Abstract
Biotrophic invasive hyphae (IH) of the blast fungus Magnaporthe oryzae secrete effectors to alter host defenses and cellular processes as they successively invade living rice (Oryza sativa) cells. However, few blast effectors have been identified. Indeed, understanding fungal and rice genes contributing to biotrophic invasion has been difficult because so few plant cells have encountered IH at the earliest infection stages. We developed a robust procedure for isolating infected-rice sheath RNAs in which ∼20% of the RNA originated from IH in first-invaded cells. We analyzed these IH RNAs relative to control mycelial RNAs using M. oryzae oligoarrays. With a 10-fold differential expression threshold, we identified known effector PWL2 and 58 candidate effectors. Four of these candidates were confirmed to be fungal biotrophy-associated secreted (BAS) proteins. Fluorescently labeled BAS proteins were secreted into rice cells in distinct patterns in compatible, but not in incompatible, interactions. BAS1 and BAS2 proteins preferentially accumulated in biotrophic interfacial complexes along with known avirulence effectors, BAS3 showed additional localization near cell wall crossing points, and BAS4 uniformly outlined growing IH. Analysis of the same infected-tissue RNAs with rice oligoarrays identified putative effector-induced rice susceptibility genes, which are highly enriched for sensor-transduction components rather than typically identified defense response genes.
INTRODUCTION
Rice blast is a significant disease that affects one of the most important food sources in the world. Each year, rice blast causes losses between 10 and 30% even though diverse cultivars expressing different resistance genes are used for cultivation (Talbot, 2003; Kawasaki, 2004; Ebbole, 2007). The causal agent, the hemibiotrophic fungus Magnaporthe oryzae (Couch et al., 2005), undergoes complex morphological development throughout its infection cycle. Many studies have focused on the process by which fungal spores produce germ tubes that differentiate into appressoria, which are specialized cells for leaf surface penetration (Howard and Valent, 1996, 2003; Dean et al., 2005; Oh et al., 2008). Less is known about the critical postpenetration stage where, in susceptible cultivars, the fungus succeeds in establishing biotrophic invasion leading to disease, or where, in resistant cultivars, recognition of avirulence (AVR) effectors by corresponding resistance gene products induces the plant's hypersensitive response and blocks disease. Fewer studies have focused on the susceptible (compatible) interaction than on understanding resistance responses in the incompatible interaction (Koga et al., 2004b; Ribot et al., 2008).
Recently, cellular analyses using optically clear rice (Oryza sativa) sheath tissues have led to new understanding of the biotrophic invasion strategy used by the rice blast fungus in a highly compatible interaction (Koga et al., 2004b; Kankanala et al., 2007). Thin filamentous primary hyphae first grow in the cell lumen after appressorial penetration of the cell wall, and these hyphae invaginate the host plasma membrane. These hyphae differentiate into bulbous invasive hyphae (IH) that become sealed in an extra-invasive hyphal membrane (EIHM) compartment and exhibit pseudohyphal growth as they fill the invaded cell. The IH search along the plant cell wall and undergo extreme constriction, possibly through plasmodesmata, to cross into live neighbor cells. To counteract plant defenses, to stimulate plant membrane dynamics, and to successively invade live cells, IH must express specialized effector genes, including those encoding cytoplasmic effectors secreted across the EIHM into the host cytoplasm.
A few genes that are specifically expressed in blast IH have been identified (Talbot, 2003; Donofrio et al., 2006). These are MIR1, encoding a nonsecreted IH nuclear protein of unknown function (Li et al., 2007), and several genes encoding secreted effectors, including AVR-Pita1 (Jia et al., 2000; Khang et al., 2008), PWL1 (Kang et al., 1995), PWL2 (Sweigard et al., 1995), and AVR-CO39 (Peyyala and Farman, 2006). Other than their properties as in planta–specific secreted proteins, the few diverse examples of blast AVR effector proteins have not provided motifs for identification of additional effectors in the M. oryzae genome (Dean et al., 2005). Catanzariti et al. (2006) showed that proteins secreted by haustoria of the flax rust fungus are highly enriched in AVR/effector proteins. Therefore, identification of IH-specific, secreted proteins and any structural motifs they may share represents a reasonable route to identification of additional blast effectors.
Fluorescently labeled blast effectors AVR-Pita1, PWL1, and PWL2 have recently been shown to be secreted by IH growing in rice cells (R. Berruyer, C.H. Khang, P. Kankanala, S.Y. Park, K. Czymmek, S. Kang, and B. Valent, unpublished data). Secreted effector fusion proteins accumulated in a novel pathogen-induced structure, the biotrophic interfacial complex (BIC), at specific locations inside the EIHM compartment enclosing the IH. For each filamentous hypha to enter a host cell, the BIC first appeared as the EIHM membranous cap at the hyphal tip (Kankanala et al., 2007). When each filamentous hypha differentiated into bulbous IH, the BIC moved beside the first IH cell and remained there as a discrete structure as IH continued to grow in the cell. The accumulation of fluorescent effectors in BICs as long as IH grew inside rice cells has led to the hypotheses that BIC localization is diagnostic of blast effectors and that BICs play a role in translocation of effectors to the rice cell cytoplasm.
Multiple studies have examined gene expression by M. oryzae during axenic (free from other organisms) growth (Irie et al., 2003; Takano et al., 2003; Gowda et al., 2006; Oh et al., 2008). For example, >28,000 ESTs were obtained from cDNA libraries representing fungal cell types that are produced in vitro (mycelia, conidia, appressoria, and perithecia), mycelium from different culture conditions, and a pmk1− nonpathogenic mutant lacking the mitogen-activated protein kinase PMK1 (Ebbole et al., 2004; Soanes and Talbot, 2005). Microarray analyses have compared gene expression in germlings growing on inductive surfaces (promoting appressorium formation) and noninductive surfaces (Oh et al., 2008) and gene expression in mycelium growing in nitrogen-rich and nitrogen-deficient media (Donofrio et al., 2006). In planta expression analyses performed following development of macroscopic symptoms identified both fungal and rice genes expressed in planta (Kim et al., 2001; Rauyaree et al., 2001; Matsumura et al., 2003). However, infected tissue with visible symptoms probably included filamentous necrotrophic hyphae in addition to IH (Berruyer et al., 2006). Large-scale EST analysis (Jantasuriyarat et al., 2005) and microarray analysis (Vergne et al., 2007) performed at early infection stages before appearance of macroscopic symptoms focused on rice gene expression because so little fungus was present in the infected leaf tissue.
Few host susceptibility genes that contribute to the pathogen's progress have been identified. Plant genes encoding enzymes of phytoalexin biosynthesis as well as defense and pathogenesis-related proteins have been shown to be upregulated during infection in diverse host pathogen systems (van Loon et al., 2006). The same genes were generally induced in incompatible and compatible interactions, although expression usually occurred later and at lower levels during compatibility (Song and Goodman, 2001; Tao et al., 2003; Vergne et al., 2007). For blast, the jasmonic acid–induced rice transcription factor gene JAMyb (AK069082) represented an example of a host gene that was more highly expressed in compatible than incompatible interactions (Lee et al., 2001).
Our goal in this study was to identify putative fungal effector genes and rice susceptibility genes contributing to biotrophic blast invasion. Using a highly compatible interaction between a strain of M. oryzae that is an aggressive rice pathogen and rice leaf sheath cells (Kankanala et al., 2007), we produced infected-tissue RNAs that were highly enriched for RNA from IH growing in first-invaded host cells. Using a M. oryzae microarray, gene expression in IH in planta was compared with expression in axenic mycelium grown in vitro. With the same samples and a rice microarray, rice gene expression in infected tissue was compared with expression in mock-inoculated rice. We identified numerous effector candidates as well as putative rice susceptibility genes. Here, we describe distinctive in planta secretion patterns of four M. oryzae biotrophy-associated secreted proteins, including one, named BAS4, which provides a valuable tool for assessing EIHM compartment integrity in individual invaded rice cells.
RESULTS
Infected Leaf Sheath Samples Enriched for Biotrophic IH
A major challenge for identifying genes expressed at early stages of fungal infection is that most plant cells have not yet encountered the fungus. We developed a reproducible procedure to obtain infected tissues enriched for rice cells containing IH and their immediate neighbors (Figure 1A). We used sheath tissues at 36 h postinoculation (hpi) because infection development at this point was relatively synchronous: most IH were growing in first-invaded cells and a few had just moved into neighbor cells. Use of a fungal strain with strong constitutive, cytoplasmic expression of enhanced yellow fluorescent protein (EYFP) allowed visualization of contaminating fungal cell types in the tissue as well as the state of development of IH. Although inoculation of conidia in gelatin solution, our standard procedure, promoted uniform distribution of infection sites, this practice also promoted growth of vegetative mycelium on the sheath surface. The first step in our procedure was to remove vegetative hyphae, appressoria, and conidia from this surface. Abundant hyphae that invaded the tissue from cut sheath ends were also discarded. Using the procedure of Kankanala et al. (2007), we next manually dissected the sheath tissue to produce pieces with the inoculated adaxial epidermal layer and approximately three underlying mesophyll cell layers, thus removing many plant cell layers that had not yet experienced fungal invasion. The last steps were rapid epifluorescence screening for selection of densely invaded sheath segments (Figure 1A, top image) and freezing of selected segments in liquid nitrogen. The process was carried to completion with a single sheath piece at a time, resulting in ∼2 min of processing time for each. To estimate the ratio of fungal-to-rice RNAs in infected sheaths, we compared RT-PCR amplification of the fungal actin gene in infected tissue to amplification in standards produced by mixing pure mycelial RNA and mock-inoculated rice RNA (see Supplemental Figure 1A online). Using this assay,∼20% of the infected tissue RNAs were fungal. With this procedure, we obtained RNAs from infected tissues that were significantly enriched both for fungal biotrophic IH content and for invaded rice cells compared with RNAs prepared from leaf samples using standard spray inoculation (see Supplemental Figure 1B online; Berruyer et al., 2006).
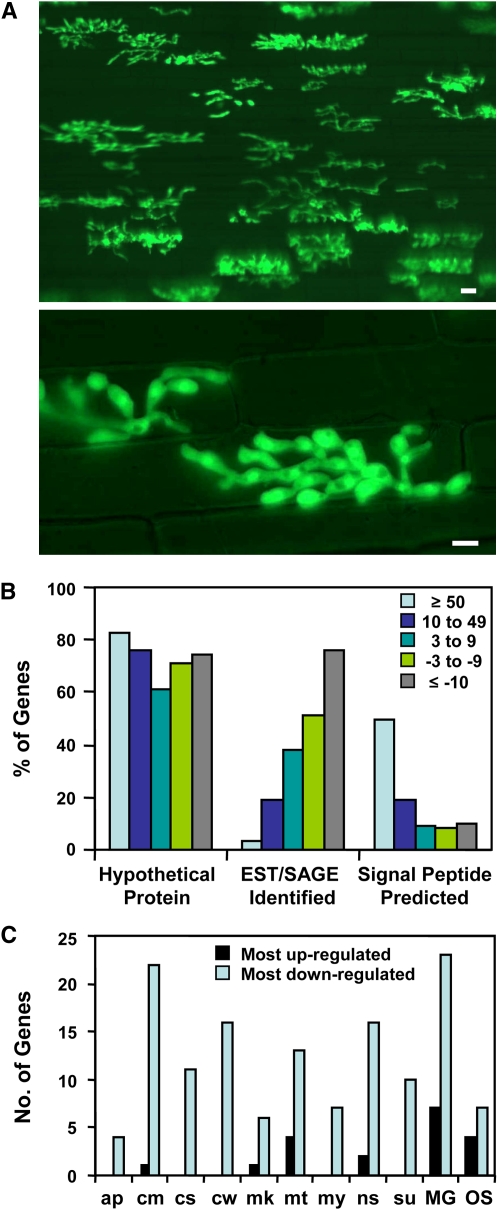
Figure 1.
