Abstract
Maize (Zea mays) has an exceptionally complex genome with a rich history in both epigenetics and evolution. We report genomic landscapes of representative epigenetic modifications and their relationships to mRNA and small RNA (smRNA) transcriptomes in maize shoots and roots. The epigenetic patterns differed dramatically between genes and transposable elements, and two repressive marks (H3K27me3 and DNA methylation) were usually mutually exclusive. We found an organ-specific distribution of canonical microRNAs (miRNAs) and endogenous small interfering RNAs (siRNAs), indicative of their tissue-specific biogenesis. Furthermore, we observed that a decreasing level of mop1 led to a concomitant decrease of 24-nucleotide siRNAs relative to 21-nucleotide miRNAs in a tissue-specific manner. A group of 22-nucleotide siRNAs may originate from long-hairpin double-stranded RNAs and preferentially target gene-coding regions. Additionally, a class of miRNA-like smRNAs, whose putative precursors can form short hairpins, potentially targets genes in trans. In summary, our data provide a critical analysis of the maize epigenome and its relationships to mRNA and smRNA transcriptomes.
INTRODUCTION
Histones are decorated by numerous epigenetic modifications, particularly at their N-terminal ends (Fuchs et al., 2006; Kouzarides, 2007). It has been proposed that combinations of different histone modifications form a histone code (Jenuwein and Allis, 2001), which extends the genetic code embedded in the DNA nucleotide sequence. Numerous studies have demonstrated that histone modifications influence gene expression genome-wide. Whereas histone acetylation generally is associated with gene activation (e.g., Wang et al., 2008), histone methylation can lead to either gene repression or activation depending on the modification site (Shi and Dawe, 2006; Barski et al., 2007; Mikkelsen et al., 2007; Zhang et al., 2007).
DNA methylation adds another layer of heritable epigenetic changes. In higher plants, methylation of cytosines is present in CG, CHG (where H is A, C, or T), and asymmetric CHH sequence contexts (Henderson and Jacobsen, 2007). Recent studies have shown that cytosines are methylated not only in plant repetitive sequences and transposable elements (TEs) but also in promoters and gene bodies and that DNA methylation is highly correlated with transcription (Rabinowicz et al., 2005; Zhang et al., 2006; Vaughn et al., 2007; Zilberman et al., 2007; Cokus et al., 2008; Li et al., 2008c; Lister et al., 2008). Epigenetic changes, such as DNA methylation and histone modifications, do not act in isolation but rather in concert with each other, allowing for complex interdependencies. For example, in Arabidopsis thaliana, CHG DNA methylation is associated with dimethylation of histone H3K9 (Bernatavichute et al., 2008), and CG DNA methylation is necessary for transgenerational epigenetic stability, including H3K9 methylation (Mathieu et al., 2007). Moreover, histone deacetylase HDA6 and histone methyltransferase KRYPTONITE are known to control DNA methylation (Aufsatz et al., 2002; Jackson et al., 2002). Other histone methylations and acetylations have been shown to be excluded by chromatin structure remodeling induced by DNA methylation (Lorincz et al., 2004; Okitsu and Hsieh, 2007). A complex interplay between DNA methylation, histone modifications, and gene expression has been reported in rice (Oryza sativa; Li et al., 2008c).
In addition, recent studies have shown that small RNAs (smRNAs) are associated with DNA methylation (Lister et al., 2008) and that small interfering RNAs (siRNAs) target epigenetic changes to specific regions of the genome (Martienssen et al., 2005). In Arabidopsis, siRNAs are highly correlated with repetitive regions (Kasschau et al., 2007). Epigenetic modifications achieve an additional layer of complexity through the involvement of TEs, whose DNA is generally highly methylated and can attract the RNA silencing machinery and interact with histone modifications (Lippman et al., 2003, 2004). Epigenetic changes of TEs are not restricted to the TEs themselves, but in turn also regulate neighboring genes, which gives TEs a key role in the genome-wide distribution of epigenetic marks and smRNAs (Slotkin and Martienssen, 2007; Weil and Martienssen, 2008). This aspect is of particular importance in maize (Zea mays), since >60% of its genome consists of TEs (Meyers et al., 2001; Haberer et al., 2005; Messing and Dooner, 2006). Moreover, although genes are estimated to make up 8 to 20% of the maize genome, we now know that they are organized in islands surrounded by TEs (Chandler and Brendel, 2002; Messing et al., 2004; Rabinowicz and Bennetzen, 2006). In early 2008, a first draft of the sequence of the maize inbred line B73 genome was released, the largest and most complex plant genome ever sequenced. Sequencing projects for Mo17, another well-studied inbred line, and a popcorn strain are also scheduled to be completed shortly (Pennisi, 2008). However, presently, the maize genome is only sparsely annotated and assembled, which hampers its full exploitation.
Here, we describe an integrated genome-wide analysis of DNA methylation, histone modifications, smRNAs, and mRNA transcriptional activity, using maize as a model. We surveyed the epigenomes of the maize inbred line B73 in shoot and root tissue by Illumina/Solexa 1G parallel sequencing after digesting genomic DNA with a methylation-sensitive restriction enzyme and after conducting chromatin immunoprecipitation (ChIP) using antibodies that target specific histone modifications (H3K4me3, H3K9ac, H3K27me3, and H3K36me3). Additionally, we profiled RNA pools (microRNA [miRNA], siRNA, and mRNA) using the same sequencing strategy. This study provides a comprehensive and integrated organ-specific analysis of diverse epigenetic marks, smRNAs, and transcriptional activity and also gives new insight into the organization of the maize genome, which will aid in its continued assembly and annotation.
RESULTS
Direct Sequence Profiling of Maize Transcripts, Epigenetically Modified Genomic Regions, and smRNAs
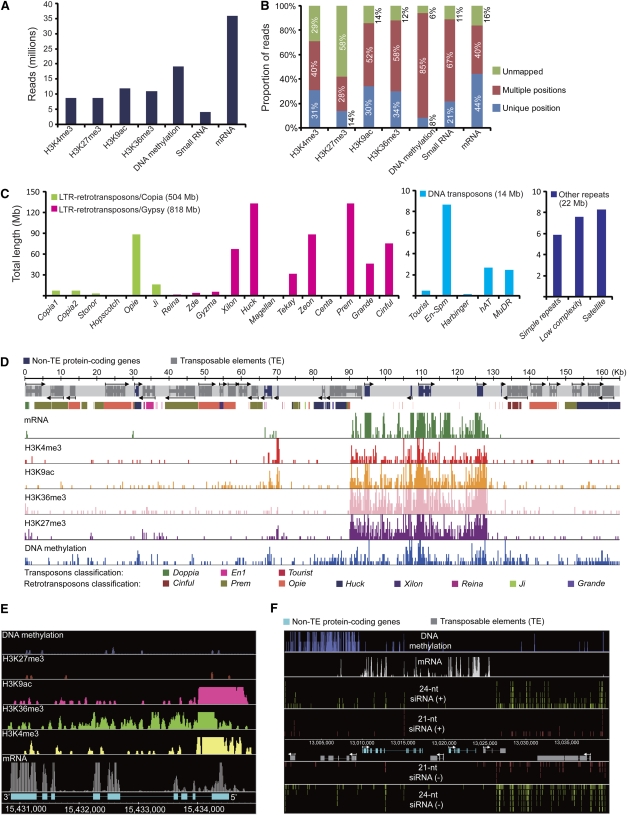
To survey the mRNA transcriptome, epigenetic landscapes, and smRNAs in a maize inbred line, we isolated total RNA and genomic DNA from shoots and roots of 14-d-old B73 seedlings. We extracted mRNA from total RNA using Dynabeads and enriched for smRNA by running total RNA on a PAGE gel for gel purification of RNAs in the 19- to 24-nucleotide size range, respectively. Methylated regions of the genome were enriched by digesting genomic DNA with the methylation-sensitive restriction enzyme McrBC. Genomic regions populated by epigenetically modified histone H3 proteins were enriched by a ChIP approach using antibodies targeting H3K4me3, H3K9ac, H3K27me3, or H3K36me3, respectively (see Methods). We used the resulting fractions to build libraries for Illumina/Solexa 1G high-throughput parallel sequencing, which generated 8.4 to 35.9 million reads for the individual libraries (Figure 1A; see Supplemental Figure 1A and Supplemental Table 1 online).
