersch et al., 2008).
Alternatively, LAP-A may influence perception of JA or JA conjugates (Chini et al., 2007; Thines et al., 2007). While jai1 mutants are female sterile (Li et al., 2004), healthy LapA-SI lines did not display discernable vegetative or reproductive phenotypes relative to wild-type plants, suggesting that LAP-A may activate signaling steps downstream or independent of JA perception. This was also supported by the fact that when germinated in the presence of JA, LapA-SI and wild-type plant root elongation was inhibited, unlike root elongation in the JA perception mutant jai1 (Li et al., 2004). An additional distinction between the jai1 mutant and the LapA-SI and LapA-AS lines was observed when expression of Pins in floral buds and after wounding was compared. The jai1 mutant suppresses both early and late wound-response gene expression in damaged leaves and the developmental regulation of Pin RNAs in floral buds (Li et al., 2004). By contrast, LapA-SI and LapA-AS plants impaired the accumulation of Pin RNAs in leaves but did not alter Pin RNA levels in floral buds. These data indicated that Pin1 RNA levels were controlled by different mechanisms in floral buds and wounded leaves, implicating a developmental regulation that was independent of LAP-A. Collectively, these data and the fact that exogenous JA does not restore wound signaling in LapA-SI or LapA-AS lines suggest that LAP acts downstream of JA biosynthesis and perception. Alternatively, LAP-A could act in a JA-independent pathway that modulates JA-regulated wound signaling. There are several other potential sites for LAP-A action. LAP-A could suppress a negative regulator of the JA pathway, such as salicylic acid or auxin (Ryan, 2000; Walling, 2000; Rojo et al., 2003), or influence the production or perception of NO and H2O2 (Orozco-Cárdenas and Ryan, 1999, 2002; Orozco-Cárdenas et al., 2001). Several experiments are in progress to elucidate the site of LAP-A action.
Both LapA-SI and LapA-OX lines displayed distinctive phenotypes when challenged by M. sexta feeding. LapA-SI plants were more susceptible to herbivory and larval masses were twofold greater than wild-type when insects consumed LapA-SI foliage. By contrast, M. sexta larvae consumed less LapA-OX foliage relative to wild-type plants. Most significantly, the insects that fed on LapA-OX plants were delayed in both growth and development. Since the LapA-OX plants did not alter LAP-N levels, these data support the idea that LAP-A, and not LAP-N, is the modulator of wound responses and insect defense.
It is important to note that LapA-OX plants were phenotypically normal with wild-type stature and fertility, suggesting there was no fitness cost associated with overexpressing LAP-A in greenhouse studies. This is important because the maintenance of constitutive proteins or the continuous mounting of defenses often has severe impacts on plant growth or fertility (Bostock, 2005). Because the overexpression of LAP-A does not impose a significant fitness cost to the plant, the LapA-OX plants are viable candidates for field trials to improve insect resistance in crop tomato.
While the step(s) in wound signaling downstream of JA perception that is controlled by LAP-A is not presently known, LAP-A's influence on insect defense was likely due to the impaired expression of the late wound-response genes that encode proteins with known antinutritive effects (Pins, PPO, arginase, and Thr deaminase). This is supported by the quantitative downregulation of wound-response RNAs in leaves (locally and systemically) after wounding and reduced levels of Pin1 and Pin2 activities M. sexta–infested leaves in LapA-SI plants relative to wild-type plants.
However, LAPs may also have a direct effect in the insect gut (Pautot et al., 1993; Chen et al., 2005). Endoproteases have important roles in defense (Schaller, 2004; van der Hoorn and Jones, 2004). For example, the Mir1 Cys endopeptidase is essential for resistance to the fall army worm (Spodoptera frugiperda) in maize (Zea mays; Pechan et al., 2000; Mohan et al., 2006). Like Mir1, LAP-A could disrupt midgut integrity and impact digestive physiology or influence the bioavailability of essential amino acids. The recent finding that LAP-A, arginase, and Thr deaminase are enriched in the food bolus in the larval midgut makes this hypothesis feasible (Chen et al., 2005, 2007). In addition, LAP-A's alkaline pH optima (Gu et al., 1999) and substrate specificity (Gu et al., 1999; Gu and Walling, 2000), activity in the food bolus and insect frass (D.N. Aromdee and L.L. Walling, unpublished results) (Chen et al., 2007) also suggest that LAP-A may directly contribute to tomato defenses in the alkaline environment of the lepidopteran gut. However, preliminary experiments with artificial diets laced with LAP-A suggest that LAP-A alone cannot directly deter insect development (D.N. Aromdee, N. Beckage, and L.L. Walling, unpublished results). Since LAP-A readily hydrolyzes substrates with N-terminal Arg, as well as Leu (Gu et al., 1999), it is possible that LAP-A acts cooperatively with arginase to deplete the levels of Arg, which is an essential amino acid for larvae (Chen et al., 2005).