Biotrophic IH Are Morphologically Distinct and They Express Many Previously Undescribed Genes.
(A) IH of strain KV1 growing in susceptible YT16 rice at 36 hpi. The fungus expressed cytoplasmic EYFP to facilitate microscopic assessment of infection site density before harvesting tissue for RNA preparation. Merged differential interference contrast (DIC) and fluorescence images illustrate a densely infected sheath segment (top image, bar = 20 μm) and typical IH in first-invaded epidermal cells (bottom image, bar = 5 μm).
(B) Properties of pathogen genes with threefold or larger differential expression ratios, considering expression in IH relative to mycelium. Bars for each expression group indicate the percentage of the total number of genes in that group with each property.
(C) Comparison of the top 50 genes expressed in IH with the bottom 50 genes for representation in EST and SAGE data sets. For each gene set, the library hits are shown for the nine genes (18%) with highest expression in IH (black bars) and the 42 genes (84%) with the lowest (light-blue bars). Many genes were identified in multiple libraries. EST libraries: ap, appressoria; cm, mycelium from complete medium; cs, conidia; cw, mycelium grown on rice cell walls; mk, pmk1− mutant; mt, mating culture; my, mycelium from minimal medium; ns, nitrogen-starved mycelium; and su, subtracted library (Ebbole et al., 2004). SAGE libraries: MG, MG_SGa, mycelium from minimal medium; OS, OSJNGg, compatible interaction at 96 hpi (Gowda et al., 2006).
Identification of the Interaction Transcriptome Using Microarray Hybridization
Samples from three independent sheath assays, performed over time, were analyzed separately using the M. oryzae genome microarray (Agilent Technologies), which contains 60-bp oligonucleotide probes for 15,152 M. oryzae genes (genome sequence version 4; Dean et al., 2005). The same samples were analyzed with the Agilent rice microarray, which contains oligonucleotides corresponding to ∼21,500 expressed rice genes (based on cDNAs from the KOME database; http://cdna01.dna.affrc.go.jp/cDNA/). Complementary RNAs from infected tissues were labeled with Cyanine-3 or Cyanine-5 and hybridized together with control RNA mixtures (20% mycelial RNA and 80% mock-inoculated rice RNA) labeled with the other dye (Hughes et al., 2001). Three biological replications were performed, each with four separate microarray hybridizations (two technical replicates and two dye swap experiments). Data were analyzed using Rosetta Resolver software (Weng et al., 2006). Analyzing the biological replicates separately, signature sequences (denoted as significantly different from the diagonal) showed correlations of >80% between replicates. This confirmed that our biological sampling procedure was robust. Technical replicates showed correlations >95%. The results detailed further were obtained by analysis of all 12 data sets together, for both the fungal and rice microarrays.
Comparing expression in IH to mycelium, the ribosomal protein genes showed expression ratios between +3 and −1. Using a threefold threshold for differential expression, 1120 fungal genes showed higher expression in IH relative to mycelium, and 781 genes showed lower expression in IH. Ninety-one percent of the overexpressed genes and 93% of the underexpressed genes had P values that were still significant (P < 10−6) after Bonferroni correction for multiple comparisons (Sokal and Rohlf, 1981). For rice, 1231 genes were overexpressed and 483 genes underexpressed in invaded tissue, again most with P < 10−6. Unless stated otherwise, we focus on fungal and rice genes with P < 10−6.
Differential expression of AVR-Pita1 and selected fungal genes was confirmed by quantitative RT-PCR (qRT-PCR) (Table 1). Fungal and rice genes with positive or negative differential expression ratios >30 were consistently confirmed as differentially expressed by RT-PCR (see Supplemental Figures 1C and 1D online). The major discrepancy seen in the validation analysis was for AVR-Pita1, which showed low levels of enhanced IH expression in the microarray (threefold higher in IH, P = 10−6) but clear differential expression by qRT-PCR (Table 1). Expression ratios for this AVR gene varied between biological replications, perhaps because the fungal strain used contained the hypervariable, telomeric allele that undergoes spontaneous deletion (Orbach et al., 2000). All experiments confirmed that we have identified hundreds of genes that are regulated during biotrophic invasion.
Table 1.
Confirmation of IH-Specific Expression of Fungal Genes Using Real-Time-RT-PCR
| Ct Valuesa
|
Calculated Fold Changeb | ||
|---|---|---|---|
| Gene (Fold Change) | Mycelium | IH at 36 hpi | |
| Actin | 24.8 ± 1.3 | 23.3 ± 1.2 | – |
| AVR-Pita1 (3) | N/D | 29.6 ± 1.4 | – |
| MGG_04795.6 (100) | N/D | 25.3 ± 3.1 | – |
| MGG_09693.6 (84) | 35.9 ± 1.2 | 25.8 ± 3.3 | 388 |
| MGG_11610.6 (71) | 33.5 ± 2.5 | 23.9 ± 2.5 | 274 |
| MGG_06224.6 (64) | 38.4 ± 1.2 | 29.3 ± 0.6 | 194 |
Ct values represent the fractional number of cycles at which the amount of the amplified target reached a fixed threshold. High Ct values indicate low abundance of the specific transcript. The Ct values represent the mean of two biological and two technical replications. Standard deviations are shown. N/D, not detected.
Relative expression (RQ value) in IH defining expression in mycelium as 1X, calculated as 2−(ΔΔCT). Ct values were normalized to the fungal actin gene. A fold change was not calculated for AVR-Pita1 and MGG_04795.6 because their expression was not detected in the mycelial samples.
Genes Upregulated in IH Are Enriched for AVR and Newly Described Genes but Not Known Pathogenicity Genes
The cRNAs from differentially labeled 36-hpi infected tissue and control mixtures were analyzed with M. oryzae microarrays. In this analysis, the in planta–specific AVR-Pita1 and PWL2 genes were highly expressed in IH (Tables 1 and 2). The AVR gene ACE1, which encodes an appressorium-specific polyketide synthase-nonribosomal peptide synthetase (Böhnert et al., 2004), showed negligible signals in both samples. Finding expected expression patterns for known AVR effectors suggested that other IH-specific secreted proteins were good candidates for being effectors (Table 2).
Table 2.
Properties of Putative Secreted Proteins with ≥50-Fold Differential Expression in IH
| Probe (Fold Change)a | Gene Nameb | AA (Cys) | M.o. Hits (E Value)c | Other Fungal Hits (Organism; E Value)d | Chr. Location/Commentse |
|---|---|---|---|---|---|
| AMG08263 (100) | MGG_14965.6 | 208 (5) | MGG_15371.6 (E = 0); MGG_10780.6 (E = 8e-23) | None | IV: 4,093,668- |
| Same as above | MGG_15371.6 | 208 (5) | MGG_14965.6 (E = 0); MGG_10780.6 (E = 8e-23) | None | VII: 234,746- Located next to AVR-Pita1 |
| AMG08261 (100) | MGG_04795.6 BAS1 | 115 (0) | None | None | IV: 1,216,801- |
| AMG08417.2 (88) | MGG_09379.6 | 121 (4) | None | None | IV: 3,567,509- |
| AMG08541 (84) | MGG_09693.6 BAS2 | 102 (6) | MGG_07969.6 (E = 2e-28); MGG_07749.6 (E = 4e-17) | P. tritici (E = 4e-21); and five other fungi | V: 5,545,247+ |
| AMG06765 (75) | MGG_15264.6 | 191 (1) | None | None | III: 6,290,613+ |
| AMG06650 (74) | MGG_07834.6 | 120 (3) | MGG_09035.6 (E = 3e-4) | None | III: 6,015,965+ |
| AMG12560 (71) | MGG_11610.6 BAS3 | 113 (10) | None | None | VI: 3,454,688+ |
| AMG07384 (71) | MGG_08657.6 | 207 (8) | None | None | IV: 195,247+ |
| AMG08859 (64) | MGG_06224.6 | 134 (4) | None | None | V: 4,531,287- |
| AMG13014 (63) | MGG_09803.6 | 125 (6) | None | None | VI: 4,659,149- |
| AMG11184 (63) | MGG_04301.6 PWL2 | 145 (2) | MGG_13863.6 (E = 0); MGG_07398.6 (E = 8e-16) | None | IV: 1,480,013- |
| Same as above | MGG_13863.6 PWL2 | 145 (2) | MGG_04301.6 (E = 0); MGG_07398.6 (E = 8e-16) | None | III: 5,594,534- |
| AMG15980 (61) | MGG_10914.6 BAS4 | 102 (8) | MGG_02154.6 (E = 2e-7) | None | VI: 236,698+ |
| AMG08432 (51) | MGG_09387.6 | 219 (0) | None | None | IV: 3,535,091+ |
| RMG00001 (3) | MGG_15370.6 AVR-Pita1 | 224 (9) | MGG_14981.6 (E = 3e-71) | H. capsulatus (E = 9e-12) | VII: 232,500- Located next to MGG_15371.6 |
Magnaporthe grisea Oryza sativa Interaction database (MGOS) gene names correspond to the probes used in version 2 of the Agilent M. oryzae microarray. They will not change with new releases of the genome sequence. MGOS names can be converted to Broad Database gene names at www.mgosdb.org. Fold changes for all genes except AVR-Pita1 (with P = 10−6) have P values = 0. Most of these genes were tested and confirmed to be differentially expressed (see Supplemental Figure 1C online). Not included in this table are the genes corresponding to AMG08160, corresponding to the chitinase MGG_04732.6, and the gene for AMG08787, which is not included in version 6.
Gene names are from the genome sequence release 6 (http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea/MultiHome.html).
BLAST similarity search against the Magnaporthe grisea proteins database (http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea/Blast.html?sp=Sblastp).
NCBI BLAST with protein sequences from the Broad Institute.
Chromosomal locations are derived from MGOS (http://www.mgosdb.org).
Fungal genes with differential expression levels ≥ ±3-fold were divided into five groups for comparison of types of encoded proteins and previous expression data (Figure 1B). The two groups with the highest levels of expression in IH were enriched in putative secreted proteins (having a predicted signal peptide): 50% of proteins in the ≥+50-fold group and 19.3% of proteins in the +10- to 49-fold group (Figure 1B, right side). The remaining three groups included ∼9% putative secreted proteins, which is comparable to the average frequency in the entire genome (Dean et al., 2005). By contrast, the proportion of genes with previous expression data increased from only 3% in the 50-fold and higher group to 76% in the −10-fold and lower group (Figure 1B, middle). We next compared the 50 genes showing the highest positive expression ratios (highest in IH) with the 50 genes with the highest negative ratios (highest in mycelium) for reported expression data. Compared with the IH-enriched genes, sequences for the mycelium-enriched genes were frequently identified from libraries representing numerous different in vitro growing conditions and cell types (Figure 1C; see Supplemental Table 1A online). Twenty-three percent of the 50 most IH-enriched genes had clues to their functions compared with 48% for the 50 most mycelium-enriched genes (see Supplemental Table 1A online). Characterization of the 59 putative secreted proteins with ≥10-fold higher expression levels in IH showed that only 15% had predicted functions (see Supplemental Table 1B online). Considering putative secreted IH proteins upregulated ≥3-fold, there was an inverse correlation between higher positive expression ratios and functional domains identified (Table 3). Among these putative secreted IH proteins, 20% appeared to be glycosyl hydrolases and 17% appeared to be proteases.
Table 3.