Figure 1.
Sequencing, Mapping, and Visualization of the Maize Transcriptome, Epigenome, and smRNAome.
(A) Counts of quality reads from Illumina/Solexa 1G sequencing.
(B) Proportions of unmapped and mapped reads with unique and multiple locations.
(C) Distribution of classified repetitive sequences in maize 2.4-Gb BAC sequences.
(D) A representative BAC (AC199189.3) showing predicted gene models with mRNA and epigenetic landscapes in shoots.
(E) Distribution of epigenetic patterns on an actively transcribed gene in shoots.
(F) The 21- and 24-nucleotide siRNAs are enriched in methylation-depleted regions in shoots.
Previous studies estimated that repetitive elements make up 80% or more of the maize genome (Chandler and Brendel, 2002; Messing et al., 2004; Rabinowicz and Bennetzen, 2006). This poses a major challenge to map Illumina/Solexa 1G sequencing reads to the maize genome accurately, since each read is usually 36 nucleotides or less in length. We used MAQ software (Li et al., 2008b; see also Supplemental Methods online) to map our 196 million sequencing reads to the currently available 2.4 Gb of B73 genome sequence represented by 16,205 BACs at http://www.maizesequence.org (dated June 4, 2008). The MAQ algorithm uses quality (MQ) scores to evaluate the reliability of a read based on both the uniqueness of the mapping position and the probability of sequencing errors. This allowed us to exploit sequencing data even for repetitive regions. A statistical model for calculating MQ scores and a detailed mapping procedure are described in Supplemental Methods online. Using MAQ, we mapped the proportion of reads corresponding to unique positions in the B73 genome as follows for shoot (root) libraries: H3K4me3, 31% (25%); H3K27me3, 14% (12%); H3K9ac, 30% (19%); H3K36me3, 34% (25%); DNA methylation, 8% (8%); smRNAs, 21% (23%); and mRNA, 44% (42%) (Figure 1B; see Supplemental Figure 1B and Supplemental Table 1 online). Using our criteria, we could map ∼85% of all mRNA reads to unique or non-unique positions. This indicates that even though the sequencing project is still ongoing, the currently available B73 genomic sequence is nearly complete. It also indicates that Illumina/Solexa 1G sequencing is a feasible alternative to previous large-scale transcriptome studies in maize (Ma et al., 2006; Fernandes et al., 2008).
Most reads that could not be correctly mapped to unique locations matched repetitive sequences, which are widespread in the maize genome. To classify recognizable repeat types, we used RepeatMasker software (http://www.repeatmasker.org) and found that 504 Mb of the B73 genome sequence were made up of long terminal repeat (LTR) retrotransposons of the Copia class, while 818 Mb were made up of LTR retrotransposons of the Gypsy class. Similarly, we found that 14 Mb of the genome sequence were occupied by DNA transposons and 22 Mb by other repeats (Figure 1C). For example, BAC AC199189.3 shows that maize genes are surrounded by a vast number of TEs, which is a key characteristic of the maize genome. As indicated for this representative BAC, we found that, in general, TE-rich regions were less commonly modified by H3K4me3, H3K9ac, H3K27me3, and H3K36me3 relative to non-TE regions and TE-free intergenic regions between non-TE genes (Figure 1D).
To visualize the epigenetic profiles of TEs and non-TE genes in more detail, we developed a pipeline to display a continuous 20-Mb stretch of the B73 genome (see Supplemental Figure 2 online). As illustrated for a representative section of this 20-Mb region, mRNA signals showed a strong correlation with predicted gene structures. Sequencing reads for the studied activating epigenetic marks (H3K4me3, H3K9ac, and H3K36me3) were generally present at high levels at transcribed genes in this region (Figure 1E). As shown for a larger region of these 20 Mb, TEs were generally heavily DNA methylated and lacked transcriptional activity, while non-TE genes were transcriptionally active and lacked significant DNA methylation (Figure 1F). Interestingly, we detected many smRNA reads at TEs whose DNA was not methylated. Conversely, we found that TEs whose DNA was highly methylated were relatively devoid of smRNAs (Figure 1F).
An Initial Estimate of Transcriptional Activity in the Maize Genome Using mRNA-seq
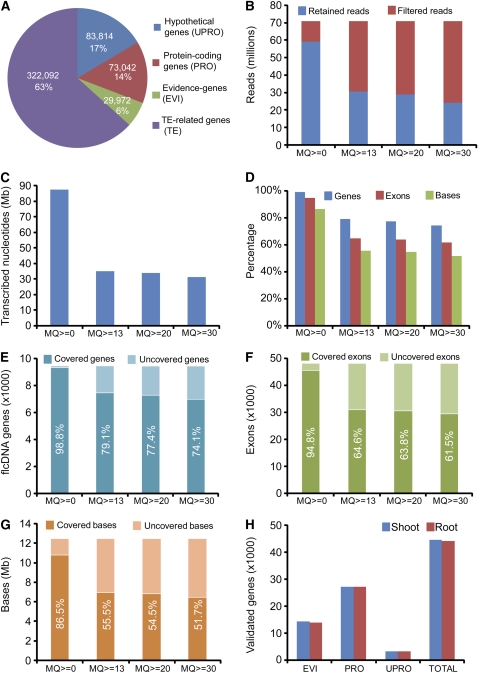
We used two gene sets for analyzing the transcriptional activity of the maize genome: a set of 11,742 full-length cDNAs (flcDNAs) obtained from http://maizecdna.org and prediction results from the FgeneSH gene finding software for 16,205 BACs obtained from http://maizesequence.org. Compiling these flcDNAs resulted in 9451 nonredundant sequences mapped to the maize genome, including 7141 flcDNAs with only one best location and 2310 with multiple best locations (see Supplemental Figure 3A online).
To estimate the transcriptional activity of the maize genome using mRNA-seq data, we developed a pipeline for de novo scanning of transcribed exons by merging overlapping Illumina/Solexa reads into contiguous regions (see Supplemental Figure 2 online). For this part of our analysis, we combined 16 lanes of mRNA reads (71 million) from both shoot and root libraries to achieve a maximum coverage. We then scanned for putative exons using MQ scores larger or equal to 0, 13, 20, and 30 (Figures 2B and 2C). We identified up to 1,122,064 putative exons representing 87,606,799 transcribed bases using our de novo scanning approach. To evaluate the coverage of mRNA-seq, we matched the de novo detected exons with flcDNAs representing bona fide genes. At MQ 0, the detected exons covered 99% at gene level, 95% at exon level, and 87% at base level, while at MQ 13 only 79, 65, and 56% were covered, respectively (Figures 2D to 2G).
Figure 2.
Validation of flcDNAs and FgeneSH-Predicted Genes.
(A) FgeneSH-predicted maize genes in different groups.
(B) Numbers of retained and filtered reads in 16 merged lanes of mRNA-seq reads using different mapping quality (MQ) scores.
(C) Total lengths of transcribed nucleotides by adding up de novo exons using different MQ scores.
(D) to (G) Percentages and numbers of validated flcDNAs at gene, exon, and base level.
(H) Numbers of validated non-TE genes in different groups.
We next matched the de novo detected exons as derived from our mRNA-seq data of shoot and root libraries with FgeneSH predicted genes. This resulted in the identification of nearly 45,000 validated protein-coding genes (Figure 2H; see Supplemental Table 2 online). Because the maize genome is not completely sequenced and because the available sequence data is marginally annotated, we were unable to estimate all transcribed regions. However, our pilot survey of transcriptional activity in maize suggests that even though the maize genome is about six times larger than the rice genome (Goff et al., 2002; Yu et al., 2002), the number of genes is likely to be similar. To complement these data, a series of protein-level comparative analyses, including functional comparisons based on pathway enrichment and Gene Ontology (The Gene Ontology Consortium, 2000) for maize, rice, and Arabidopsis, were performed (see Supplemental Figures 4 to 7 and Supplemental Data Sets 1 and 2 online). This analysis assigned the products of ∼20,000 genes to known Gene Ontology pathways.