In conclusion, tomato LAP-A is the first plant aminopeptidase that modulates a signal transduction pathway in plants. Despite the conserved Zn-ion binding sites and highly conserved catalytic domains in LAPs from plants, animals, and prokaryotes (Gu and Walling, 2000, 2002), LAPs serve very different roles in each of these organisms. It is unclear if the tomato LAP-A is bifunctional like the E. coli XerB or if its regulatory activity is mediated solely by its aminopeptidase domain. With the discovery of >28 aminopeptidases in the Arabidopsis genome (Walling, 2006), it is clear that our understanding of the role of aminopeptidases in plant growth, development, and defense is in its infancy. The discovery that LAP-A modulates the signaling of late-wound response genes, illustrates that this category of enzymes holds potential for important future discoveries.
METHODS
Plant Materials and Growth Conditions
Solanum lycopersicum (formerly Lycopersicon esculentum) UC82 (wild-type), P35S:LapA-SI, and P35S:LapA-AS plants were grown in a growth chamber with an 18-h (28°C)/6-h (24°C) light (300 μE)/dark cycle. The P35S:LapA transgene was constructed as a transcriptional fusion. This transgene fused the 35S promoter from Cauliflower mosaic virus, the 5′-untranslated region and coding region of the LapA1 cDNA, and the 3′-untranslated region of nopaline synthase. This was achieved by digestion of pBlapA1 (Gu et al. 1996a) with SpeI and blunt end cloning into SmaI-digested pBI121 (Clonetech). Tomato transformation methods were previously described (Chao et al., 1999). Kanamycin-resistant plants were regenerated and P35S:LapA plants identified by genomic PCR using LapA primer pairs that selectively amplified the transgenic LapA gene (Lap5, 5′-AGAATTCCGTTGCTGTCG-3′ and Lap6, 5′-CATCGGTGGAAGCAAGTG-3′). The Lap5 forward primer was designed in the EcoRI adapter region of this chimeric gene and allowed the specific amplification of the transgene only. LapA-SI lines were identified by assessing levels of LapA RNAs and proteins by RNA and protein blots (Gu et al., 1996b; Chao et al., 1999). Homozygous P35S:LapA-SI lines were identified by following transgene segregation in the T2 and T3 generations using Lap5 and Lap6 primers. Four independent homozygous P35S:LapA-SI lines with varying levels of LapA expression were identified and characterized. The P35S:LapA-SI lines 28 and 29 had the strongest silencing phenotypes and were characterized further. P35S:LapA-AS plants that partially suppressed Lap gene expression were previously described (Pautot et al., 2001); LapA-AS line 4 was used for these studies. Homozygous P35S:LapA-OX lines were identified in the same manner. Six independent P35S:LapA-OX lines with varying levels of LapA expression were identified and characterized. Data for P35S:LapA-OX line 11 is presented. Other P35S:LapA-OX lines silenced their transgenes in T2 and T3 generations.
Wound and JA Treatments
Three-week-old plants were used in the wounding time-course studies. Plants were wounded by crushing the distal end of each leaflet of two lower leaves with a pair of needle-nosed pliers. Wounded leaves (local response) and all leaflets (typically six to eight leaflets) from apical leaves (systemic response) were collected at designated times. The leaves of five plants at each time point were pooled together for RNA extractions. For qPCR experiments, the lower leaf of 2-week-old tomato plants was wounded twice with a hemostat across the midvein of all leaflets. The wounded and apical unwounded leaves were collected at designated times after wounding. Leaves from three plants at each time point were pooled together, and the experiment was repeated twice. For exogenous JA treatments, shoots were excised from 3-week-old tomato plants with a razor blade; shoots were incubated in flasks with 0.005% ethanol (control) or 10 μM JA and 0.005% ethanol in a closed environment for 18 h as described by Chao et al. (1999). Unopened, 8-mm floral buds were collected from mature greenhouse-grown wild-type, LapA-SI (line 28), and LapA-OX (line 11) plants. Leaves and buds were excised with a razor blade and deposited immediately into liquid nitrogen; frozen tissues were stored until use.