Putative IH-Upregulated, Secreted Proteins with Hits in the Pfam Database
| Genea | Fold Up (P Value) | Pfamb | Description (E Value) |
|---|---|---|---|
| MGG_04732.6 (AMG08160) | 58 (0) | Glyco_hydro_18 | Glycosyl hydrolases family 18 (3.6e-59) |
| AMG14799 | 49 (0) | DNA_methylase | C-5 cytosine-specific DNA methylase (1.5e-19) |
| MGG_05790.6 (AMG05133) | 46 (0) | Cu-oxidase_3 | Multicopper oxidase (1e-49) |
| MGG_09035.6 (AMG13593.2) | 36 (0) | zf-C2H2 | Zinc finger, C2H2 type (0.15) |
| MGG_13327.6 (AMG16197) | 36 (0) | RnaseH | RNase H (7e-25) |
| MGG_09801.5 (AMG02457) | 28 (0) | COesterase | Carboxylesterase (7.2e-13) |
| MGG_09073.6 (AMG12553) | 19 (0) | Peptidase_S8 | Subtilase family (2.4e-16) |
| MGG_07556.6 (AMG04878) | 17 (0) | DPBB_1 | Rare lipoprotein A (RlpA)-like double-ψ β-barrel (1.6e-05) |
| MGG_05038.6 (AMG06036) | 14 (0) | Exo_endo_phos | Endonuclease/exonuclease/phosphatase family (5.7e-13) |
| MGG_05599.6 (AMG04353) | 9 (0) | Glyco_hydro_3_C | Glycosyl hydrolase family 3 C terminal domain (2.6e-62) |
| MGG_10445.6 (AMG00014) | 8 (2E-14) | Peptidase_S8 | Subtilase family (2e-43) |
| AMG16270 | 8 (5E-21) | HET | Heterokaryon incompatibility protein (5.2e-22) |
| MGG_03995.6 (AMG10755) | 8 (0) | Peptidase_S10 | Ser carboxypeptidase (2e-77) |
| MGG_05694.6 (AMG09398) | 7 (0) | Glyco_hydro_18 | Glycosyl hydrolases family 18 (1e-30) |
| MGG_09030.6 (AMG13583) | 7 (1E-27) | DUF1680 | Putative glycosyl hydrolase: unk. function (6e-194) |
| MGG_03671.6 (AMG10290) | 7 (0) | Hydrophobin | Fungal hydrophobin (0.75) |
| MGG_07709.6 (AMG06488) | 7 (0) | Glyco_hydro_16 | Glycosyl hydrolases family 16 (0.00046) |
| MGG_04546.6 (AMG07869) | 6 (2E-33) | Ogr_Delta | Ogr/δ-like zinc finger (0.44) |
| MGG_00973.6 (AMG11537) | 6 (5E-25) | FAD_binding_4 | FAD binding domain (6.3e-29) |
| MGG_10585.6 (AMG13523) | 5 (0) | Tyrosinase | Common central domain of tyrosinase (2e-06) |
| MGG_10423.6 (AMG09849) | 5 (1E-10) | Cellulase | Cellulase (glycosyl hydrolase family 5) (0.0014) |
| MGG_10260.5 (AMG04914) | 5 (2E-10) | Peptidase_S28 | Ser carboxypeptidase S28 (5.1e-10) |
| MGG_08096.6 (AMG01465) | 4 (0) | EMP24_GP25L | emp24/gp25L/p24 family/GOLD (5.3e-10) |
| MGG_10811.6 (AMG15832) | 4 (2E-8) | Acid_phosphat_A | Histidine acid phosphatase (9.7e-49) |
| MGG_01945.6 (AMG07158.1) | 4 (4E-5) | Lipoxygenase | Lipoxygenase (5.3e-09) |
| MGG_02849.6 (AMG07436) | 4 (7E-10) | Asp | Eukaryotic aspartyl protease (7.4e-38) |
| AMG12824 | 3 (2E-13) | Peptidase_S24 | Peptidase S24-like (0.00055) |
| MGG_09087.6 (AMG02047) | 3 (2E-9) | PLA2_B | Lysophospholipase catalytic domain (2.2e-229) |
| MGG_08415.6 (AMG02456) | 3 (0) | Peptidase_S8 | Subtilase family (9.5e-26) |
| MGG_04439.6 (AMG07730) | 3 (3E-12) | EMP24_GP25L | emp24/gp25L/p24 family/GOLD (2.2e-17) |
| MGG_09398.6 (AMG08447) | 3 (2E-14) | peroxidase | Peroxidase (2.6e-10) |
| MGG_11036.6 (AMG16160) | 3 (1E-8) | CBM_1 | Fungal cellulose binding domain (2.4e-10) |
| MGG_12539.5 (AMG16053) | 3 (8E-8) | Chromo | Chromatin organization modifier domain (1.2e-11) |
| MGG_10710.6 (AMG15699.3) | 3 (1E-40) | DAO | FAD-dependent oxidoreductase (6.4e-09) |
| MGG_09098.6 (AMG13694) | 3 (2E-4) | Glyco_hydro_43 | Glycosyl hydrolases family 43 (5.2e-70) |
| MGG_04015.6 (AMG10782) | 3 (1E-5) | Glyco_hydro_76 | Glycosyl hydrolase family 76 (1.1e-177) |
| AMG02226 | 3 (3E-3) | Nop | Putative snoRNA binding domain (3.6e-79) |
| MGG_08319.6 (AMG02322) | 3 (3E-9) | Peptidase_M28 | Peptidase family M28 (6.4e-57) |
| MGG_05529.6 (AMG04253) | 3 (2E-7) | Tannase | Tannase and feruloyl esterase (2e-88) |
| MGG_05531.6 (AMG04255) | 3 (5E-4) | CFEM | CFEM domain (4.4e-09) |
| MGG_04756.6 (AMG08194) | 3 (2E-20) | Amidase | Amidase (1e-50) |
All genes are from M. oryzae genome version 6 except those labeled with “.5”. In this case, the corresponding version 5 gene was deleted from version 6. Genes labeled only with the probe name were defined in genome version 4 but dropped from version 5.
Domain with the best score from predicted amino acid sequences analyzed for Pfam matches (http://pfam.sanger.ac.uk).
Expression of known pathogenicity genes was unchanged or downregulated in IH (see Supplemental Table 1C online). Only the adenylate cyclase-interacting protein ACI1, which is involved in signal transduction during appressorium formation, reached our +3-fold differential expression threshold. Most genes with known roles in appressorium formation and function were not differentially regulated in IH and mycelium, including PMK1 (Zhao et al., 2007), which also has a role in IH colonization. The most dramatic examples of differential expression corresponded to lower expression in IH. PTH11, encoding the G protein–coupled receptor involved in surface sensing and appressorium formation, showed fivefold lower expression levels in IH. The MPG1 hydrophobin gene, the RSY1, BUF1, and 4HNR melanin biosynthesis genes, and the alternative oxidase gene AOX showed dramatically lower expression in IH (≥20-fold).
In summary, the most abundant IH mRNAs often corresponded to secreted proteins identified as expressed for the first time, while mycelium-enriched genes encoded fewer secreted proteins, and these genes were highly represented in expression libraries from fungus in axenic culture. These results validate our hypothesis that IH express many specialized genes during biotrophic invasion.
In Vitro Growth Conditions Do Not Mimic Biotrophic Invasion
The M. oryzae genome contains numerous genes for enzymes that degrade plant cell walls (Dean et al., 2005), but their role in biotrophic invasion is not understood. These genes are expressed in planta after macroscopic symptoms develop (Wu et al., 2006), and they are induced in mycelium grown on nutrient medium containing isolated plant walls as the major carbon source (Wu et al., 2006). The sequenced cDNAs from mycelium grown on cell wall medium identified more secreted proteins than cDNAs from other sources (Ebbole et al., 2004). One of the >10-fold upregulated secreted protein genes (see Supplemental Table 1B online) encodes a putative cellulase. However, out of five characterized xylanases (Wu et al., 2006) represented in the microarray, none were upregulated in IH. ESTs from cell wall–grown mycelium were well represented among transcripts that were underexpressed in IH (Figure 1C; see Supplemental Table 1A online) but not among transcripts for the >10-fold induced IH genes (see Supplemental Table 1B online). Therefore, growth on medium containing plant cell walls does not mimic the intracellular environment experienced by IH during biotrophic invasion of rice.
Genes required for pathogenicity in several fungal pathogen systems are induced during growth in media lacking sufficient nitrogen, suggesting that the plant environment might be nitrogen deficient for the pathogen (Donofrio et al., 2006). However, our results on expression in IH differed from results of expression analysis for mycelium grown under nitrogen starvation conditions, also performed using the M. oryzae microarray (Donofrio et al., 2006). Of the five pathogenicity genes that were upregulated by nitrogen starvation, PTH11, MPG1, 4HNR, and AOX were downregulated in IH, and NTH1 was unchanged in its expression level (see Supplemental Table 1C online). Only three of the top 55 genes upregulated by nitrogen starvation were also upregulated in IH. Therefore, it appears that expression in response to nitrogen starvation has relevance to the prepenetration phase when the fungus is growing on the plant surface (Soanes et al., 2002; Donofrio et al., 2006, and references therein) but not to early biotrophic invasion stages of growth inside the plant.
Candidate Rice Susceptibility Genes Expressed during Biotrophic Invasion
Infected tissue samples that are enriched for IH mRNAs must also be enriched in rice mRNAs from cells impacted by IH. Therefore, the same IH-enriched RNAs were used for analysis with the Agilent rice microarray. Among the group with highest expression during biotrophic invasion (>50-fold rice group, Table 4, P values still significant after Bonferroni correction), genes for mitogen-activated protein kinase kinase kinases (MAPKKKs), for transcription factors, and for cytochrome P450 proteins were represented at least three times. According to PSORT, other genes encoded proteins with predicted membrane or nuclear localization. By contrast, rice genes with higher expression in mock-inoculated rice included PSORT-predicted extracellular and cytoplasmic proteins (>-50-fold rice group, Table 4). Defense-related genes that have been previously reported to be highly expressed in blast-infected tissues were induced at low to moderate levels during biotrophic invasion (see Supplemental Table 2 online). The unique highly expressed gene set induced during biotrophic invasion was distinct from the gene set induced by wounding cuts on rice leaves (see Supplemental Table 3 online), showing that the wounding of our leaf sheath pieces was not an issue (Katou et al., 2007).
Table 4.