Epigenetic Marks Differ in Their Absolute and Relative Distributions on a Whole-Genome and Gene Level
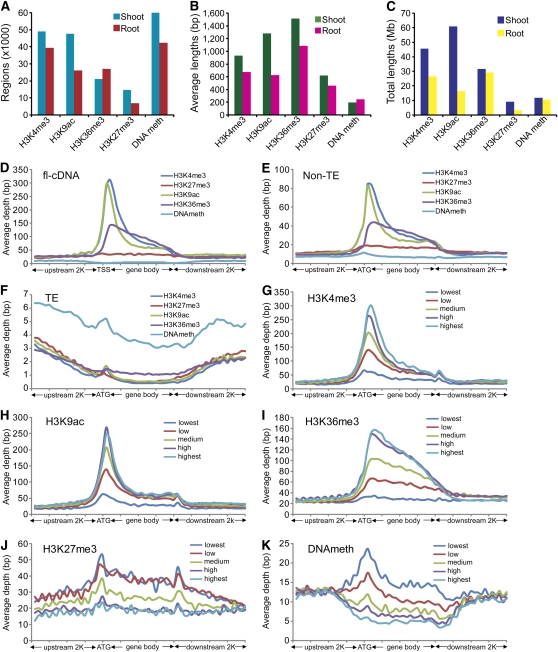
To analyze the extent of epigenetic modifications on a whole-genome level, we determined how many regions were covered by DNA methylation, H3K4me3, H3K9ac, H3K27me3, or H3K36me3 (Figure 3A) using MACS software (Zhang et al., 2008; see Supplemental Methods online). We found that DNA methylation was the most prevalent modification in both shoots and roots, covering ∼60,000 regions in shoots and 40,000 regions in roots, respectively. Two of the studied activating histone modifications, H3K4me3 and H3K9ac, were also found at high frequencies. Interestingly, the number of regions modified by H3K9ac or H3K27me3 was almost twice as high in shoots compared with roots, which might indicate genome-wide tissue-specific epigenetic alterations. The length and frequency of modified regions varied dramatically. DNA methylation was found at more regions than any other modification, but the average length of the affected genomic regions was only ∼200 bp, by far the shortest of all modifications studied. Conversely, H3K36me3 was present at relatively few regions, but their average length was almost 1600 bp; significantly longer than any other modification (Figure 3B). Similar conclusions can be drawn when the total lengths of modified regions are considered rather than the average lengths or number of regions (Figure 3C).
Figure 3.
Genome-Wide and Genic Distribution Patterns of Epigenetic Modifications.
(A) to (C) Numbers, average lengths, and total lengths of epigenetically modified regions detected by MACS software.
(D) to (F) Distribution of H3K4me3, H3K27me3, H3K9ac, H3K36me3, and DNA methylation levels within flcDNAs, predicted TE-related, and non-TE genes aligned from TSSs and ATG, respectively. The y axis shows the average depth, which is the frequency of piled-up reads at each base divided by the bin size. The x axis represents the aligned genes that were equally binned into 40 portions, including 2K up- and downstream regions.
(G) to (K) Distribution of H3K4me3, H3K27me3, H3K9ac, H3K36me3, and DNA methylation within five groups of genes with different expression levels summarized from validated non-TE genes.
To study the level of epigenetic modifications in different regions of genes and TEs, we aligned all flcDNAs at their transcript start site (TSS) and all predicted non-TE genes and TEs at their start codon (ATG). We defined the region of a gene or TE as its body (annotated transcribed region) plus 2 kb upstream. We observed no significant differences in the distributions of the epigenetic marks on aligned genes when we compared flcDNAs and predicted non-TE genes (Figures 3D and 3E; see Supplemental Figures 8A and 8B online). H3K4me3 and H3K9ac formed a strong peak at or near the TSS or ATG, respectively, and were present at relatively low levels in the gene body. By contrast, H3K36me3 was found throughout the gene body in shoots, but formed a more distinct peak at the TSS or ATG in roots (Figures 3D and 3E; see Supplemental Figures 8A and 8B online). As expected, DNA methylation was present at very low levels in genes but was the most prevalent modification in TEs (Figure 3F; see Supplemental Figure 8C online).
To determine the effect of individual epigenetic modifications on transcriptional activity, we sorted all protein-coding genes (∼45,000) as identified above based on their expression levels derived from mRNA-seq reads using percentile grouping. The top ∼9000 most highly expressed genes were labeled “highest,” the next ∼9000 genes “high,” the next ∼9000 genes “medium,” etc., such that five groups of equal size were obtained, for each of which we analyzed the distribution of each epigenetic modification of interest. We found that in both shoots and roots, the genes with the highest expression levels showed the highest amounts of H3K4me3, H3K9ac, or H3K36me3 (Figures 3G to 3I; see Supplemental Figures 8D to 8F online). By contrast, genes with the lowest expression levels had the highest amounts of H3K27me3 or DNA methylation in both tissue types (Figures 3J and 3K; see Supplemental Figures 8G and 8H online). Whereas H3K27me3 was present throughout the gene body, DNA methylation peaked at the ATG for genes in the lowest expression group. In addition, we determined the average levels of all four histone modifications of interest relative to the expression levels of genes (see Supplemental Figure 9 online). We found that in shoots and roots, genes with the highest expression levels tended to have the most H3K4me3, H3K9ac, or H3K36me3. By contrast, genes with the lowest expression levels tended to have the most H3K27me3, albeit at markedly lower levels relative to activating histone marks in highly expressed genes.
Epigenetic Modifications Show Differential Targeting of Genes and TEs and Display Combinatorial Effects in Maize Shoots and Roots
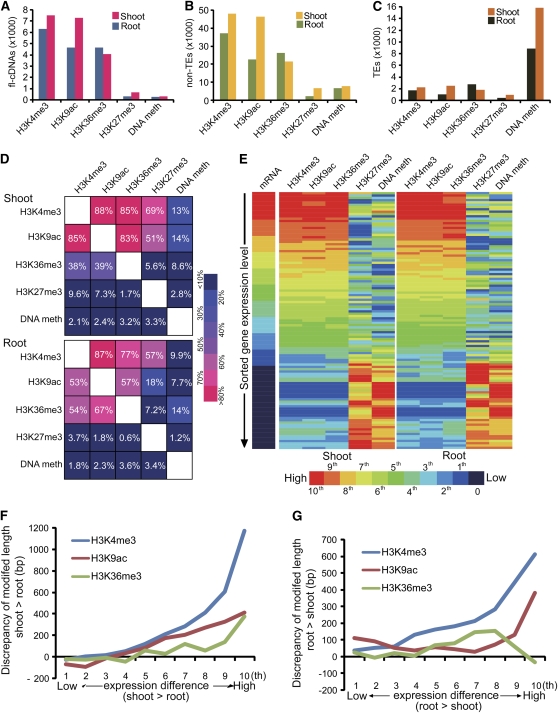
To analyze whether epigenetic modifications target genes and TEs differentially, we determined how many flcDNAs, predicted non-TE genes, and TEs show specific epigenetic modifications. We found that for both shoots and roots, genes (represented by either a flcDNA or as predicted non-TE gene) were less commonly affected by H3K27me3 or DNA methylation than by H3K4me3, H3K9ac, or H3K36me3 (Figures 4A and 4B). By contrast, TEs were epigenetically modified by DNA methylation up to 8 times more often than by modification of histone H3 (Figure 4C).
Figure 4.
Combinatory Modifications and Correlation with Gene Expression.
(A) to (C) Numbers of modified flcDNAs, non-TEs, and TEs by H3K4me3, H3K9ac, H3K36me3, H3K27me3, and DNA methylation in shoot and root.
(D) Frequencies of concurrent modifications on genes. Above the diagonal, numbers indicate the percentage of genes modified by X also have modification Y, while below the diagonal, percentages indicate how many genes were modified by Y and also modified by X.
(E) Heat maps of epigenetic modification levels on ∼60,000 genes sorted by their expression measured by mRNA-seq. Gene expression levels and modifications levels were transformed to 100 percentiles, and each bar represents the averaged level of ∼600 genes within each percentile.