Wild-type (UC82b), LapA-SI (line 28), Castlemart (wild-type control), jai1/jai1 homozygous, and Jai1/jai1 heterozygous seeds were germinated in the presence of 0, 1, 10, 100, and 1000 μM JA for 5 d as described by Li et al. (2004). The jai1/jai1 root elongation phenotype was only observed in seeds germinated in the presence of 1 mM JA. The inherent asynchrony of seedling growth did not allow discrimination of wild-type, heterozygous, or homozygous jai1 plants at 1 to 100 μM JA.
RNA and Protein Blots
Total leaf protein was extracted as previously described (Wang et al., 1992a). Protein concentrations were determined using a modified Bradford assay with BSA as a standard. Proteins were fractionated by two-dimensional PAGE and gels were stained with Coomassie Brilliant Blue R 250 or electrotransferred to nitrocellulose filters (Wang et al., 1992b; Gu et al., 1996b). Protein blots were incubated with a LAP-A polyclonal antiserum (1:500) and goat anti-rabbit IgG (1:3000) conjugated to alkaline phosphatase; antigen antibody complexes were visualized using 5- bromo-4-chloro-3-indolyl phosphate substrate and nitrotetrazolium (Wang et al., 1992b; Gu et al., 1996b). RNAs were extracted using a hot phenol method and blotted to a nylon membrane as previously described (Pautot et al., 2001). RNA gel blots were hybridized with 32P-labeled probes generated using a Prime-a-Gene random labeling kit (Promega); templates included the following S. lycopersicum cDNAs: LapA1 (U50151), Pin1 (K03290), Pin2 (K03291), PPO-F (AI490781), AOS (AJ271093), LoxD (U37840), and elongation factor 1α (EF1-α) (X14449) cDNA clones. RNA gel blots were exposed to Hyperfilm MP (Amersham) with an intensifying screen.
Quantitative PCR and Data Analysis
Total RNA was extracted from wounded and systemic leaves using the hot-phenol method (Pautot et al., 2001) and quantified using a Nano-Drop ND-1000 spectrophotometer. High RNA quality was inferred from 260/280-nm absorbance ratios higher than 1.8 and by the presence of intact rRNA bands in denaturing 1.5% formaldehyde gels. Genomic DNA was removed with RQ1 DNase (Promega), followed by heat inactivation for 10 min at 70°C. A minus reverse transcriptase (RT) control was performed with reference gene primers to assure cDNAs were free of genomic DNA. First-strand cDNA was synthesized from 1 μg total RNA using the ImpromII reverse transcriptase (Promega) and oligo-dT (25-mer) primers as previously described (Zarate et al., 2007). cDNAs were diluted with water 10 to 100 times for qPCR analysis.
Tomato translation EF1α, ubiquitin (Ubi3), and housekeeping gene 4 (HKG4; a hypothetical protein) were used as internal reference genes for qPCR analysis. Primers for EF1-α and Ubi3 have been previously reported (Rotenberg et al., 2006). Analysis of tomato microarray data sets identified HKG4 as encoding an RNA that is present at similar levels in flowers and fruit, after JA treatment of roots and wounding of leaves (M.A. Smith and L.L. Walling, unpublished results). Transcript stability for the three reference genes during the wound-induction time course was determined using the BestKeeper program (Pfaffl et al., 2004). qPCR primers for reference gene HKG4 and the wound-inducible genes Lox-D, AOS, LapA, Pin1, Pin2, and PPO-F were designed based on tomato cDNA sequences obtained from GenBank, using the Vector NTI Advance 10 software package (Invitrogen). Primers were designed to avoid secondary structures and primer dimer formation (Bio-Rad Laboratories). Annealing temperatures and PCR efficiencies were experimentally determined for each primer set using the MyiQ real-time detection system as indicated by the manufacturer (Bio-Rad Laboratories). All primers sequences, and their corresponding annealing temperatures and amplicon sizes for each gene, are shown in Supplemental Table 1 online.