Rice Genes Induced or Repressed >50-Fold in Infected Tissue
| Sequence IDa | Annotation (E Value) | Fold Change (P Value) | Commentsb |
|---|---|---|---|
| Induced | |||
| AK071227 | Unknown expressed protein | 99 (0) | Chr. 8; not predicted; none |
| AK105196 | Zea mays NPK1-related protein kinase (MAPKKK1) mRNA (E value = 7e-72) | 96 (0) | Chr. 1; microbody |
| AK109702 | Z. mays NPK1-related protein kinase-like (MAPKKK1) mRNA (E value = 1e-62) | 86 (0) | Chr. 5; nucleus |
| AK061237 | Arabidopsis mRNA, clone RAFL25-06-N10 (E value = 4e-79) | 83 (0) | Chr. 1; cytoplasmic; phosphatase |
| AK071585 | Triticum aestivum mRNA wdi1c.pk004.j19:fis (E value = 8e-94) | 77 (0) | Chr. 1; nucleus; related to NPK1 MAPKKK |
| AK100808 | Z. mays inward rectifying shaker K+ channel mRNA, complete CDS (E value = 0) | 77 (2.8E-15) | Chr. 2; microbody |
| AK062422 | O. sativa putative DRE binding protein 1B mRNA (E value = 0) | 76 (0) | Chr. 9; microbody |
| AK106404 | Z. mays clone EL01N0511B03.d mRNA sequence (E value = 0) | 70 (0) | Chr.11; mitochondrial inner membrane; cytochrome P450 |
| AK071546 | Lolium rigidum Lol-5-v putative cytochrome P450 mRNA (E value = 1e-163) | 68 (0) | Chr. 4; mitochondrial inner membrane |
| AK111076 | Unknown expressed protein | 66 (2.6E-25) | Chr. 4; nucleus; none |
| AK073848 | O. sativa mRNA for OsNAC4 transcription factor (E value = 1e-174) | 64 (0) | Chr. 1; microbody |
| AK064287 | Z. mays clone EL01N0511B03.d mRNA sequence (E value = 1e-163) | 59 (0) | Chr. 12; endoplasmic reticulum; cytochrome P450 |
| AK101957 | Arabidopsis At2g46890 mRNA for unknown protein, clone: RAFL17-06-H20 (E value = 1e-100) | 59 (5.4E-19) | Chr. 4; endoplasmic reticulum; endomembrane system, integral to membrane |
| AK062882 | O. sativa AP2 domain-containing protein AP29 mRNA (E value = 2e-16) | 58 (0) | Chr. 8; nucleus; ethylene responsive element binding factor (1E-12) |
| AK067516 | Unknown expressed protein | 58 (1.1E-20) | Chr. 1; nucleus; none |
| AK063042 | Unknown expressed protein | 58 (0) | Chr. 3; nucleus; transcription factor |
| AK111091 | Unknown expressed protein | 57 (0) | Chr. 1; chloroplast stroma; none |
| Repressed | |||
| AK107088 | Arabidopsis At2g46930/F14M4.24 mRNA (E value = 1e-134) | −51 (8.5E-10) | Chr. 1; extracellular; pectin acetyl esterase |
| AK072459 | Malus domestica unknown mRNA (E value = 0) | −59 (8.9E-11) | Chr. 10; extracellular; methyl transferase |
| AK105875 | Unknown expressed protein | −69 (8.7E-19) | Chr. 1; nucleus; disease resistance kinase |
| AK065689 | Arabidopsis chloroplast carotenoid epsilon-ring hydroxylase (LUT1) mRNA, nuclear gene for chloroplast product (E value = 0) | −70 (2.2E-12) | Chr. 10; plasma membrane; cytochrome P450 |
| AK067229 | O. sativa alkaline α-galactosidase mRNA (E value = 0) | −72 (1.5E-8) | Chr. 8; endoplasmic reticulum |
| AK105369 | Unknown expressed protein | −72 (1.6E-10) | Chr. 7; cytoplasm; none |
| AK107138 | M. truncatula triacylglycerol/steryl ester lipase-like protein mRNA (E value = 1e-107) | −75 (1.7E-14) | Chr. 8; extracellular |
Rice gene names are from the KOME database (http://cdna01.dna.affrc.go.jp/cDNA/).
Chromosome number; PSORT localization prediction; function predicted by protein homology or Gene Ontology annotation designations in the KOME database.
The three putative MAPKKKs (Table 4; see Supplemental Figure 1D online) had homology to the NPK1-related protein kinase from Zea mays (Shou et al., 2004). NPK1, originally described from tobacco (Nicotiana tabacum), is involved in responses to abiotic stresses drought, cold, and high salt (Nakagami et al., 2005). Three more NPK-related kinases were upregulated between 44- and 26-fold (see Supplemental Table 2 online). By contrast, the better-studied rice MAPKKK Os ERD1 (AK111595), which plays a role in defense/stress signaling and development (Nakagami et al., 2005), was expressed in both samples at similar levels. The MAP kinases that have been characterized for response in blast disease were not highly induced in our study (Reyna and Yang, 2006). A wall-associated kinase (AK067041) and a leucine-rich repeat kinase (AK111536) that each showed the highest level of induction in a compatible blast interaction at 24 hpi (Vergne et al., 2007) were not differentially regulated in ours.
Additionally, a transcription factor in the Os DREB family (dehydration response element binding protein), which regulates genes expressed in response to drought, cold, and high salt (Dubouzet et al., 2003), was upregulated 75-fold in infected tissue (Table 4; see Supplemental Figure 1D online). The transcription factor Os NAC4 (AK073848), which plays a role in initiation of hypersensitive cell death induced by flagellin recognition in rice (Kaneda et al., 2007), was 64-fold upregulated (see Supplemental Figure 1D online). A transcription factor (AK062882) of the APETALA2/Ethylene-Responsive Element Binding Protein family was 58-fold upregulated. The rice transcription factor JAMyb (AK069082) previously associated with compatibility (Lee et al., 2001) was ninefold upregulated (P value = 0; see Supplemental Figure 1D online). Overall, these results are consistent with extensive reprogramming of rice cell processes during biotrophic invasion. Genes that are highly expressed during biotrophic rice invasion may be involved in effector-triggered susceptibility (Jones and Dangl, 2006).
Biotrophy-Associated Secreted Proteins Exhibit Different Localization Patterns in Planta
Properties of characterized AVR effectors suggest that additional effectors will be specifically expressed and secreted by IH. As the best candidates, we focused on putative secreted protein genes with >50-fold higher expression in IH (Table 2). With the notable exception of AVR-Pita1 (MGG_15370.6) and MGG_15371.6 (100-fold up), these genes were widely dispersed on the fungal chromosomes (Table 2). In strain 70-15, PWL2 and genes corresponding to the probe AMG08263 were duplicated at different chromosomal locations. Several genes encoded small, Cys-rich polypeptides, and most had no related M. oryzae genes or orthologs in other fungi. Interproscan analysis yielded no functional clues. Four genes validated as IH-specific by qRT-PCR (Table 1) and by RT-PCR (see Supplemental Figure 1C online) were chosen for promoter expression analysis (Figure 2), for secretion analysis (Figure 3; see Supplemental Figure 2 online), and for functional analysis (see Supplemental Figure 3 online). Fluorescence secretion patterns are based on microscopy visualization of >100 individual rice sheath infection sites per gene at each time point. All four genes were verified to encode biotrophy-associated secreted (BAS) proteins.
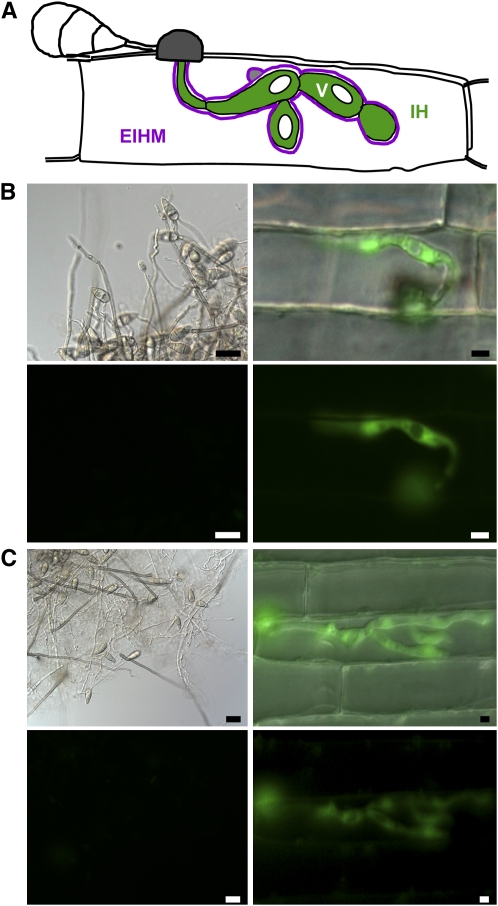
Figure 2.
Fungal Promoters from Upregulated Genes Are Specifically Expressed in IH.
(A) Illustration showing expected IH cytoplasmic expression pattern with EGFP excluded from vacuoles (V).
(B) In vitro and in planta expression of EGFP using BAS1 promoter region.
(C) In vitro and in planta expression of EGFP using BAS3 promoter region.
For (B) and (C), EGFP was expressed with a 1-kb promoter fragment from each gene. Left panels show phase contrast (top) and EGFP images (bottom, 3-s exposure) of mycelium, conidia, and conidiophores from agar plates (bars = 20 μm). Right panels show merged DIC and EGFP images (top) and the EGFP fluorescence alone (bottom, 3-s exposure) of IH inside rice sheath cells at 30 hpi (bars = 5 μm).
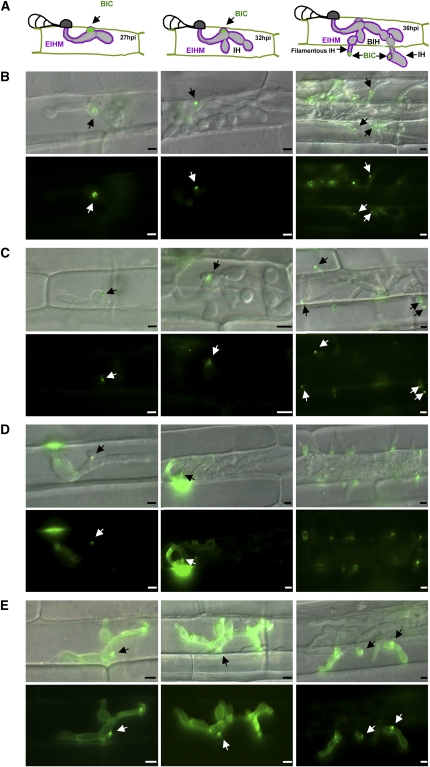
Figure 3.
M. oryzae BAS Proteins Are Secreted to Distinct Locations in Sheath Epidermal Cells.
(A) Illustrations showing the BIC localization pattern characteristic of AVR effectors at 27, 32, and 36 hpi. The first stage of BIC development, secretion into EIHM membranous caps at the tips of primary hyphae, is not represented. BIH, bulbous IH.
(B) to (E) The promoter and coding sequence for each BAS gene (BAS1-4) was cloned with EYFP as a C-terminal translational fusion. Fungal transformants expressing BAS:EYFP fusions were observed in susceptible YT16 rice. Merged DIC and EYFP images (top) and EYFP fluorescence alone (bottom) are shown. Time points are: 27 hpi (left), 32 hpi (middle), and 36 hpi (right). BICs are indicated by arrows. Bars = 5 μm.
(B) Secretion of BAS1:EYFP into BICs.
(C) Secretion of BAS2:EYFP into BICs.
(D) Secretion of BAS3:EYFP at 27 hpi. Note a faint BIC and fluorescence at the penetration site and outlining the primary hypha. At 32 hpi, multiple fluorescent spots were dispersed around the IH. At 36 hpi, fluorescence was near the cell wall crossing points and not in BICs at the filamentous hyphal tips.
(E) Secretion of BAS4:EYFP. Fluorescence uniformly outlined the IH. Some fluorescence was also observed in BICs.
BAS1 (MGG_04795.6)
BAS1 was 100-fold upregulated in IH (P value = 0). This gene on chromosome IV encodes a secreted protein with 115 amino acids, but no Cys residues (Table 2). No paralogs occur in genome version 6 (http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea/MultiHome.html), and no orthologs occur in other organisms. For functional analysis, we analyzed two independent knockout mutants of BAS1 for growth and sporulation in vitro, for appressorium formation and function, and for pathogenicity using the sheath assay, drop inoculation, and whole-plant infection assays. There was not a major pathogenicity phenotype, although three out of six independent whole-plant assays showed quantitative decreases in lesion numbers and lesion sizes in mutants relative to the wild-type strain and ectopic transformants. When transformants producing the BAS1:EYFP fusion protein invaded rice sheath cells, the fluorescence was secreted in a classical BIC pattern (Figure 3A). That is, EYFP fluorescence was first seen in BICs at primary hyphal tips and then moved with the BIC to the side of the first IH cell (Figure 3B, 27 hpi). Fluorescence remained in the BIC as IH continued to grow in the cell (Figure 3B, 32 hpi). Each biotrophic hypha that moved into a neighbor cell formed a secondary BIC, first at the filamentous IH tip and then beside the enlarged IH cell (Figure 3B, 36 hpi). To confirm secretion into BICs, we demonstrated that BAS1 fluorescent protein failed to colocalize with a cytoplasmic fluorescence reporter in IH but that it did colocalize with a fluorescent AVR-Pita1 reporter protein previously shown to localize to BICs (Figures 4A and 4B). Therefore, BAS1 encodes a small unique protein that is secreted into BICs.