(F) and (G) Correlation of differential modifications and differential gene expression in shoot and root. The y axis shows differences in the modification level of shoot higher than root and vice versa. The x axis shows the difference in the expression level of shoot higher than root and vice versa.
Furthermore, we analyzed whether different epigenetic marks showed distinct combinatorial effects. We found that in both shoots and roots, a significant and similar proportion of regions that were modified by one of the activating marks studied (H3K4me3, H3K9ac, and H3K36me3) were also modified by a second activating epigenetic mark (Figure 4D). While most pairwise combinations of activating epigenetic marks did not differ drastically between shoots and roots, 51% of all shoot-derived regions that were modified by H3K27me3 were comodified by H3K9ac, while only 18% of root-derived regions showed the same comodification pattern.
Additionally, we analyzed the influence of various combinations of epigenetic marks on the mRNA level of such modified genes. We observed that while all three activating epigenetic modifications under study were cooperatively present in genes with high mRNA levels and lacking in genes with low mRNA levels, the two repressive marks showed a mutually exclusive pattern (Figure 4E). In both shoots and roots, genes with low mRNA levels were marked with either H3K27me3 or methylated DNA, but genes marked with one of these modifications had low levels of the other, indicating a mutually exclusive effect between these two modifications. The mutually exclusive effect of those two repressive marks could also be observed for genes with high mRNA levels.
We observed that H3K9ac was more enriched in shoots than in roots (see Supplemental Figure 10 online). To analyze tissue-specific epigenetic effects in more detail, we grouped all non-TE genes into 10 percentiles based on their mRNA levels and plotted them against differences in the respective epigenetic modifications in shoots and roots (Figures 4F and 4G; see Supplemental Figure 11 online). We observed that H3K4me3, H3K9ac, and H3K36me3 were all correlated with tissue-specific gene expression, albeit to different degrees. Whereas a very distinct trend could be determined for H3K4me3, which was positively correlated with expression levels, H3K36me3 was less correlated with differential gene expression between shoots and roots. The different degrees of correlation with gene expression between these two activating histone modifications are unclear at this point. Interestingly, H3K36me3 continued to increase in the highest expression percentiles for genes that were more strongly expressed in shoots than in roots, but in contrast, it dramatically dropped in the highest gene expression percentiles for genes that were more strongly expressed in roots than in shoots (Figures 4F and 4G). Neither H3K27me3 nor DNA methylation displayed a clear trend like the activating epigenetic marks of interest, which indicates that in our study, neither H3K27me3 nor DNA methylation had a clear effect on differential expression of genes in maize shoots and roots at the genome scale (see Supplemental Figure 11 online).
Changes in smRNA Populations Follow mop1 Gene Expression
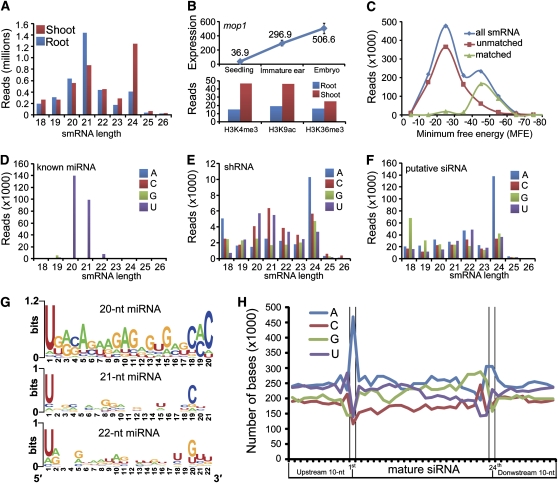
To profile smRNA populations in maize seedling shoot and root tissue, we generated smRNA libraries for Illumina/Solexa 1G sequencing. After removing reads that likely originated from rRNA or tRNA contamination, we obtained 4,406,055 adaptor-trimmed sequences representing 1,639,984 unique smRNAs from shoots and 3,960,345 sequences representing 709,440 unique smRNAs from roots, respectively (see Supplemental Figure 12 online). We noted a tissue-specific smRNA size distribution: 24-nucleotide smRNAs were the predominant size class in shoots, whereas the predominant smRNAs in roots were 21 nucleotides (Figure 5A). This observation indicates that in maize, miRNAs, most of which are 20 to 22 nucleotides in length, are relatively enriched in roots, while siRNAs, which are mostly 24 nucleotides long, are relatively more prevalent in shoots. Interestingly, we did not observe a dramatic enrichment of 24-nucleotide siRNAs, as recently reported for maize flower organs (Nobuta et al., 2008) and for Arabidopsis immature floral tissue (Lister et al., 2008). It has been previously described (Henderson and Jacobsen, 2007) that in Arabidopsis the endogenous siRNA biogenesis pathway requires RNA-dependent RNA polymerase-2 (RDR2). In maize, MOP1 is homologous to RDR2, and it has been shown that a loss of function of RDR2 and MOP1 caused dramatic reduction of 24-nucleotide siRNAs in Arabidopsis and maize, respectively (Nobuta et al., 2008; Woodhouse et al., 2006). To determine whether differences in mop1 expression levels could explain the different compositions of smRNA populations in maize seedling and floral tissue, we examined the mop1 expression level across different organs using published microarray data (Stupar and Springer, 2006) and our mRNA-seq reads for shoots and roots. We found that mop1 expression in seedlings was significantly lower than in immature ears and embryos (Figure 5B), confirming previous findings for maize (Woodhouse et al., 2006) and for RDR homologs in rice (Kapoor et al., 2008). In fact, when examining the mRNA-seq data from our study, we found that only ∼40 reads, mostly from shoots, mapped to the mop1 gene, which indicates a very low expression level in seedling tissues. Moreover, we found that the three activating epigenetic marks H3K4me3, H3K9ac, and H3K36me3 were slightly more abundant for mop1 in shoots compared with roots (Figure 5B). In summary, these findings suggest that decreasing mop1 expression leads to a concomitant decrease of 24-nucleotide siRNAs relative to 21-nucleotide miRNAs in a tissue-specific manner progressing from floral organs, to shoots, to roots.
Figure 5.
In Silico Classification Indicates Dynamic smRNA Populations in Maize Shoots and Roots.
(A) smRNA length distributions in shoots and roots.
(B) Tissue-specific expression and epigenetic modification of maize mop1 gene.
(C) Distribution of smRNAs and matched and unmatched known miRNAs in miRBase within different MFE bins.
(D) to (F) Length distributions of known miRNA, shRNAs, and putative siRNAs with different 5′ terminal nucleotides.
(G) Sequence motifs of 20-, 21-, and 22-nucleotide miRNAs analyzed by WebLogo (Crooks et al., 2004).
(H) Nucleotide composition of mature 24-nucleotide putative siRNAs.
Classification of smRNAs Based on Secondary Structure Predictions of Precursors
The smRNA population within a cell is composed of miRNA and natural antisense transcript-derived miRNA (Lu et al., 2008) as well as several classes of endogenous siRNAs, including repeat-associated RNA, natural antisense transcript-derived siRNA, and trans-acting siRNA (Bonnet et al., 2006; Ramachandran and Chen, 2008). To separate miRNAs from siRNAs, we aligned all smRNA reads with known miRNA sequences from miRBase (Griffiths-Jones et al., 2006). Since sequence similarity alone does not necessarily guarantee that the smRNA in question is a miRNA, we next determined whether the respective smRNA precursor sequences were able to form a stem-loop structure indicative of miRNAs, which are derived from short hairpin structures, whereas siRNAs generally form from long double-stranded RNA (dsRNA) molecules. To determine putative precursor sequences, we adopted a more stringent mapping method using SOAP software (Li et al., 2008a) to retrieve all perfectly mapped locations for each smRNA and then extended 20 nucleotides at the 5′-end and 70 nucleotides at the 3′-end (see Supplemental Methods online). Using this approach, we obtained 37,763,920 and 18,734,677 putative precursor sequences from 2,890,098 smRNAs in shoots and 1,650,153 smRNAs in roots, respectively. We employed RNAfold (Hofacker et al., 1994) to calculate a minimum free energy (MFE) for each putative precursor. The lower the MFE, the higher the possibility that a precursor can form a stem-loop structure (Hofacker et al., 1994). To determine the minimum threshold, we compared the MFE for the smRNAs that matched known miRNAs in miRBase and those with unmatched sequences (Figure 5C; see Supplemental Figure 14A and Supplemental Tables 3 and 4 online). For the overall set of smRNAs, we observed two distinct peaks at −25 and −45, indicating a mixture of miRNAs and siRNAs, while for the matched and the unmatched smRNAs, single peaks were found to center at −45 and −25, respectively. Therefore, we set the MFE minimum threshold at −40 to determine the ability of a smRNA's precursor to form a hairpin structure.