All reactions were performed in triplicate using iQ SYBRGreen Supermix (Bio-Rad Laboratories) and 200 nM of gene-specific qPCR primers. Reactions (25-μL) were performed using the MyiQ real-time detection system using a two-step amplification program as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30 s and 55-63°C for 45 s. A standard melting curve protocol began immediately after amplification to determine the quality of the PCR products and allow detection of primer-dimers (Bio-Rad Laboratories). Background-subtracted, fluorescent values were imported into an Excel file, processed, and analyzed using a real-time PCR miner program (Zhao and Fernald, 2005), which allowed calculation of efficiencies and Ct values and associated standard errors and variance coefficients of individual reactions. Average Ct values of replicate samples and the average efficiency of genes were used to calculate mRNA levels of reference and wound-inducible genes. Relative mRNA level of each wound-inducible gene at each time point was obtained after normalization against the average mRNA levels of the three reference genes for each time point. The results are presented as the RNA levels (relative fold change) relative to the designated control (LapA-SI RNA levels at 0 h).
Insect Infestations and Proteinase Inhibitor Assays
Manduca sexta were reared on artificial diet (Bell and Joachim, 1976). Eighteen 1st-instar M. sexta larvae were placed on a canopy of 24 4-week-old tomato plants of each line for 11 d (wild-type and LapA-SI line 28) or 14 d (wild type and LapA-OX line 11). The caterpillars were removed, weighed, and the masses analyzed using a Student's t test. Larvae were chilled at 4°C for 2 h to limit movement, and capsule diameters were measured using an ocular micrometer and analyzed using a Student's t test. Proteinase inhibitor levels in leaves were measured using a radial immunodiffusion assay in damaged leaves from five plants infested with M. sexta for 24 h (Hare and Walling, 2006). Developmentally matched leaves from five noninfested plants served as controls. The experiment was repeated once. Pin1 and Pin2 polyclonal antisera and Pin standards were kindly supplied by Clarence Ryan (Washington State University, Pullman, WA),
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. LapA and Pin1 RNAs in Wild-Type and LapA-OX Plants from 24 to 96 h.
Supplemental Table 1. Quantitative PCR Primers.
Supplementary Material
Acknowledgments
We thank Melissa Smith for review of the manuscript, Nancy Beckage for supplying M. sexta larvae, Michael Adams for use of his ocular micrometer, S.-Y. Park for partial characterization of LapA-SI lines, Ana C. Morillo for aide with RNA isolation and qPCR reactions, and members of the Walling and Kaloshian labs for helpful conversations. This work was supported by National Science Foundation grants (IBN-0077862) and (IOS-0725093) to L.L.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Linda L. Walling (linda.walling@ucr.edu).
Online version contains Web-only data.
References
- Bell, R.A., and Joachim, F.G. (1976). Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms. Ann. Entomol. Soc. Am. 69 365–373. [Google Scholar]
- Beninga, J., Rock, K.L., and Goldberg, A.L. (1998). Interferon-γ can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 273 18734–18742. [DOI] [PubMed] [Google Scholar]
- Bostock, R.M. (2005). Signal crosstalk and induced resistance: Straddling the line between cost and benefit. Annu. Rev. Phytopathol. 43 545–580. [DOI] [PubMed] [Google Scholar]
- Broadway, R.M., and Duffey, S.S. (1986). Plant proteinase inhibitors mechanism of action and effect on the growth and digestive physiology of larval Heliothis zea and Spodoptera exigua. J. Insect Physiol. 32 827–834. [Google Scholar]
- Chao, W.S., Gu, Y.-Q., Pautot, V., Bray, E.A., and Walling, L.L. (1999). Leucine aminopeptidase RNAs, proteins, and activities increase in response to water deficit, salinity, and the wound signals systemin, methyl jasmonate, and abscisic acid. Plant Physiol. 120 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, W.S., Pautot, V., Holzer, F.M., and Walling, L.L. (2000). Leucine aminopeptidases: The ubiquity of LAP-N and the specificity of LAP-A. Planta 210 563–573. [DOI] [PubMed] [Google Scholar]
- Chen, H., Gonzales-Vigil, E., Wilkerson, C.G., and Howe, G.A. (2007). Stability of plant defense proteins in the gut of insect herbivores. Plant Physiol. 143 1954–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Wilkerson, C.G., Kuchar, J.A., Phinney, B.S., and Howe, G.A. (2005). Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 102 19237–19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J.M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671. [DOI] [PubMed] [Google Scholar]
- Colloms, S.D. (2004). Leucyl aminopeptidase PepA. In Handbook of Proteolytic Enzymes, A.J. Barrett, N.D. Rawlings, and J.F. Woesner, eds (San Diego, CA: Elsevier Academic Press), pp. 905–911.