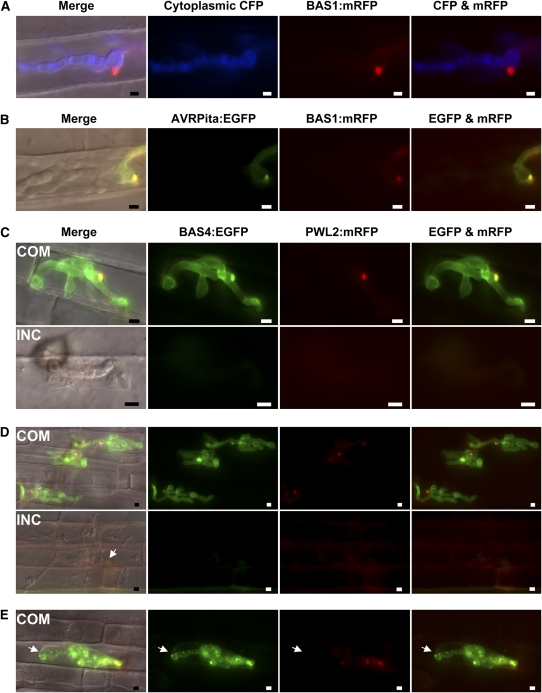
Figure 4.
Fluorescent Effector PWL2 and BAS Proteins Accumulate in Susceptible YT16 but Not in Resistant Yashiro-mochi Rice.
(A) BAS1:mRFP (red) is secreted into BICs and fails to colocalize with enhanced cyan fluorescent protein (CFP, blue) in the fungal cytoplasm (24 hpi in YT16). Exposure time for mRFP was 1 s and that for ECFP was 0.2 s. “Merge” corresponds to merged DIC, ECFP, and mRFP channels. Bars = 5 μm.
(B) Secreted BAS1:mRFP (red) colocalizes (yellow) with AVRPita:EGFP (green) in BICs and around BIC-associated hyphal cells (YT16 at 32 hpi). Exposure times for mRFP and EGFP are 2 s. “Merge” shows DIC, EGFP, and mRFP together. Bars = 5 μm.
(C) to (E) Transformants of strain O-137 (AVR-Pita1+) expressing PWL2:mRFP (red) and BAS4:EGFP (green) were inoculated on YT16 rice lacking Pi-ta (compatible [COM]) and on Yashiro-mochi containing Pi-ta (incompatible [INC]). “Merge” is DIC, EGFP, and mRFP channels together. Rice cells in (D) and (E) were plasmolyzed in 0.75 M sucrose. Exposure times: 1.5 s for mRFP and EGFP. Bars = 5 μm.
(C) A COM infection site at 24 hpi has an apparently healthy IH secreting PWL2:mRFP into BICs and BAS4:EGFP around the IH. An INC site in the same experiment has a short hypha in an unhealthy rice cell. Neither fluorescent protein was observed, even with longer exposure times to detect faint signals.
(D) In three COM infection sites (33 to 34 hpi), IH in plasmolyzed rice cells were secreting both PWL2:mRFP and BAS4:EGFP. In an INC site (same experiment), thin hyphae from the first-invaded cell (arrow) had moved to neighbor cells. Cytoplasm in all invaded cells was severely disrupted. Faint PWL2:mRFP fluorescence was observed, but BAS4:EGFP was not.
(E) At a COM infection site (33 to 34 hpi), an IH in a plasmolyzed rice cell secreted PWL2:mRFP and BAS4:EGFP, but showed nonuniform BAS4:EGFP labeling around the IH and weak mRFP fluorescence in the BIC. BAS4:EGFP appeared to have spilled into the surrounding rice cytoplasm (arrow).
BAS2 (MGG_09693.6)
This gene was 84-fold upregulated in IH (P value = 0). BAS2 on chromosome V encodes a conserved hypothetical protein with 102 amino acids. The encoded protein has similarity to proteins from two M. oryzae genes that are located ∼800 kb apart on chromosome III. One of these, MGG_07969.6, encodes a potentially secreted, conserved hypothetical protein with 101 amino acids (E = 2e-28). The other homolog, MGG_07749.6, encodes a larger, secreted protein, 198 amino acids, with homology at the N terminus (E = 4e-17). Both related genes have corresponding EST and SAGE hits, and they have fold-change levels between 1 and 2 in our analysis, suggesting they are expressed in fungal cell types other than IH. The BAS2 protein has homology to predicted proteins from other fungal pathogens with the highest level to a predicted protein from the wheat tan spot pathogen Pyrenophora tritici-repentis (E = 4e-21). BAS2 and all related polypeptides have six conserved Cys residues. Gene replacement analysis failed to show phenotypes in pathogenicity, mycelial growth, sporulation, or appressorium formation and function. BAS2 is a small Cys-rich secreted protein that localizes preferentially to BICs (Figure 3C).
BAS3 (MGG_11610.6)
This gene was 71-fold upregulated in IH (P value = 0). The gene resides on chromosome VI and encodes a secreted protein with 113 amino acids, including 10 Cys residues. BAS3 has two distant relatives in the blast genome. The closest of these, MGG_05895, which is not in genome version 6, has conserved Cys residues (E = 2e-05), and it was 25-fold upregulated in IH. Gene replacement functional analysis failed to identify an associated phenotype. Microscopy of the secreted BAS3:EYFP protein showed strong fluorescence at the appressorial penetration site and outlining the primary hyphae (Figure 3D). Low levels of fluorescence were observed in BICs in first-invaded cells (Figure 3D). After the IH had grown for some time in the cell, fluorescence faintly outlined the IH and formed focused fluorescent spots associated with individual IH cells (Figure 3D, 32 hpi). Fluorescent BICs were not observed at the tips of IH that invaded neighbor cells (Figure 3D, 36 hpi). Instead, fluorescence accumulated at the point where each IH had crossed the cell wall and sometimes surrounded the IH where they had crossed. We conclude that BAS3 is a small Cys-rich secreted protein with a localization pattern suggestive of a function in rice cell wall crossings.
BAS4 (MGG_10914.6)
This gene, 61-fold upregulated in IH (P value = 0), resides on chromosome VI (∼200 kb from one end) and encodes a secreted protein with 102 amino acids, including eight Cys residues. The BAS4 protein has homology to a conserved hypothetical M. oryzae protein, MGG_02154.6 (E = 1.9e-7), which is also a small secreted protein with eight conserved Cys residues. This latter gene is on chromosome II, ∼1139 kb from one end, and shows threefold higher expression in IH. The MGG_02154.6 protein has homology to proteins from other filamentous fungi, with the highest similarity to a small Cys-rich protein from the wheat scab pathogen, Gibberella zeae (E = 9e-24). BAS4 is quite diverged from the G. zeae protein. Thus, the MGG_02154.6 protein is more diverged from its IH-specific paralog BAS4 than from its orthologs in other filamentous fungi. BAS4 showed a secretion pattern that was clearly distinct from those of the other proteins (Figure 3E). Bright fluorescence uniformly outlined the IH that were actively growing in a rice cell. Fluorescence also occurred in BICs but not preferentially there, as seen for known AVR effectors. Gene replacement mutants have not yet been obtained for this gene. However, BAS4 appears to encode a small Cys-rich interfacial matrix protein.
BAS Protein Accumulation in Compatible and Incompatible Interactions
Using fungal transformants containing a native AVR-Pita1 gene, we compared secretion of the known effector PWL2 and the BAS1, BAS3, and BAS4 proteins in susceptible YT16 (pi-ta−) and in resistant Yashiro-mochi (Pi-ta+) rice. In the compatible interaction with YT16, high levels of PWL2 and BAS protein fluorescence occurred in 98% of infection sites, with >300 infection sites examined per protein. By contrast, in the incompatible interaction with Yashiro-mochi, fluorescence from PWL2 and the BAS proteins was absent or, at 10% of ∼50 infection sites per protein, severely attenuated. Typical results are illustrated for a transformant expressing both PWL2 fused to monomeric red fluorescent protein (PWL2:mRFP) and BAS4 fused to enhanced green fluorescent protein (BAS4:EGFP). Both fluorescent proteins were highly expressed at infection sites (>100 observed) in the compatible interaction (Figure 4C). However, in incompatible sites (>50 observed), the fungus generally stopped growing in the first-invaded cell, and neither mRFP nor EGFP fluorescence was observed (Figure 4C). Occasionally in the incompatible interaction, faint PWL2:mRFP fluorescence occurred without BAS4:EGFP fluorescence. BAS4:EGFP fluorescence alone was never observed. In one example, thin hyphae in damaged first-invaded Yashiro-mochi rice cells had spread to neighboring cells that also appeared damaged (Figure 4D). Only weak PWL2 fluorescence was observed. Thin hyphae in dying plant cells were never outlined with BAS4 reporter protein. Secretion of low levels of effector proteins in incompatible infection sites is consistent with the role of effectors in inducing R gene–mediated resistance. Clearly, BAS protein accumulation is characteristic of IH during compatible, but not during incompatible interactions.
Although IH exhibited bright BAS4:EGFP fluorescence at most compatible infection sites, IH in some cells lacked uniform outlining (Figure 4E). At these infection sites, the host cells appeared less healthy and many failed to plasmolyze. Additionally, fluorescent protein appeared to have leaked into the host cell. For example (Figure 4E), an IH in a plasmolyzed host cell lacked uniform outlining, and EGFP fluorescence was observed in the host cytoplasm. Apparently, EGFP had leaked from the EIHM matrix. These results indicated that not all IH in the compatible interaction were healthy. We conclude that BAS4 reporter provides a valuable indicator of EIHM integrity at individual infection sites during live cell imaging of biotrophic invasion.
DISCUSSION
An in-Depth View of the Blast Biotrophic Interaction Transcriptome
Biotrophic colonization of plant cells is barely explored at the molecular level for either the pathogen or the host. We report a procedure for purification of infected tissue RNA that is highly enriched for RNAs from biotrophic IH and from invaded rice cells or their immediate neighbors. Using this procedure, we identified 262 fungal genes and 210 rice genes that are ≥10-fold induced as IH are colonizing susceptible rice cells and secreting AVR effectors into BICs. The following results confirm that we have gained new insight on fungal and host gene expression during biotrophic invasion. First, known blast AVR genes and many M. oryzae genes previously only predicted by genome sequencing were highly expressed in our samples (Figure 1B; see Supplemental Table 1B online), and many genes previously identified from in vitro–grown fungal cell types were not expressed (Figure 1C; see Supplemental Table 1A online). Second, the hydrophobin gene MPG1, the most highly expressed fungal gene from other large-scale in planta expression analyses (Kim et al., 2001; Rauyaree et al., 2001; Matsumura et al., 2003), was downregulated in ours (see Supplemental Tables 1A and 1C online). Our results were consistent with the original report that MPG1 expression was high before penetration and at the later colonization stage when symptoms developed (Talbot et al., 1993). Third, our biotrophic fungal expression profile is quite distinct from the expression profiles corresponding to proposed in vitro pathogenicity models, growth on nitrogen-deficient medium or on medium with isolated rice cell walls. Fourth, on the host side, certain rice kinase and transcription factor genes were highly upregulated (≥50-fold) in tissue enriched for IH compared with expression levels for rice basal defense response genes (Table 4; see Supplemental Tables 1A and 1C online). In previous studies of compatible and incompatible blast interactions, the most highly induced host genes were in the defense response gene category (Kim et al., 2001; Rauyaree et al., 2001; Matsumura et al., 2003; Jantasuriyarat et al., 2005; Vergne et al., 2007). Together, these results confirm the importance of our enrichment strategy and the unique insights on IH-specific invasion this enrichment has allowed.