Based on these criteria, we categorized all smRNAs into three groups (see Supplemental Figure 13 online). Group I, “known miRNAs” with matches in miRBase and MFE < −40, consisted of 526,961 reads representing 155 unique sequences from shoots and 252,505 reads representing 126 unique sequences from roots. Group II, “small hairpin RNAs (shRNAs)” without matches in miRBase but MFE < −40, consisted of 120,227 reads representing 10,314 unique sequences from shoots and 131,553 reads representing 31,856 unique sequences from roots. This group might include unidentified miRNAs and other smRNA species. Group III consisted of all remaining smRNAs whose precursors could not form hairpins. We classified all smRNAs in group III as “putative siRNAs,” consisting of 1,768,555 reads representing 984,890 unique sequences from shoots and 800,094 reads representing 379,199 unique sequences from roots, respectively. Interestingly, these three groups of smRNAs had distinctly different average frequencies with ∼3400 copies for known miRNA, ∼120 copies for shRNAs, and ∼1.8 copies for putative siRNAs.
Three Groups of smRNAs Exhibited Distinct Signatures of 5′ Terminal Nucleotide Identities and Overall Nucleotide Compositions
It has been shown that in Arabidopsis, the 5′ terminal nucleotide is a key characteristic to direct distinct smRNA classes to different Argonaute (AGO) complexes (Mi et al., 2008). Therefore, we examined the size distributions of smRNAs in these three groups based on their 5′ terminal nucleotides. We found that virtually all known miRNAs (Group I) had a 5′ U, the signature of canonical miRNAs (Figure 5D; see Supplemental Figure 14B online), while most 24-nucleotide putative siRNAs (Group III) had a 5′ A, a signature feature of canonical siRNAs (Figure 5F; see Supplemental Figure 14D online).
Unexpectedly, smRNAs in Group II demonstrated a more complex distribution (Figure 5E). Within this group, a large number of 20-, 21-, and 22-nucleotide smRNAs had a 5′ terminal U, indicative of canonical miRNAs. However, an equally large number of smRNAs in these size classes also had a 5′ terminal C, which might represent either a novel group of miRNAs or unknown small hairpin siRNAs. Furthermore, this group of smRNAs also contained a large number of 24-nucleotide siRNAs with a 5′ A, suggesting that certain siRNA species need a hairpin precursor state for processing through DICER. The complex composition of this group of smRNAs, which most likely includes miRNAs and siRNAs as well as potentially other unknown smRNA species, led us to classify these smRNAs collectively as shRNAs. In shoots, 20- to 22-nucleotide smRNAs with a 5′ terminal C were not detected, indicating that 5′ C shRNAs might potentially represent a group of uncharacterized tissue-specific smRNAs (see Supplemental Figure 14C online).
To further characterize the sequence patterns of these three groups of smRNAs and to explore smRNAs in irregular lengths other than 21 and 24 nucleotides, we calculated the frequencies for each nucleotide within the mature smRNA and extended the mature RNA by 10 nucleotides at both ends. For the known miRNAs in lengths of 20, 21, and 22 nucleotides, sequence motifs were analyzed by WebLogo (Crooks et al., 2004). The sequence motifs reflected the most enriched miRNA families (Figure 5G; see Supplemental Figures 15A and 15B online). Overall, we observed a high frequency of upstream As and Us for half of the putative siRNA group and a sharp peak for 5′ terminal A (Figure 5H). This result is congruent with sequence patterns found in Arabidopsis (Lister et al., 2008). However, the relative enrichment of 3′ Gs seems to be a unique feature of maize when compared with Arabidopsis. For putative 20- to 26-nucleotide siRNAs (excluding the 24-nucleotide class), we observed a relatively high frequency of As up to two nucleotides upstream of the 5′ terminus as well as for the 3′ terminal nucleotide (see Supplemental Figure 16 online). This result indicates that the siRNA of other lengths could be variations of canonical siRNAs. Overall, the nucleotide composition of the shRNA group showed the highest amount of GC from −10 nucleotides to +10 nucleotides in the mature smRNAs, indicating distinct differences in the nature of shRNA compared with miRNAs and siRNAs (see Supplemental Figure 17 online).
22-Nucleotide siRNAs Are Differentially Enriched in Long Hairpin dsRNAs
In both shoots and roots, we found that siRNA populations were enriched primarily in 24-nucleotide and secondarily in 22-nucleotide species (see Supplemental Figures 13E and 13F online). A recent study showed that 22-nucleotide siRNAs were specifically enriched in maize compared with other plants, which led to the hypothesis that this size class might potentially represent a new species of smRNA in addition to the canonical 21- and 24-nucleotide siRNA (Nobuta et al., 2008). It is possible that a yet to be identified siRNA biogenesis pathway exists in maize (Nobuta et al., 2008). Two other recent reports summarizing work in mouse delivered evidence that siRNAs found in naturally formed endogenous long hairpin dsRNA molecules are responsible for generating a certain class of smRNAs (Tam et al., 2008; Watanabe et al., 2008). It has also been shown in maize that smRNAs produced from a hairpin version of MuDR, Muk, are not lost in a mop1 mutant background (Woodhouse et al., 2006). Taken together, these findings led us to explore whether long hairpin dsRNAs are the sources of 22-nucleotide siRNAs in maize because a naturally formed RNA duplex could be independent of mop1, whose expression we found to be very low in seedling tissues.
We performed de novo scanning of 2.4-Gb maize BACs using the einverted program (see Supplemental Methods online) and identified 1086 long hairpin dsRNAs with a stem length of at least 1000 nucleotides and at least 90% base pair complementation within the stem sequence. By mapping the putative siRNAs onto long hairpin dsRNAs, we indeed observed a higher relative enrichment of 22-nucleotide compared with 24-nucleotide siRNAs in both shoots and roots (Figures 6A to 6D), which differed from the siRNAs mapped onto LTR-TEs (Figures 6E to 6G). A detailed comparison of siRNAs derived from long hairpin dsRNAs or LTR-TEs revealed more unique features of this novel siRNA species. First, we found that these siRNAs had a higher copy number (305,288 reads representing 58,210 unique sequences from shoots and 238,313 reads representing 30,138 unique sequences from roots). Second, we identified shorter siRNAs (18 to 22 nucleotides), which were replicated in even higher frequencies (e.g., 30 times for 20-nucleotide siRNAs in roots). Third, 19-, 20-, 21-, and 24-nucleotide siRNAs bore a signature 5′ terminal A, whereas 22-nucleotide siRNAs had approximately equal amounts of 5′ A and 5′ U. In summary, our observations indicate that siRNAs derived from long hairpin dsRNA might be a miRNA-like species, even though they bear canonical siRNA features.
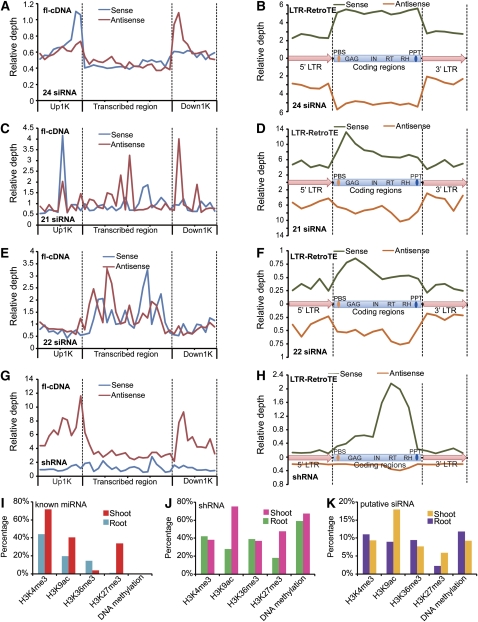
Figure 6.