- Felton, G.W., Donato, K., Del Vecchio, R.J., and Duffey, S.S. (1989). Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. J. Chem. Ecol. 15 2667–2694. [DOI] [PubMed] [Google Scholar]
- Gidda, S.K., Miersch, O., Levitin, A., Schmidt, J., Wasternack, C., and Varin, L. (2003). Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278 17895–17900. [DOI] [PubMed] [Google Scholar]
- Gu, Y.-Q., Chao, W.S., and Walling, L.L. (1996. a). Localization and post-translational processing of the wound-induced leucine aminopeptidase proteins of tomato. J. Biol. Chem. 271 25880–25887. [DOI] [PubMed] [Google Scholar]
- Gu, Y.-Q., Holzer, F.M., and Walling, L.L. (1999). Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato. Eur. J. Biochem. 263 726–735. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., Pautot, V., Holzer, F.M., and Walling, L.L. (1996. b). A complex array of proteins related to the multimeric leucine aminopeptidase of tomato. Plant Physiol. 110 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.Q., and Walling, L.L. (2000). Specificity of the wound-induced leucine aminopeptidase (LAP-A) of tomato - Activity on dipeptide and tripeptide substrates. Eur. J. Biochem. 267 1178–1187. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., and Walling, L.L. (2002). Identification of residues critical for activity of the wound-induced leucine aminopeptidase (LAP-A) of tomato. Eur. J. Biochem. 269 1630–1640. [DOI] [PubMed] [Google Scholar]
- Hare, J.D., and Walling, L.L. (2006). Constitutive and jasmonate-inducible traits of Datura wrightii. J. Chem. Ecol. 32 29–47. [DOI] [PubMed] [Google Scholar]
- Hartl, M., Merker, H., Schmidt, D.D., and Baldwin, I.T. (2008). Optimized virus-induced gene silencing in Solanum nigrum reveals the defensive function of leucine aminopeptidase against herbivores and the shortcomings of empty vector controls. New Phytol. 179 356–365. [DOI] [PubMed] [Google Scholar]
- Howe, G.A. (2004). Jasmonates as signals in the wound response. J. Plant Growth Regul. 23 223–237. [Google Scholar]
- Johnson, R., Narvaez, J., An, G., and Ryan, C. (1989). Expression of proteinase inhibitors I and II in transgenic tobacco plants: Effects on natural defense against Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 86 9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jwa, N.-S., and Walling, L.L. (2001). Influence of elevated CO2 concentration on disease development in tomato. New Phytol. 149 509–518. [DOI] [PubMed] [Google Scholar]
- Kang, J.H., and Baldwin, I.T. (2006). Isolation and characterization of the threonine deaminase promoter in Nicotiana attenuata. Plant Sci. 171 435–440. [DOI] [PubMed] [Google Scholar]
- Li, L., Zhao, Y.F., McCaig, B.C., Wingerd, B.A., Wang, J.H., Whalon, M.E., Pichersky, E., and Howe, G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, M., Fowler, J.H., and Walling, L.L. (2006). Leucine aminopeptidases: diversity in structure and function. Biol. Chem. 387 1535–1544. [DOI] [PubMed] [Google Scholar]
- Miersch, O., Neumerkel, J., Dippe, M., Stenzel, I., and Wasternack, C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177 114–127. [DOI] [PubMed] [Google Scholar]
- Mohan, S., Ma, P.W.K., Pechan, T., Bassford, E.R., Williams, W.P., and Luthe, D.S. (2006). Degradation of the S. frugiperda peritrophic matrix by an inducible maize cysteine protease. J. Insect Physiol. 52 21–28. [DOI] [PubMed] [Google Scholar]
- Narváez-Vásquez, J., and Ryan, C.A. (2004). The cellular localization of prosystemin: A functional role for phloem parenchyma in systemic wound signaling. Planta 218 360–369. [DOI] [PubMed] [Google Scholar]
- Narváez-Vásquez, J., Tu, C.J., Park, S.Y., and Walling Linda, L. (2007). Targeting and localization of wound-inducible leucine aminopeptidase A in tomato leaves. Planta 227 341–351. [DOI] [PubMed] [Google Scholar]
- Nijhout, H.F., and Williams, C.M. (1974). Control of molting and metamorphosis in the tobacco hornworm Manduca sexta cessation of juvenile hormone secretion as a trigger for pupation. J. Exp. Biol. 61 493–502. [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M., and Ryan, C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M.L., Nárvaez-Vásquez, J., and Ryan, C.A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13 179–191. [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M.L., and Ryan, C.A. (2002). Nitric oxide negatively modulates wound signaling in tomato plants. Plant Physiol. 130 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot, V., Holzer, F.M., Chaufaux, J., and Walling, L.L. (2001). The induction of tomato leucine aminopeptidase genes (LapA) after Pseudomonas syringae pv. tomato infection is primarily a wound response triggered by coronatine. Mol. Plant-Microbe Interact. 14 214–224. [DOI] [PubMed] [Google Scholar]
