The age of omics is in full swing. Full genome sequences and the use of DNA microarrays to monitor gene expression on a genome-wide scale arrived on the scene a little over a dozen years ago, and today major developments in DNA sequencing technology have ushered in a new era in the study of gene regulation. Around the turn of the century, the introduction of massively parallel next-generation DNA sequencing technology allowed the development of new assays for analyzing complex nucleic acid samples based on highly parallelized sequencing of vast numbers of DNA molecules in a complex sample, combined with matching to a reference genome (reviewed in Morozova and Marra, 2008; Wold and Myers, 2008).
Massively parallel signature sequencing (MPSS), the first of the next-generation sequencing technologies, involved cloning cDNA fragments onto microbeads and a fluorescence-based sequencing method that generated a unique 17-base signature sequence for each mRNA at a specific site upstream from its poly(A) tail (Brenner et al., 2000; Reinartz et al., 2002). The original MPSS platform (from the former Lynx Therapeutics, which became Solexa and now Illumina) has since been replaced by an improved sequencing by synthesis (SBS) method, based on detecting the identity of a nucleotide immediately after its incorporation into a growing strand of newly synthesized cDNA. Other next-generation approaches include 454 Life Sciences' SBS pyrosequencing method, Applied Biosystems' SOLiD sequencing by ligation system, Helicos Biosciences' single-molecule synthesis platform, and several other single-molecule synthesis platforms that will be available soon (Schuster, 2008; Gupta, 2009). These sequencing technologies are often referred to as “deep sequencing” because of their ability to generate huge numbers of sequencing reads per experiment or instrument run. For example, as early as 2005, it was shown that a small-sized bacterial genome could be sequenced with nearly complete coverage and accuracy in a single instrument run using the pyrosequencing technology of the 454 Life Sciences platform (Margulies et al., 2005).
These sequencing-based approaches have some distinct advantages over microarray-based approaches for genome-wide transcriptomics (the study of gene expression) and epigenomics (the study of chromatin organization and dynamics), such as avoiding complex intermediate cloning and microarray construction steps and the ability to generate a massive amount of sequence quickly. Using these approaches, gene expression is assayed by directly sequencing cDNA molecules obtained from an mRNA sample and simply counting the number of molecules corresponding to each gene to assess transcript abundance.
The power of next-generation sequencing approaches for transcriptomics was demonstrated in Arabidopsis by Meyers et al. (2004) and Lu et al. (2005), using the MPSS approach, and by numerous others since then in Arabidopsis and other organisms. Weber et al. (2007) provided another important proof-of-concept transcriptome analysis in plants, with a comparison of conventional EST sequencing and microarray transcript profiling to sampling of the Arabidopsis transcriptome through pyrosequencing. This study (similar to several that have been conducted in other organisms) highlighted some of the advantages of direct sequencing methods and showed that they have the potential for greater sensitivity (allowing for detection of more novel and rare-abundance transcripts) than conventional EST sequencing or microarray analysis.
More than identifying genes and discovering when and where they are expressed, the ultimate goal of functional genomics is a complete understanding of the regulatory networks that control genome activity, drive organismal development and environmental responses, and give rise to the evolution of species. Crucial to this goal is an understanding of the epigenome and the small regulatory RNA component of the transcriptome, and direct sequencing methods are proving particularly useful in exploring these realms of the genome. Lu et al. (2005) first demonstrated the utility of direct sequencing technology for analyses of small RNA on a genomic scale, using MPSS in Arabidopsis. More recently, Nobuta et al. (2008) used Illumina SBS technology to analyze the size distribution of small RNA in maize. These and other researchers have used MPSS and the newer next generation technologies to provide significant new insights on the identification, biogenesis, and function of small RNAs in a number of plant and animal species.
Direct sequencing methods also are being employed to begin mapping the epigenome of a number of organisms and to correlate this information with transcriptome data sets. Cokus et al. (2008) used Illumina's SBS technology to map cytosine methylation in Arabidopsis. They analyzed DNA methylation sequence composition and distribution, and they described the effect of various DNA methylation mutants on genome-wide methylation patterns. They were also able to assess methylation on previously inaccessible components of the genome. Lister et al. (2008) also used this approach to map cytosine methylation patterns in Arabidopsis floral tissues and further to integrate this data with direct sequencing analysis of the mRNA and small RNA transcriptomes. They discovered extensive previously undetected DNA methylation and a direct relationship between DNA methylation and the location of small RNAs in the genome, yielding insight into RNA-directed DNA methylation. Both of these studies confirmed the broad patterns of methylation shown in previous studies (e.g., a high degree of methylation in heterochromatin and more dispersed pattern in euchromatin) and also provided significant new insights into the dynamics of the epigenome.