22-Nucleotide siRNAs Are Differentially Enriched in Long Hairpin dsRNAs Rather Than in LTR-TEs.
(A) to (C) Length distributions of putative siRNAs mapped on long hairpin dsRNAs. (A) Count of unique sequences; (B) and (C) total reads.
(D) An example of a long hairpin dsRNAs generating more 22-nucleotide siRNAs than 24-nucleotide siRNAs. The loop region of ∼500 bp is not shown, and paired regions in stem are 99% in identity. Bubbles indicate unmatched nucleotides.
(E) to (G) Length distributions of putative siRNAs mapped on full-length LTR-retrotransposons. (E) Count of unique sequences; (F) and (G) total reads.
smRNAs Target Distinct Regions in Genes and Full-Length LTR-RetroTEs
Traditional annotation of TEs is based on open reading frame predictions followed by comparison with known repeat types in public databases. However, TEs predicted following this strategy cannot represent a complete unit, especially in the case of LTR-retrotransposons, which have a complicated architecture. Therefore, we used a program called LTR-finder (Zhao and Wang, 2007) and identified 75,015 full-length LTR retrotransposons de novo, representing 880 Mb of DNA sequence (see Supplemental Methods online). By mapping putative siRNAs to LTR-TEs, we found 753,512 siRNA reads representing 314,044 unique sequences from shoots and 455,881 reads representing 138,853 unique sequences from roots, respectively. When we analyzed the distribution of the 5′ terminal nucleotides for siRNAs matching LTR-TEs, we found that in both shoots and roots, most 24-nucleotide siRNAs had the characteristic 5′ terminal A, but that 22-nucleotide siRNAs started with an A or U in about equal proportions (Figures 6E to 6G). This result might indicate different mechanisms in 22- and 24-nucleotide siRNA biogenesis as well as tissue-specific siRNA populations.
siRNAs have two main known functions. The majority of repeat-associated 24-nucleotide siRNAs contribute to the formation of DNA methylation, while a small portion of siRNAs including 21- and 24-nucleotide classes contribute to the RNA interference machinery targeting genes and TEs either in trans-acting or natural-antisense-transcript mode (Bonnet et al., 2006). Therefore, we analyzed the distributions of the respective siRNA classes surrounding and within genes and LTR-TEs. Since most siRNAs are associated with repetitive sequences, keeping a randomly selected subset of all siRNAs would lead to a significant bias. For this reason, we adopted a method based on a best possible compromise using the following formula: coverage of one siRNA divided by all the locations this siRNA could be mapped to in the genome (see Supplemental Methods online). As a basic classification, we assumed here that if a smRNA is mapped to the sense strand of a genomic locus, this smRNA might originate from this site, while a smRNA mapped on the antisense strand of a locus might indicate that this smRNA targets this site. However, this approximation does not take other, more complicated scenarios into account (e.g., origination of siRNAs from antisense mRNAs and base-pairing of siRNAs to genomic DNA in addition to sense mRNAs). To determine whether different size classes of siRNAs and shRNAs target different regions in genes or LTR-TEs, we examined the distribution of the respective smRNAs over gene regions and LTR-TEs in shoots and roots (Figures 7A to 7H; see Supplemental Figures 18A to 18H online).
Figure 7.
Origin and Target Sites on Genes and LTR-TEs for Different Classes of Putative siRNAs.
(A), (C), (E), and (G) The 24-, 21-, and 22-nucleotide siRNAs and shRNAs on flcDNA genes show significant strand bias on different positions in originating and targeting strands.
(B), (D), (F), and (H) The 24-, 21-, and 22-nucleotide siRNAs and shRNAs on LTR-TEs. Calculation of relative depth and de novo identification of LTR-TEs is described in the supplemental data online.
(I) to (K) Percentages of unique smRNA loci situated in epigenetic regions of H3K4me3, H3K9ac, H3K36me3, H3K27me3, and DNA methylation.
Interestingly, each class of siRNAs exhibited a distinct pattern on genes and LTR-TEs. For the 24-nucleotide siRNAs on flcDNA genes, we observed a distinct bias toward the sense and antisense strand in specific regions. A sharp 5′ peak indicated that a group of 24-nucleotide siRNAs originated from the immediate upstream 100- to 200-bp region on the sense strand of genes, while another equal amount of 24-nucleotide siRNAs targeted the immediate downstream 100 to 200 bp, which would still be within the 3′ untranslated regions. This group of 24-nucleotide siRNAs potentially represents natural antisense transcript-derived siRNAs. For the 24-nucleotide siRNAs on LTR-TEs, we found that the overall distribution on the sense strand was mirrored on the antisense strand and that the transcribed regions had a higher proportion of 24-nucleotide siRNAs than the 5′ and 3′ LTR regions (Figures 7A and 7B; see Supplemental Figures 18A and 18B online).
The 21-nucleotide siRNAs exhibited a similar pattern compared with 24-nucleotide siRNAs in genes, but differed in their origin regions. On the LTR-TEs, the distribution of 21-nucleotide siRNAs on the sense and antisense strands was dissimilar. We found more 21-nucleotide siRNAs on the sense strand at the 5′ end, indicating more origin sites, while we observed more 21-nucleotide siRNAs on the antisense strand at the 3′ end, indicating more targeting sites in this region (Figures 7C and 7D; see Supplemental Figures 18C and 18D online).
Similarly, 22-nucleotide siRNAs exhibited a strand-specific distribution in the transcribed region of genes (Figure 7E; see Supplemental Figure 18E online). However, the origins of 22-nucleotide siRNAs were biased toward the 3′ end, while the targeting sites were biased toward the 5′ end within the transcribed regions. The 22-nucleotide siRNAs showed a similar pattern on LTR-TEs (Figure 7F; see Supplemental Figure 18F online). This pattern indicates 22-nucleotide siRNAs might fulfill their silencing function in a different way compared with 21- and 24-nucleotide siRNAs.
Interestingly, shRNAs were extremely strand-specific in both flcDNAs and LTR-TEs (Figures 7G and 7H; see Supplemental Figures 18G and 18H online). Virtually all shRNAs mapped to the antisense strand in both 5′ and 3′ regions of flcDNAs, indicating that shRNAs could function in a trans-acting fashion. In LTR-TEs, almost all shRNAs mapped to the sense strand in the 3′ coding region.
Overall, our findings indicate that different siRNA classes target different regions in genes and LTR-TEs and target different transposon classes (see Supplemental Figure 19 online), pointing at specialized regulatory roles during epigenetic regulation of these siRNAs (Figures 7I to 7K).
DISCUSSION
Using maize, we have generated an integrated genome-wide and organ-specific survey of epigenetic marks together with transcriptional outputs. Our results show that Illumina/Solexa 1G sequencing and read mapping are feasible with high accuracy even in large and repeat-rich plant genomes, opening the door to exploring similarly complex genomes in the future.
Epigenetic changes, including histone modifications and DNA methylation, have a profound impact on gene regulation. We observed that H3K4me3, H3K9ac, and H3K36me3 were associated with transcriptionally active genes, while H3K27me3 and DNA methylation were predominantly found in transcriptionally inactive genes and repetitive elements, supporting the findings of previous studies in other organisms (e.g., Martens et al., 2005; Zhang et al., 2006; Barski et al., 2007; Zilberman et al., 2007; Li et al., 2008c; Wang et al., 2008). Interestingly, we found that genic DNA methylation patterns in maize are very similar to rice, but very different from Arabidopsis. While in maize and rice, genic DNA methylation peaks around the ATG (Figure 3K; Li et al., 2008c), it is most prevalent in the transcribed region in Arabidopsis genes (Zhang et al., 2006; Zilberman et al., 2007). Moreover, we found that the differential accumulation of distinct epigenetic marks in genes and repetitive elements was reflected in the proportion of reads mapped to unique or nonunique positions in the genome. As expected for strongly repeat-associated modifications, we only identified a small number of unique genome positions for H3K27me3 and DNA methylation (Figure 1B; see Supplemental Figure 1B online). Interestingly, we found that while multiple activating epigenetic marks tended to occur together, the two repressive marks under study, H3K27me3 and DNA methylation, were more likely to exclude each other at the same loci (Figures 4D and 4E). This supports similar findings for genome-wide studies in Arabidopsis (Mathieu et al., 2005; Zhang et al., 2007) and for a locus-specific analysis in mouse (Lindroth et al., 2008). For example, in Arabidopsis, <10% of H3K27me3-covered regions overlapped with DNA methylation (Zhang et al., 2007). Even though the reason behind this antagonism is unclear, it suggests a very different mode of action compared with activating epigenetic marks, which generally do not seem to be mutually exclusive. H3K27me3 is regarded as a mark of transcriptional quiescence, but a recent study (Riclet et al., 2009) showed that in mouse, upon loss of heterochromatin protein 1 on the mesoderm-specific transcript promoter, H3K27me3 associates with gene activation and correlates with DNA hypomethylation. In animals, H3K27me3 regions typically form large domains (>5 kb) and include multiple genes (Bernstein et al., 2006; Schwartz et al., 2006). In plants, it covers much shorter regions (typically <1 kb), and it tends to be restricted to the coding region of single genes (Figures 3B and 3J; Zhang et al., 2007). Taken together, these results suggest that H3K27me3 might be based on different spreading and maintenance mechanisms and that it might also have different functions in gene activation and gene repression in plants and animals.