Our transcriptome analysis provides an in-depth molecular view of pathogen and host gene expression in the same biological system recently used to understand the cellular invasion strategies of the fungus (Kankanala et al., 2007). The lack of induction of plant cell wall degrading enzymes in IH was consistent with biotrophic IH appearing to cross plant cell walls at pit fields and with the general lack of visible degradation of plant cell walls during this early stage (Kankanala et al., 2007). Dramatically lower expression of melanin biosynthesis genes by IH (see Supplemental Tables 1A and 1C online) was consistent with reports that melanin-deficient IH colonize rice cells normally after entering the plant through damaged cuticle (Kankanala et al., 2007). The dramatic downregulation of the alternative oxidase gene AOX in IH (see Supplemental Table 1C online) was consistent with reports that AOX is repressed during invasive growth and that it may only play a role during oxidative stress generated by respiration-inhibitor fungicides (Avilla-Adams and Köller, 2002, and references therein). Clearly, our data provide entry into many unknown molecular mechanisms behind the cellular biology of biotrophic blast invasion.
On the host side, we have identified candidates for future investigations of host effector-triggered susceptibility genes that are recruited to promote success of the pathogen. Association of drought-response genes, the NPK1-related MAPKKK and Os DREB genes, with biotrophic invasion is interesting in the context of the field biology of blast disease. That is, drought stress and cold stress make rice more susceptible to blast (Kawasaki, 2004; Koga et al., 2004a). These results are consistent with a report that the rice MAP kinase Os MAPK5 positively regulates drought, salt, and cold tolerance and negatively regulates PR gene expression and resistance to blast (Xiong and Yang, 2003). Understanding the role of the drought-associated MAPKKK and Os DREB in biotrophic invasion takes on practical significance because both classes of genes are being used in transgenic strategies to confer drought resistance in rice and maize (Dubouzet et al., 2003; Shou et al., 2004).
Biotrophy-Associated Secreted Proteins in Rice Blast Disease
Half of the M. oryzae genes that are ≥50-fold upregulated in IH (Figure 1B) and nearly a quarter of the genes that are ≥10-fold upregulated encode putative secreted proteins (Figure 1B, Table 2). This contrasts with an average 9% of genes encoding putative secreted proteins in the fungal genome (Figure 1B). Enrichment for secreted protein genes in the biotrophic interaction transcriptome is consistent with the recent report that mutation of the M. oryzae endoplasmic reticulum chaperone gene LHS1 disproportionately impacts biotrophic invasion in the compatible interaction, as well as induction of plant defenses in the incompatible interaction (Yi et al., 2009). The M. oryzae LHS1 chaperone is required for proper processing and secretion of pathogen proteins. Mutants that lack LHS1 grow normally in axenic culture and on the plant surface, but they are severely impaired in biotrophic colonization and sporulation. Consistent with the report of Yi et al. (2009) that proper secretion is required for the AVR-Pita1–mediated incompatible interaction, we found that secretion of low levels of fluorescent effector/BAS proteins can be detected even in the incompatible interaction (Figure 4D). These results highlight the importance of studying the proteins that IH secrete in planta as well as the secretion mechanisms involved.
Our results have greatly increased the number of IH-specific, secreted (BAS) proteins identified for rice blast, and many of these are small and Cys rich. Secreted Cys-rich polypeptides have been identified as effectors in other host-pathogen systems (Catanzariti et al., 2006; Kamoun 2007). For rice blast, Dean et al. (2005) identified three Cys-rich protein families in the M. oryzae genome. Although it was suggested that these might play a role as blast effectors, functional analyses had not been performed. Among these, family 180, including 10 Cys-rich polypeptides (with ∼150 amino acids and 6 to 10 Cys residues) had five members that were upregulated in IH in our analysis, by 3-, 5-, 5-, 5-, and 25-fold. The BAS2, BAS3, and BAS4 proteins belong in family 180, although they were not included among the original members (Dean et al., 2005). No previously reported members of the two larger families of Cys-rich proteins (180 to 220 amino acids) were differentially expressed in our study, although one putative BAS protein, MGG_08657.6 (Table 2), belongs in this group.
Although it is likely that BAS proteins play key roles during biotrophic invasion, targeted gene replacements of BAS1, BAS2, and BAS3 did not result in reproducible pathogenicity phenotypes. It remains possible that there are minor plant colonization phenotypes, especially with BAS1, that could be confirmed with improvements in blast pathogenicity assays, which are extremely sensitive to environmental and physiological conditions. However, the lack of phenotypes is consistent with the general lack of phenotypes associated with known AVR effector genes (Kamoun, 2007) and with the general failure to identify genes with IH-specific phenotypes through classical mutational analyses (Talbot, 2003; Ebbole 2007). Together, these results suggest extensive functional redundancy associated with biotrophic invasion. Among these BAS genes (Table 2), BAS1 has no candidate for a functional paralog in the sequenced genome, BAS2 has two candidates (neither gene is upregulated in IH), and BAS3 has one candidate (subsequently deleted from genome version 6). So, in some cases, assaying double or triple mutants might provide clues to function. Additional insight to BAS protein function could come from determining if IH-specific proteins interact physically with induced rice proteins or, as we demonstrate, from studying in planta secretion patterns.
The role that BAS4 plays in the EIHM matrix is unknown. However, our finding that BAS4:EGFP uniformly outlines growing IH confirms the report by Kankanala et al. (2007) that IH are sealed within a plant membrane compartment inside living host cells. We have not observed BAS4:EGFP outlining of hyphae in the incompatible interaction (Figure 4D). Therefore, BAS4 expression might be repressed resulting from recognition of AVR-Pita1, BAS4 might be expressed but not concentrated inside the EIHM, or both. We suggest that uniform outlining of IH as they grow in a living cell indicates that healthy IH inside an EIHM compartment have established a successful biotrophic interaction. This information is very important for cell biological studies of biotrophic invasion. For example, our results on leaking of BAS4 (Figure 4E) raise an important caution to researchers studying translocation of blast effectors into the host cytoplasm. Breakage of the EIHM membrane enclosing the fungus would result in artifactual dispersal of EIHM matrix proteins to the surrounding rice cytoplasm. Effector translocation studies in blast disease must account for EIHM integrity at individual infection sites, and fluorescent BAS4 reporter proteins provide a means to do this.
The hypothesis that blast IH co-opt host plasmodesmata for crossing to living neighbor cells (Kankanala et al., 2007) could be confirmed by identification of fungal proteins involved in recognizing and manipulating plasmodesmata. Fluorescent BAS3 reporter protein accumulates at rice wall crossing points, suggesting that BAS3 might play a role in cell-to-cell movement of IH. However, deletion of the BAS3 gene produced no phenotype to aid in discovery of its function. Identification of additional proteins with the BAS3 localization pattern is a high priority. Deletion of two or more components participating in the same cellular process might produce a dramatic phenotype, even though the fungus grows normally after mutation of each component individually (Tong et al., 2001). High-throughput in planta secretion analysis for all M. oryzae secreted proteins is underway (M.L. Farman, M. Goodin, and B. Valent, unpublished data). These experiments should identify additional BAS proteins that localize at cell wall crossing points.
BAS Proteins as Putative Effectors
The known AVR gene products AVR-Pita1, PWL1, and PWL2 are BAS proteins, suggesting that BAS proteins are a rich source for additional effectors. Identifying the entire set of rice blast effectors, including the subset of AVR effectors corresponding to rice blast resistance genes, remains an important challenge. Presently, the few known blast AVR effectors have not provided bioinformatic handles for identifying effector candidates among secreted protein genes in the M. oryzae genome. Clues might have come from identification of large effector gene families as seen with oomycetes (Kamoun, 2007) or from genome clusters of in planta–specific, secreted protein genes as seen for the maize smut pathogen Ustilago maydis (Kämper et al., 2006). Neither strategy has proven useful from our analysis. Additional blast cytoplasmic effectors could be identified through bioinformatics if they contained membrane translocation motifs, such as the RXLR motif found in oomycete effectors (Kamoun, 2007). Oomycete pathogens deliver their cytoplasmic effectors across plant membrane by a mechanism that requires this amino acid motif following the classical signal peptide (Whisson et al., 2007; Dou et al., 2008). We searched sequences for blast effectors and for proven and putative BAS proteins for potential amino acid translocation motifs, but without success.
Another strategy for bioinformatic identification of effector candidates involves identifying cis-elements in promoters mediating IH-specific transcription and use of these elements for identifying additional coregulated genes. AVR-Pita1, PWL1, PWL2, and other BAS genes should share cis-elements for in planta–specific transcription factors. To discover these, we performed bioinformatic searches on the 500-bp upstream regions of effector and BAS genes using various programs, including MEME (http://meme.sdsc.edu/meme/meme-output-example.html), again without success. Further refinement of in planta time course studies might identify coordinately regulated gene clusters that would provide better groupings for promoter element searches. Indeed, reported accumulation of effectors in BICs would be consistent with highly regulated effector gene expression in the BIC-associated hyphal cells (R. Berruyer, C.H. Khang, P. Kankanala, S.Y. Park, K. Czymmek, S. Kang and B. Valent, unpublished data). Further refinement of in planta–specific gene expression is needed.
The discovery of BICs presents a third strategy for identifying effectors, based on the hypothesis that the specific BIC localization pattern is characteristic of effectors that are delivered to the host cytoplasm (R. Berruyer, C.H. Khang, P. Kankanala, S.Y. Park, K. Czymmek, S. Kang, and B. Valent, unpublished data). Among our four analyzed BAS proteins, BAS1 and BAS2 show the same BIC-specific localization pattern as known effectors. The BIC localization seen for BAS3 and BAS4 is probably passive compared with targeted localization for BAS1 and BAS2, since BICs appear to be expanded, differentiated regions within the EIHM matrix. BAS1 and BAS2 might represent structural BIC components, or they might represent candidate effectors that are translocated across the EIHM into the rice cell. Preliminary evidence suggests that, indeed, the effector protein that accumulates in BICs is translocated into the cytoplasm of living rice cells (C.H. Khang and B. Valent, unpublished data). Our analysis has identified many more potential players for continued investigation of roles for effectors and for BICs during biotrophic invasion.
METHODS
Preparation of Infected Tissue and Control Samples for Microarray Analysis
Magnaporthe oryzae strain KV1 (Kankanala et al., 2007) expressing constitutive EYFP was derived from O-137, a highly aggressive field isolate collected from rice (Oryza sativa) in China. The fungus was maintained in frozen storage and cultured on oatmeal agar plates at 24°C under continuous light (Valent et al., 1991). Rice sheath inoculations were performed as described (Kankanala et al., 2007). Briefly, 5-cm-long sheath pieces from 3-week-old plants were placed in Petri dishes with wet filter papers to maintain high humidity. Sheaths were placed in wire supports to avoid contact with the wet paper and to hold them horizontally flat for even inoculum distribution. A spore suspension (1 × 105 spores/mL in 0.25% gelatin; Sigma-Aldrich) was injected in one end of the sheath using a 1-mL pipette. At 36 hpi, 0.5-cm pieces were removed from the incubated sheath ends to eliminate fungus that grew into injured tissue. Each sheath segment was cleaned using a wet sterile swab to remove spores, appressoria, and mycelium on the surface. Each segment was trimmed and immediately scanned for infection site density using epifluorescence microscopy. Heavily infected samples were frozen in liquid nitrogen and stored at −80°C. Sheath pieces were processed one at a time to minimize time between trimming and freezing. For mock-inoculated controls, sheaths were inoculated with gelatin solution, incubated, and processed identically to inoculated pieces.