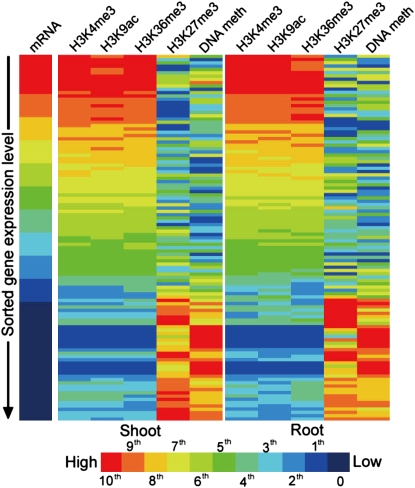
In this issue of The Plant Cell, Wang et al. (pages 1053–1069) extend the use of direct sequencing methods to the maize epigenome using the Illumina SBS platform to provide a comprehensive analysis of DNA methylation and histone modifications in relation to expression patterns of both the mRNA and small RNA subsets of the trancriptome in maize. This study largely confirms epigenomic patterns that have been established in other species (Arabidopsis and other organisms), such as, for example, H3K4me3, H3K9ac, and H3K36me3 being associated with transcriptionally active genes, and H3K27me3 and DNA methylation predominantly being found in transcriptionally inactive genes and repetitive elements (see figure). However, it also substantially extends the available resources for maize and provides new insights into chromatin dynamics and small RNA biology. The data set is remarkably comprehensive, including separate analyses from shoot and root tissue for DNA methylation (assessed using the methylation-sensitive restriction enzyme McrBC), four specific histone modifications (methylation or acetylation of specific lysine residues of histone H3, assessed using antibodies specific for H3K4me3, H3K9ac, H3K27me3, and H3K36me3), and separation of the transcriptome into mRNA and small RNA components, including microRNA and small interfering RNA (siRNA).
Figure 1.
Heat Maps of Epigenetic Modification Levels on ∼60,000 Genes Sorted by Their Expression Measured by Deep Sequencing.
Gene expression levels and modifications levels were transformed to 100 percentiles, and each bar represents the averaged level of nearly 600 genes within each percentile. (From Figure 4 of Wang et al. [2009].)
An interesting feature of the maize epigenomic landscape that was found is that the genic DNA methylation pattern is similar to that of rice in that it shows a peak around ATG sites but differs substantially from that of Arabidopsis, which shows higher DNA methylation in transcribed regions. In addition, it was found that whereas the histone modifications associated with gene activation tend to occur together, the two gene-repressive histone marks under study, H3K27me3 and DNA methylation, tend to exclude each other at the same locus. Intriguing features of the small RNA landscape include an organ-specific (shoot versus root) distribution of small RNAs, indicative of their tissue-specific biogenesis. Nobuta et al. (2008) previously found that the maize genome contains a class of 22-nucleotide siRNA that is lacking in a number of other monocots and introduced the possibility of two distinct biogenesis pathways for siRNA in maize. Wang et al. provide evidence that these 22-nucleotide siRNAs might be generated from long double-stranded RNA precurors, offering additional support for the existence of at least two pathways for the biogenesis of siRNAs. Further information is gleaned on the biogenesis and activity of other size classes of small RNA, for example, indicating the possibility of siRNA involvement in tissue-specific and targeted paramutation.
Understanding the regulation of genome activity is a daunting task, in particular for organisms such as maize that contain an enormous quantity of repetitive sequence and non-protein-coding DNA. Methods based on various platforms of microarrays and conventional sequencing will undoubtedly continue to contribute critical information, at least in the near future. However, studies such as that of Wang et al. show that direct sequencing technology has clearly proven its value. Further improvements to cost and efficiency as well as to the computational and bioinformatic analysis of sequence data will allow us to make even greater advancements in mapping genomic and epigenomic landscapes and understanding the intricacies of their function.
References
- Brenner, S., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18 630–634. [DOI] [PubMed] [Google Scholar]
- Cokus, S.J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C.D., Pradhan, S., Nelson, S.F., Pellegrini, M., and Jacobsen, S.E. (2008). Shotgun bisulfite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.K. (2009). Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 26 602–611. [DOI] [PubMed] [Google Scholar]
- Lister, R., O'Malley, R.C., Tonti-Filippini, J., Gregory, B.D., Berry, C.C., Millar, A.H., and Ecker, J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Margulies, M., et al. (2005). Genome sequencing in microfabricated high-density picoliter reactors. Nature 437 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Tej, S.S., Vu, T.H., Haudenschild, V.A., Edberg, S.B., Ghazal, H., and Decola, S. (2004). The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 14 1641–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova, O., and Marra, M.A. (2008). Application of next-generation sequencing technologies in functional genomics. Genomics 92 255–264. [DOI] [PubMed] [Google Scholar]
- Nobuta, K., et al. (2008). Distinct size distribution of endogenous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinartz, J., Bruyns, E., Lin, J.-Z., Burcham, T., Brenner, S., Bowen, B., Kramer, M., and Woychik, R. (2002). Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief. Funct. Genomic. Proteomic. 1 95–104. [DOI] [PubMed] [Google Scholar]
- Schuster, S.C. (2008). Next generation DNA sequencing transforms today's biology. Nat. Methods 5 16–18. [DOI] [PubMed] [Google Scholar]
- Wang, X., Elling, A.A., Li, X., Li, N., Peng, Z., He, G., Sun, H., Qi, Y., Liu, X.S., and Deng, X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.P.M., Weber, K.L., Wilkerson, C., and Ohlrogge, J.B. (2007). Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 144 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold, B., and Myers, R.M. (2008). Sequence census methods for functional genomics. Nat. Methods 5 19–21. [DOI] [PubMed] [Google Scholar]