smRNAs have been increasingly recognized as key regulators of gene activity that can have major effects. For example, a recent study has shown that miRNAs were involved in the domestication of maize (Chuck et al., 2007). Whereas most endogenous plant siRNAs are 21 to 24 nucleotides long (Ramachandran and Chen, 2008), maize possesses an additional class of 22-nucleotide siRNAs. Interestingly, other monocots, such as wheat (Triticum aestivum), barley (Hordeum vulgare), or rice, lack 22-nucleotide siRNAs (Nobuta et al., 2008). To elucidate the biogenesis of this 22-nucleotide class, we examined potential sources of these siRNAs and found that they might be generated from long dsRNAs. We hypothesize that the respective long dsRNAs might be encoded by pseudogenes similar to those found in mouse, where duplexes formed by their sense and antisense transcripts have been shown to produce siRNAs without requiring RdRP activities (Tam et al., 2008; Watanabe et al., 2008). Canonical 24-nucleotide siRNAs bear a 5′ terminal A, which is recognized by AGO2 and AGO4 (Mi et al., 2008). Interestingly, we found that 22-nucleotide siRNAs matching long dsRNAs bear all four nucleotides at their 5′ end, which indicates the involvement of other AGO proteins or potentially non-AGO proteins during 22-nucleotide siRNA-mediated silencing processes. We observed marked differences in the distributions of siRNAs derived from long hairpin dsRNAs compared with those derived from LTR-TEs. For example, long hairpin dsRNA-derived siRNAs were relatively enriched for small sizes (18 to 22 nucleotides) and had a high copy frequency (Figures 6B and 6C), while for LTR-TE–derived siRNAs, the copy frequency was relatively low. These differences might indicate two distinct siRNA biogenesis pathways in maize, in which RdRP is necessary to generate siRNAs from LTR-TEs but not from long hairpin dsRNAs. We found that the expression level of one RdRP gene, mop1, correlated with a decrease of 24-nucleotide siRNAs relative to 21-nucleotide miRNAs in a tissue-specific manner progressing from floral organs to shoots and roots. Intriguingly, mop1 also seems to be involved in a tissue-specific regulation of paramutation and silencing at the p1 locus in maize (Sidorenko and Chandler, 2008), which opens the possibility that siRNAs might be involved in tissue-specific and targeted paramutation.
Maize was one of the first model organisms for biological research and has a rich history in the study of epigenetics, plant domestication, and evolution. With the recent release of its first draft genomic sequence, it is once again taking center stage in both plant biology and crop improvement. We hope that the epigenetic and transcriptomic survey we have described here will aid in further annotating and understanding the maize genome. It will also be useful for exploring epigenetic principles and even more complex smRNA biology, as well as the interplay between epigenomes and transcriptomes. In summary, we hereby have delivered a critical analysis of the overall landscapes of epigenetic histone marks and DNA methylation, together with mRNA and smRNA transcriptomes in maize.
METHODS
Plant Growth Conditions
Maize (Zea mays) inbred line B73 was obtained from the USDA–Agricultural Research Service North Central Regional Plant Introduction Station in Ames, IA. Seeds were planted in individual pots containing a mixture of three parts soil (Premier Pro-Mix Bx Professional; Premier Horticulture) and two parts vermiculite (D3 Fine Graded Horticultural Vermiculite; Whittemore). Plants were grown under controlled environmental conditions (15 h light/25°C, 9 h dark/20°C) in a growth chamber, and the soil mixture was kept moist by watering the pots with 0.7 mM Ca(NO3)2. Seedlings were harvested after 14 d, separated into shoots and roots, frozen in liquid nitrogen, and stored at −80°C or processed directly after harvesting for ChIP.
Sample Preparation and Solexa Library Construction
Maize tissue from 10 different seedlings was ground in liquid nitrogen, and genomic DNA was extracted from 1 g pooled tissue using a Qiagen DNeasy plant maxi kit. To enrich for methylated genomic DNA, 20 μg genomic DNA were digested with 200 units McrBC (New England Biolabs) overnight, and fragments 500 nucleotides and smaller were gel purified and used for library construction following the manufacturer's instructions, but adding a final gel purification step. To enrich for histone-modified regions, ChIP was conducted using 5 g fresh maize tissue from 10 seedlings following a previously described procedure (Lee et al., 2007). The following antibodies were used: H3K9ac (Upstate; 07-352), H3K27me3 (Upstate; 07-449), H3K4me3 (Abcam; ab8580), and H3K36me3 (Abcam; ab9050). For each 1-mL ChIP reaction, 5 μL antibody were added. The ChIPed DNA from three reactions was pooled to construct Solexa libraries essentially following the manufacturer's standard protocol but running 18 PCR cycles before gel purification of the samples. Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer's instructions. mRNA was extracted from total RNA using Dynabeads Oligo(dT) (Invitrogen Dynal) following the manufacturer's directions. After elution from the beads, first- and second-strand cDNA was generated using SuperscriptII reverse transcriptase (Invitrogen), and the standard Solexa protocol was followed thereafter to create mRNA libraries. smRNA was extracted by running total RNA on a 15% PAGE gel and cutting out bands in the ∼19- to 24-nucleotide size range. Libraries for smRNAs were constructed following previously published procedures (Mi et al., 2008; see Supplemental Methods online for details). All samples were prepared for sequencing following the manufacturer's standard protocol.
Sequence Data
The data for this article have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE15286. All data also can be freely accessed at http://plantgenomics.biology.yale.edu.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequencing and Mapping of mRNA Transcripts, H3K4me3, H3K9ac, H3K36me3, H3K27me3, DNA Methylation, and smRNAs in Maize Roots.
Supplemental Figure 2. Detection of Individual Transcribed Exons by de Novo Scanning of mRNA-seq Reads across the Maize Genome Sequence.
Supplemental Figure 3. Mapping of 11,742 Maize flcDNAs to the Maize Genome Sequence.
Supplemental Figure 4. Comparison of Gene Homology in Maize, Rice, and Arabidopsis.
Supplemental Figure 5. Pathway Annotation of Maize Genes.
Supplemental Figure 6. Comparison of Pathway Enrichment in Maize versus Rice and Maize versus Arabidopsis.
Supplemental Figure 7. Comparison of Gene Ontology Enrichment between Maize/Rice and Maize/Arabidopsis.
Supplemental Figure 8. Distribution of Epigenetic Patterns within Maize Genes in Roots.
Supplemental Figure 9. Effect of Modification Levels on Gene Expression.
Supplemental Figure 10. A Differentially Expressed Gene Shows a Different Epigenetic Pattern.
Supplemental Figure 11. Correlation of Differential Modifications of H3K27me3 and DNA Methylation with Differential Gene Expression in Shoots and Roots.
Supplemental Figure 12. Length Distributions of Removed smRNA Reads Matched with tRNA and rRNA Sequences.
Supplemental Figure 13. Length Distribution of Three Groups of smRNAs.
Supplemental Figure 14. Classification of smRNA Population in Shoots.