For preparation of mycelium for control samples, a 1-cm2 piece of agar containing KV1 mycelium was excised from the surface of an oatmeal agar plate, placed in 25 mL of 3,3,3 medium (3 g/L of glucose, 3 g/L of casamino acids, and 3 g/L of yeast extract), and fragmented in a blender. The suspension of mycelial fragments was mixed with 225 mL of fresh medium in a 500-mL flask and incubated at 24°C with continuous rotation (120 rpm). After 24 h, mycelium was collected by filtration and the blending treatment was repeated. After three rounds of growth, the mycelium was collected, dried with paper towels, and stored at −80°C for RNA extraction. For assessing the ratio of fungal-to-rice RNAs in the infected tissue by RT-PCR, control samples were prepared by mixing mycelial RNA with RNA from mock-inoculated rice plants to produce mixtures with 10, 20, 30, 40, and 50% fungal RNA.
RNA Extraction, cDNA Preparation, and RT-PCR
Total RNAs from mycelium and rice tissues were extracted using a Trizol method (Invitrogen). Briefly, 100 mg of tissue was ground using a mortar and pestle with liquid nitrogen, and the resulting powder was suspended in 1 mL of trizol. After 5 min of incubation, 0.2 mL of chloroform was added, and samples were mixed manually for 15 s and then incubated for 3 min. After centrifugation (rcf 9300g) for 15 min at 4°C, the aqueous layer was recovered and mixed with 0.25 mL of 3 M sodium acetate, pH 5.2, and 0.25 mL of isopropanol. A pellet was obtained by centrifugation and washed twice with 75% ethanol. RNA quantity was measured using a NanoDrop Spectrophotometer (Nano-Drop Technologies). For microarray hybridizations, RNA quality was determined using an Agilent Bioanalyzer (Agilent Technologies). To obtain control RNAs with similar fungal and plant content, we produced a mixture of 20% mycelial RNA and 80% mock-inoculated rice sheath RNA. Five hundred nanograms of total RNA were used for cDNA synthesis using the SuperScript first-strand kit (Invitrogen) according to manufacturer's instructions. For RT-PCR validation of differentially expressed genes, cDNAs from four sources, including mycelium, 36-hpi mock-inoculated sheaths, 36-hpi inoculated sheaths, and mycelium/mock mixtures, were used as templates for amplification. The quality of the cDNA was tested using M. oryzae actin primers that spanned an intron to differentiate cDNA from genomic sequences. The expected actin fragment was amplified from mycelium and infected tissue (see Supplemental Figure 1C online). Amplification was not seen in negative controls lacking reverse transcriptase (data not shown). Two microliters of cDNA were used for PCR amplification. Primers are listed in Supplemental Table 4 online. When possible, primers spanned introns to differentiate genomic and cDNAs. For comparison, 27 rounds of PCR amplification were used in all expression validation experiments (see Supplemental Figure 1 online).
Microarray Hybridization and Data Analysis
Total RNA (500 ng) was labeled using a Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies). Typical cRNA yields after one round of amplification were 10 to 15 μg. Briefly, first and second cDNA strands were synthesized using reverse transcriptase and a primer containing poly(dT) and T7 polymerase promoter sequences. The cRNAs were synthesized and labeled with Cyanine-3 (Cy3) CTP or Cyanine-5 (Cy5) CTP using T7 RNA polymerase according to the manufacturer's specifications. Fluorescent cRNAs were purified and quantified using a NanoDrop Spectrophotometer (NanoDrop Technologies), and 1-μg aliquots of the labeled cRNAs were hybridized to M. oryzae microarray slides, version 2 (G4137B, Agilent Technologies), and to rice microarray slides (G4138A, Agilent Technologies). For hybridization, the labeled samples (1 μg each Cy3- and Cy5-labeled cRNA) were fragmented by the addition of 25× Agilent Fragmentation buffer and incubated for 30 min at 60°C. The sample was adjusted to a final volume of 450 μL with formamide-containing hybridization buffer (Hughes et al., 2001) and then added to the microarray slides. Slides were incubated for 18 h with continuous rotation at 40°C. After hybridization, slides were washed in 6× SSPE (1 × SSPE is 0.15 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 0.005% sarcosyl for 1 min, in 0.06× SSPE for 30 s, in water for 30 s, and then air dried. Slides were scanned on an Agilent G2565BA DNA microarray scanner, and TIFF images were extracted using the Agilent Feature Extraction software (version 8.5). Resultant.xml and.jpeg files were imported into Rosetta Resolver software (Rosetta Biosoftware). Biological replicates (four hybridizations for each) were first analyzed separately to determine reproducibility in our biological process. To obtain the numbers reported in this article, all 12 data sets for each microarray were analyzed together. Signature sequences were identified by Resolver and exported to Excel. P values were calculated using the Rosetta Resolver error model (Weng et al., 2006). The Bonferroni correction for multiple comparisons (Sokal and Rohlf, 1981) was adopted for significance: P < 3.2 × 10−6 (rounded to 10−6) for M. oryzae genes on the microarray, and P < 2.2 × 10−6 (rounded to 10−6) for rice genes.
Quantitative Real-Time RT-PCR
cDNA was synthesized using 1 to 2 μg of total RNA extracted from infected tissue or mycelium grown for 2 weeks on oatmeal agar. Infected tissue and mycelium were processed similarly using the Trizol method (Invitrogen). As a control housekeeping gene, the M. oryzae actin gene (MGG_03982) was amplified using MgActinF 5′-AGCGTGGTATCCTCACTTTGC-3′ and MgActinR 5′-ATCTTCTCTCGGTTGGACTTGG-3′ primers. Primers AVR-PitaF 5′-TGCCCTCCTTTCTTCAACAAC-3′ and AVR-PitaR 5′-CCCATTCGTAACCATAATCTTTCC-3′ were used to amplify the M. oryzae AVR-Pita1 gene. Both primers and templates were first tested using a regular RT-PCR assay. Real-time RT-PCR was performed using the following protocol: Cycle 1(1×); step 1, 95.0°C for 05:00; Cycle 2 (40×); step 1, 95.0°C for 00:20; step 2, 54.0°C for 00:30; step 3, 72.0°C for 00:45. Data collection and real-time analysis were enabled. Cycle 3 (1×); step 1, 95.0°C for 01:00; Cycle 4 (1×); step 1, 55.0°C for 01:00; Cycle 5 (80×); step 1, 55.0°C for 00:10. Set point temperature was increased by 0.5°C after cycle 2. Each reaction was set to 25 μL of final volume containing 12.5 μL of 2X iQ SYBR Green Supermix, 1 μL of 10 μM of each primer, and 10.5 μL of cDNA. Four dilutions of all cDNAs samples were used to test primer efficiency with the housekeeping gene primers. Reactions were run in an iCycler machine (Bio-Rad). The sample with the lowest concentration (highest Ct value) was used to adjust the concentration of the other samples using the following formula: dilution factor = 2 − (CtA-CtB), where CtA is the Ct value of sample A, and CtB is the Ct value of the sample with the lowest concentration. Ct values reported are the mean of four measurements, two technical replications for each of two biological replications.
Vector Construction, Fungal Transformation, and DNA Gel Blot Analysis
To observe fungal cell type–specific expression, BAS promoters were subcloned in a transcriptional fusion with EGFP (effector promoter:EGFP). Unless noted otherwise, transformation cassettes to observe secretion in rice cells were constructed containing the entire protein coding sequence (including the predicted signal peptide) with its native promoter (1 kb) in a translational fusion with EYFP, EGFP, or mRFP. The constructs for AVR-Pita:EGFP used in Figure 4B and BAS4:EGFP used in Figures 4C to 4E contained only the native promoter and signal peptide fused to EGFP. The EGFP expressed from both constructs behaved identically to EGFP expressed from constructs including the entire AVR-Pita and BAS4 coding sequences. The EGFP, EYFP, and ECFP genes were obtained from Clontech, and the mRFP gene was from Campbell et al. (2002). Primers used for amplification of each gene are listed in Supplemental Table 4 online. Each cassette was cloned into pBHt2 binary vector for transformation by Agrobacterium tumefaciens (Khang et al., 2005), with selection for hygromycin resistance. Details of plasmid construction and corresponding fungal transformants used in this study are listed in Supplemental Tables 5 and 6 (Borett et al., 2002) online, respectively. M. oryzae field isolates O-137 (Orbach et al., 2000), Guy11 (Leung et al., 1988), and laboratory strain CP987 (Orbach et al., 2000) were used as recipients, and 10 to 12 independent transformants were analyzed per gene. For gene replacement transformation, cassettes were constructed by amplifying ∼1.0 kb of 5′- and 3′-flanking regions for each predicted coding sequence. The hygromycin phosphotransferase (hph) gene from pCSN43 (Sweigard et al., 1995) was cloned between the two flanking regions using a fusion PCR strategy. The three pieces together were cloned first into the pGEMT-T vector (Promega) for sequence analysis and later into binary vector pGKO2 (Khang et al., 2005) using a restriction ligation strategy. KV1 spores were transformed using A. tumefaciens (Khang et al., 2005). After two rounds of selection in TB3 media containing 250 μg/mL of hygromycin, 50 to 150 independent fungal transformants were analyzed for gene replacement events by PCR amplification. Those that showed no amplification of the coding sequence were further tested for presence of the hygromycin resistance gene using hph-specific primers. Gene replacement events were confirmed by DNA gel blot analysis using the AlkPhos Direct Labeling Kit for nonradioactive labeling of DNA probes (Amersham RPN3690).
Assays for Growth, Sporulation, Appressorium Formation, and Plant Infection
Fungal growth and sporulation was observed on oatmeal agar plates (Valent et al., 1991). Appressorium formation, penetration, and biotrophic invasion were observed in the leaf sheath assay described above. Drop inoculation and whole-plant spray inoculation assays have been described (Berruyer et al., 2006). For whole-plant assays, 3-week-old YT16 rice plants were inoculated with spore suspensions (5 × 104 spores/mL in 0.25% gelatin) and evaluated 7 d later (Valent et al., 1991).
Microscopy
DIC and epifluorescence imaging was performed with a Zeiss Axioplan 2 IE MOT microscope and a 63X (numerical aperture 1.2) C-Apochromat water immersion objective. Fluorescence was observed with a 100 Watt FluoArc or a X-Cite 120 (EXFO Life Sciences) mercury lamp source. Filter sets used were as follows: GFP (excitation 480 ± 10 nm, emission 510 ± 10 nm, filter set 41020; Chroma Technology); YFP (excitation 500 ± 12.5 nm, emission 535 ± 15 nm, filter set 46HE); mRFP (excitation 535 ± 25 nm, emission 610 ± 32 1/2 nm); and ECFP (excitation 436 ± 10 nm, emission 480 ± 20 nm, filter set 47). Images of fungus in vitro were obtained using an EC Plan Neofluar ×40/0.75 Ph2 objective. Images were obtained with an Axiocam HRc camera and Axiovision software version 3.1. Unless stated otherwise, microscopy components were obtained from Carl Zeiss. Viability of infected cells was assessed by plasmolysis in 0.75 M sucrose.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: AF207841.1 for AVR-Pita, U26313.1 for PWL2, U36923.1 for PWL1, FJ807764 for BAS1, FJ807765 for BAS2, FJ807766 for BAS3, FJ807767 for BAS4, and XM_361508 for actin. Accession numbers for known genes shown to be differentially expressed are noted in the Supplemental Tables online. Data sets for the M. oryzae and rice microarrays can be accessed through NCBI GEO superSeries accession number GSE8670 (www.ncbi.nlm.nih.gov/geo). All M. oryzae genes can be accessed from the Broad Institute's Magnaporthe grisea Genome Database (http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea/MultiHome.html) or the MGOS Database (http://www.mgosdb.org/; Soderlund et al., 2006). The rice genes can be accessed from the KOME database (http://cdna01.dna.affrc.go.jp/cDNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Experiments to Assess Fungal RNA Content in Infected Tissue Samples and to Confirm Invasion-Specific Expression of Fungal and Rice Genes.