- Pautot, V., Holzer, F.M., Reisch, B., and Walling, L.L. (1993). Leucine aminopeptidase: An inducible component of the defense response in Lycopersicon esculentum (tomato). Proc. Natl. Acad. Sci. USA 90 9906–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechan, T., Cohen, A., Williams, W.P., and Luthe, D.S. (2002). Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc. Natl. Acad. Sci. USA 99 13319–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechan, T., Ye, L., Chang, Y.-m., Mitra, A., Lin, L., Davis, F.M., Williams, W.P., and Luthe, D.S. (2000). A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortes, H., Willmitzer, L., and Sanchez-Serrano, J.J. (1991). Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell 3 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W., Tichopad, A., Prgomet, C., and Neuvians, T.P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 509–515. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Solano, R., and Sanchez-Serrano, J.J. (2003). Interactions between signaling compounds involved in plant defense. J. Plant Growth Regul. 22 82–98. [Google Scholar]
- Rotenberg, D., Thompson, T.S., German, T.L., and Willis, D.K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 138 49–59. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (1967). Quantitative determination of soluble cellular proteins by radial diffusion in agar gels containing antibodies. Anal. Biochem. 19 430–440. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477 112–121. [DOI] [PubMed] [Google Scholar]
- Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J.W., and Fluhr, R. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A. (2004). A cut above the rest: The regulatory function of plant proteases. Planta 220 183–197. [DOI] [PubMed] [Google Scholar]
- Shrimpton, C.N., Smith, A.I., and Lew, R.A. (2002). Soluble metalloendopeptidases and neuroendocrine signaling. Endocr. Rev. 23 647–664. [DOI] [PubMed] [Google Scholar]
- Srivastava, R., Liu, J.-X., and Howell, S.H. (2008). Proteoltyic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A. (1996). Aminopeptidases, occurrence, regulation and nomenclature. In Aminopeptidases, A. Taylor, ed (Austin, TX: R. G. Landes Company), pp. 1–20.
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G.H., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Tu, C.J., Park, S.Y., and Walling, L.L. (2003). Isolation and characterization of the neutral leucine aminopeptidase (LapN) of tomato. Plant Physiol. 132 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L., and Jones, J.D. (2004). The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 7 400–407. [DOI] [PubMed] [Google Scholar]
- Walling, L.L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19 195–216. [DOI] [PubMed] [Google Scholar]
- Walling, L.L. (2004). Leucyl aminopeptidase (plant). In Handbook of Proteolytic Enzymes, A.J. Barrett, N.D. Rawlings, and J.F. Woesner, eds (San Diego, CA: Elsevier Academic Press), pp. 901–904.
- Walling, L.L. (2006). Recycling or regulation? The role of amino-terminal modifying enzymes. Curr. Opin. Plant Biol. 9 227–233. [DOI] [PubMed] [Google Scholar]
- Wang, C.S., Walling, L.L., Eckard, K.J., and Lord, E.M. (1992. a). Patterns of protein accumulation in developing anthers of Lilium longiflorum correlate with histological events. Am. J. Bot. 79 118–127. [Google Scholar]
- Wang, C.S., Walling, L.L., Eckard, K.J., and Lord, E.M. (1992. b). Immunological characterization of a tapetal protein in developing anthers of Lilium longiflorum. Plant Physiol. 99 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., Stenzel, I., Hause, B., Hause, G., Kutter, C., Maucher, H., Neumerkel, J., Feussner, I., and Miersch, O. (2006). The wound response in tomato - Role of jasmonic acid. J. Plant Physiol. 163 297–306. [DOI] [PubMed] [Google Scholar]
- Zarate, S.I., Kempema, L.A., and Walling, L.L. (2007). Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S., and Fernald, R. (2005). Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.