Supplemental Figure 15. Nucleotide Composition of Known miRNAs.
Supplemental Figure 16. Nucleotide Composition of Putative siRNAs.
Supplemental Figure 17. Nucleotide Composition of shRNAs.
Supplemental Figure 18. Origin and Target Sites on Genes and LTR-TEs for Different Classes of Putative siRNAs in Roots.
Supplemental Figure 19. Percentages of Different Types of Repeats Generating smRNAs in Different Lengths.
Supplemental Figure 20. Length Distribution of Unmapped smRNAs Classified by 5′ Terminal Nucleotides.
Supplemental Table 1. Solexa Sequencing and Mapping Statistics.
Supplemental Table 2. Validation Statistics of FgeneSH Predicted Genes.
Supplemental Table 3. Number of smRNAs Hitting Known miRNAs in Different Minimum Free Energy Ranges.
Supplemental Table 4. Solexa Sequencing Reads for miRNAs.
Supplemental Data Set 1. Comparisons of Maize and Rice Pathways.
Supplemental Data Set 2. Comparisons of Maize and Arabidopsis Pathways.
Supplemental Methods.
Supplementary Material
Acknowledgments
The Deng laboratory at Yale University was supported by grants from the National Institutes of Health (GM047850), the National Science Foundation Plant Genome Program (DBI0421675), and the National Science Foundation 2010 program. Studies conducted at the National Institute of Biological Sciences were supported by special funds from the Ministry of Science and Technology of China and Beijing Commission of Science and Technology. We thank the Maize Genome Sequencing Consortium (http://www.maizesequence.org) for communicating maize sequence information before publication and Will Terzaghi and Tim Nelson for critical reading of the manuscript. A.A.E. was supported by a Yale University Brown-Coxe Postdoctoral Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
References
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J.M. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Schones, D.E., Wang, Z., Wei, G., Chepelev, I., and Zhao, K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Bernatavichute, Y.V., Zhang, X., Cokus, S., Pellegrini, M., and Jacobsen, S.E. (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3 e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B.E., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315–326. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Van de Peer, Y., and Rouzé, P. (2006). The small RNA world of plants. New Phytol. 171 451–468. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Brendel, V. (2002). The maize genome sequencing project. Plant Physiol. 130 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Cigan, A.M., Saeteurn, K., and Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39 544–549. [DOI] [PubMed] [Google Scholar]
- Cokus, S.J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C.D., Pradhan, S., Nelson, S.F., Pellegrini, M., and Jacobsen, S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, J., Morrow, D.J., Casati, P., and Walbot, V. (2008). Distinctive transcriptome responses to adverse environmental conditions in Zea mays L. Plant Biotechnol. J. 6 782–798. [DOI] [PubMed] [Google Scholar]
- Fuchs, J., Demidov, D., Houben, A., and Schubert, I. (2006). Chromosomal histone modification patterns - from conservation to diversity. Trends Plant Sci. 11 199–208. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Grocock, R.J., van Dongen, S., Bateman, A., and Enright, A.J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G., al. (2005). Structure and architecture of the maize genome. Plant Physiol. 139 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., and Jacobsen, S.E. (2007). Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, S., Tacker, M., and Schuster, P. (1994). Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie 125 167–188. [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kapoor, M., Arora, R., Lama, T., Nijhawan, A., Khurana, J.P., Tyagi, A.K., and Kapoor, S. (2008). Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008. a). SOAP: Short oligonucleotide alignment program. Bioinformatics 24 713–714. [DOI] [PubMed] [Google Scholar]
- Li, H., Ruan, J., and Durbin, R. (2008. b). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., et al. (2008. c). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20 259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M., Park, Y.J., McLean, C.M., Dokshin, G.A., Persson, J.M., Herman, H., Pasini, D., Miró, X., Donohoe, M.E., Lee, J.T., Helin, K., and Soloway, P.D. (2008). Antagonism between DNA methylation and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 4 e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., May, B., Yordan, C., Singer, T., and Martienssen, R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R., O'Malley, R.C., Tonti-Filippini, J., Gregory, B.D., Berry, C.C., Millar, A.H., and Ecker, J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz, M.C., Dickerson, D.R., Schmitt, M., and Groudine, M. (2004). Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11 1068–1075. [DOI] [PubMed] [Google Scholar]
- Lu, C., et al. (2008). Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 105 4951–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Morrow, D.J., Fernandes, J., and Walbot, V. (2006). Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Biol. 7 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, J.H.A., O'Sullivan, R.J., Braunschweig, U., Opravil, S., Radolf, M., Steinlein, P., and Jenuwein, T. (2005). The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A., Doerge, R.W., and Colot, V. (2005). Epigenomic mapping in Arabidopsis using tiling microarrays. Chromosome Res. 13 299–308. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., Probst, A.V., and Paszkowski, J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., Reinders, J., Čaikovsky, M., Smathajitt, C., and Paszkowski, J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130 851–862. [DOI] [PubMed] [Google Scholar]
- Messing, J., Bharti, A.K., Karlowski, W.M., Gundlach, H., Kim, H.R., Yu, Y., Wei, F., Fuks, G., Soderlund, C.A., Mayer, K.F., and Wing, R.A. (2004). Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing, J., and Dooner, H.K. (2006). Organization and variability of the maize genome. Curr. Opin. Plant Biol. 9 157–163. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S., et al. (2008). Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, T.S., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta, K., et al. (2008). Distinct size distribution of endogenous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu, C.Y., and Hsieh, C.L. (2007). DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 27 2746–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. (2008). Corn genomics pops wide open. Science 319 1333. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., and Bennetzen, J.L. (2006). The maize genome as a model for efficient sequence analysis of large plant genomes. Curr. Opin. Plant Biol. 9 146–156. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Citek, R., Budiman, M.A., Nunberg, A., Bedell, J.A., Lakey, N., O'Shaughnessy, A.L., Nascimento, L.U., McCombie, W.R., and Martienssen, R.A. (2005). Differential methylation of genes and repeats in land plants. Genome Res. 15 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, V., and Chen, X. (2008). Small RNA metabolism in Arabidopsis. Trends Plant Sci. 13 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riclet, R., Chendeb, M., Vonesch, J.-L., Koczan, D., Thiesen, H.-J., Losson, R., and Cammas, F. (2009). Disruption of the interaction between transcriptional intermediary factor 1β and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol. Biol. Cell 20 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, Y.B., Kahn, T.G., Nix, D.A., Li, X.-Y., Bourgon, R., Biggin, M., and Pirrotta, V. (2006). Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38 700–705. [DOI] [PubMed] [Google Scholar]
- Shi, J., and Dawe, R.K. (2006). Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 172 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko, L., and Chandler, V. (2008). RNA-dependent RNA polymerase is required for enhancer-mediated transcriptional silencing associated with paramutation at the maize p1 gene. Genetics 180 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R.K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8 272–285. [DOI] [PubMed] [Google Scholar]
- Stupar, R.M., and Springer, N. (2006). Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, O.H., Aravin, A.A., Stein, P., Girard, A., Murchison, E.P., Cheloufi, S., Hodges, E., Anger, M., Sachidanandam, R., Schultz, R.M., and Hannon, G.J. (2008). Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2000). Gene Ontology: Tool for the unification of biology. Nat. Genet. 25 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5 e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Zang, C., Rosenfeld, J.A., Schones, D.E., Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Peng, W., Zhang, M.Q., and Zhao, K. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., et al. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453 539–544. [DOI] [PubMed] [Google Scholar]
- Weil, C., and Martienssen, R. (2008). Epigenetic interactions between transposons and genes: Lessons from plants. Curr. Opin. Genet. Dev. 18 188–192. [DOI] [PubMed] [Google Scholar]
- Woodhouse, M.R., Freeling, M., and Lisch, D. (2006). Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4(10): e339 (online). doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y.V., Pellegrini, M., Goodrich, J., and Jacobsen, S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W.L., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, J.R., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nussbaum, C., Myers, R.M., Brown, M., Li, W., and Liu, X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9 R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and Wang, H. (2007). LTR-FINDER: An efficient tool for the prediction of full-length LTR retrotransposons (2007). Nucleic Acids Res. 35 W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.