Supplemental Figure 2. Fluorescence Expression Patterns Are Conserved for Independent Fungal Transformants.
Supplemental Figure 3. Gene Replacement Analysis of BAS2.
Supplemental Table 1. M. oryzae Upregulated Genes, Down-regulated Genes, and Known Pathogenicity Genes.
Supplemental Table 2. Rice Gene Categories with Members That Were Up- or Downregulated More Than Threefold.
Supplemental Table 3. Comparison of Rice Gene Expression during Biotrophic Invasion in Our Study with Expression during Wounding in the study of Katou et al. (2007).
Supplemental Table 4. Primers Used in This Study.
Supplemental Table 5. Plasmids Used in This Study.
Supplemental Table 6. Fungal Transformants Used in This Study.
Supplementary Material
Acknowledgments
We acknowledge Melinda Dalby for excellent technical assistance. We thank X. Tang and F. White (Kansas State University) for valuable comments on the manuscript and P. Kankanala, G. Valdovinos, and M. Dalby for useful discussions and criticism. We thank K. Greer and C. Soderlund (University of Arizona) for comments and help with microarray data publication. This material is based upon work supported by the National Science Foundation under Grant 0446315 and by US Department of Agriculture-National Research Initiative Grant 2006-35319-17296. Additional support came from Kansas National Science Foundation Experimental Program to Stimulate Competitive Research/Kansas Technology Enterprise Corporation. This is contribution 07-276J from the Kansas State University Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Barbara Valent (bvalent@ksu.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Avilla-Adams, C., and Köller, W. (2002). Disruption of the alternative oxidase gene in Magnaporthe grisea and its impact on host infection. Mol. Plant Microbe Interact. 15 493–500. [DOI] [PubMed] [Google Scholar]
- Berruyer, R., Poussier, S., Kankanala, P., Mosquera, G., and Valent, B. (2006). Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96 346–355. [DOI] [PubMed] [Google Scholar]
- Böhnert, H.U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J.-L., and Lebrun, M.-H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourett, T.M., Sweigard, J.A., Czymmek, K.J., Carroll, A., and Howard, R.J. (2002). Reef coral fluorescent proteins for visulaizing fungal pathogens. Fungal Genet. Biol. 37 211–220. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.-M., Dodds, P.N., Lawrence, G.J., Ayliffe, M.A., and Ellis, J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch, B.C., Fudal, I., Lebrun, M.-H., Tharreau, D., Valent, B., van Kim, P., Notteghem, J.-L., and Kohn, L.M. (2005). Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 980–986. [DOI] [PubMed] [Google Scholar]
- Donofrio, N.M., Oh, Y., Lundy, R., Pan, H., Brown, D.E., Jeong, J.S., Coughlan, S., Mitchell, T.K., and Dean, R.A. (2006). Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 43 605–617. [DOI] [PubMed] [Google Scholar]
- Dou, D., Kale, S.D., Wang, X., Jiang, R.H.Y., Bruce, N.A., Arredondo, F.D., Zhang, X., and Tyler, B.M. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet, J.G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E.G., Miura, S., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activatiors that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. [DOI] [PubMed] [Google Scholar]
- Ebbole, D.J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45 437–456. [DOI] [PubMed] [Google Scholar]
- Ebbole, D.J., Jin, Y., Thon, M., Pan, H., Bhattarai, E., Thomas, T., and Dean, R. (2004). Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: Analysis of expressed sequence tags. Mol. Plant Microbe Interact. 17 1337–1347. [DOI] [PubMed] [Google Scholar]
- Gowda, M., et al. (2006). Deep and comparative analysis of the mycelium and appressorium transcriptomes of Magnaporthe grisea using MPSS, RL-SAGE, and oligoarray methods. BMC Genomics 7 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, R.J., and Valent, B. (1996). Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50 491–512. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., et al. (2001). Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19 342–347. [DOI] [PubMed] [Google Scholar]
- Irie, T., Matsumura, H., Terauchi, R., and Saitoh, H. (2003). Serial analysis of gene expression (SAGE) of Magnaporthe grisea: Genes involved in appressorium formation. Mol. Genet. Genomics 270 181–189. [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat, C., et al. (2005). Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol. 138 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2007). Groovy times: Filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10 358–365. [DOI] [PubMed] [Google Scholar]
- Kämper, J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444 97–101. [DOI] [PubMed] [Google Scholar]
- Kaneda, T., Fujiwara, S., Takai, R., Takayama, S., Isogai, A., and Che, F.-S. (2007). Identification of genes involved in induction of plant hypersensitive cell death. Plant Biotechnol. 24 191–200. [Google Scholar]
- Kang, S., Sweigard, J.A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8 939–948. [DOI] [PubMed] [Google Scholar]
- Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou, S., Kuroda, K., Seo, S., Yanagawa, Y., Tsuge, T., Yamazaki, M., Miyao, A., Hirochika, H., and Ohashi, Y. (2007). A calmodulin-binding mitogen-activated protein kinase phosphatase is induced by wounding and regulates the activities of stress-related mitogen-activated protein kinases in rice. Plant Cell Physiol. 46 332–344. [DOI] [PubMed] [Google Scholar]
- Kawasaki, S. (2004). Proceedings of the 3rd International Rice Blast Conference: Rice Blast: Interaction with Rice and Control. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Khang, C.H., Park, S.-Y., Lee, Y.-H., and Kang, S. (2005). A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet. Biol. 42 483–492. [DOI] [PubMed] [Google Scholar]
- Khang, C.H., Park, S.-Y., Lee, Y.-H., Valent, B., and Kang, S. (2008). Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant Microbe Interact. 21 658–670. [DOI] [PubMed] [Google Scholar]
- Kim, S., Il-Pyung, A., and Lee, Y.-H. (2001). Analysis of genes expressed during rice-Magnaporthe grisea interactions. Mol. Plant Microbe Interact. 14 1340–1346. [DOI] [PubMed] [Google Scholar]
- Koga, H., Dohi, K., and Mori, M. (2004. a). Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiol. Mol. Plant Pathol. 65 3–9. [Google Scholar]
- Koga, H., Dohi, K., Nakayachi, O., and Mori, M. (2004. b). A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol. Mol. Plant Pathol. 64 67–72. [Google Scholar]
- Lee, M.-W., Qi, M., and Yang, Y. (2001). A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant Microbe Interact. 14 527–535. [DOI] [PubMed] [Google Scholar]
- Leung, H., Borromeo, S., Bernardo, M.A., and Notteghem, J.L. (1988). Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78 1227–1233. [Google Scholar]
- Li, L., Ding, S.-L., Sharon, A., Orbach, M., and Xu, J.-R. (2007). Mir1 is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol. Plant Microbe Interact. 20 448–458. [DOI] [PubMed] [Google Scholar]
- Matsumura, H., Reich, S., Ito, A., Saitoh, H., Kamoun, S., Winter, P., Kahl, G., Reuter, M., Kruger, D.H., and Terauchi, R. (2003). Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc. Natl. Acad. Sci. USA 100 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami, H., Pitzschke, A., and Hirt, H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10 339–346. [DOI] [PubMed] [Google Scholar]
- Oh, Y., Donofrio, N., Pan, H., Coughlan, S., Brown, D., Meng, S., Mitchell, T., and Dean, R. (2008). Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 9 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach, M.J., Farrall, L., Sweigard, J.A., Chumley, F.G., and Valent, B. (2000). The fungal avirulence gene AVR-Pita determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyyala, R., and Farman, M.L. (2006). Magnaporthe oryzae isolates causing gray leaf spot of perennial ryegrass possess a functional copy of the AVR1-CO39 avirulence gene. Mol. Plant Pathol. 7 157–165. [DOI] [PubMed] [Google Scholar]
- Rauyaree, P., Choi, W., Fang, E., Blackmon, B., and Dean, R. (2001). Genes expressed during early stages of rice infection with the rice blast fungus Magnaporthe grisea. Mol. Plant Pathol. 2 347–354. [DOI] [PubMed] [Google Scholar]
- Reyna, N.S., and Yang, Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 19 530–540. [DOI] [PubMed] [Google Scholar]
- Ribot, C., Hirsch, J., Balzergue, S., Tharreau, D., Nottéghem, J.-L., Lebrun, M.-H., and Morel, J.-B. (2008). Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 165 114–124. [DOI] [PubMed] [Google Scholar]
- Shou, H., Bordallo, P., and Wang, K. (2004). Expression of the Nicotiana protein kinase NPK1 enhanced drought tolerance in transgenic maize. J. Exp. Bot. 55 1013–1019. [DOI] [PubMed] [Google Scholar]
- Soanes, D.M., Kershaw, M.J., Cooley, R.N., and Talbot, N.J. (2002). Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15 1253–1267. [DOI] [PubMed] [Google Scholar]
- Soanes, D.M., and Talbot, N.J. (2005). A bioinformatic tool for analysis of EST transcript abundance during infection-related development by Magnporthe grisea. Mol. Plant Pathol. 6 503–512. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Haller, K., Pampanwar, V., Ebbole, D., Farman, M., Orbach, M., Wang, G.-L., Wing, R., Xu, J.-R., Brown, D., Mitchell, T., and Dean, R. (2006). MGOS: A resource for studying Magnaporthe grisea and Oryza sativa interactions. Mol. Plant Microbe Interact. 19 1055–1061. [DOI] [PubMed] [Google Scholar]
- Sokal, R.R., and Rohlf, F.J. (1981). Biometry. (New York: W.H. Freeman).
- Song, F., and Goodman, R.M. (2001). Molecular biology of disease resistance in rice. Physiol. Mol. Plant Pathol. 59 1–11. [Google Scholar]
- Sweigard, J.A., Carroll, A.M., Kang, S., Farrall, L., Chumley, F.G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, Y., Choi, W., Mitchell, T.K., Okuno, T., and Dean, R.A. (2003). Large scale parallel analysis of gene expression during infection-related morphogenesis of Magnaporthe grisea. Mol. Plant Pathol. 4 337–346. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57 177–202. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J., Ebbole, D.J., and Hamer, J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.-S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H.Y., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- Valent, B., Farrall, L., and Chumley, F.G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C., Rep, M., and Pieterse, C.M.J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162. [DOI] [PubMed] [Google Scholar]
- Vergne, E., Ballini, E., Marques, S., Sidi Mammar, B., Droc, G., Gaillard, S., Bourot, S., DeRose, R., Tharreau, D., Notteghem, J.-L., and Morel, J.-B. (2007). Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytol. 174 159–171. [DOI] [PubMed] [Google Scholar]
- Weng, L., Dai, H., Zhan, Y., He, Y., Stepaniants, S.B., and Bassett, D.E., Jr. (2006). Rosetta error model for gene expression analysis. Bioinformatics 22 1111–1121. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450 115–118. [DOI] [PubMed] [Google Scholar]
- Wu, S.-C., Halley, J.E., Luttig, C., Fernekes, L.M., Gutiérrez-Sanchez, G., Darvill, A.G., and Albersheim, P. (2006). Identification of an endo-beta-1,4-D-xylanase from Magnaporthe grisea by gene knockout analysis, purification and heterologous expression. Appl. Environ. Microbiol. 72 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Yang, Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, M., Chi, M.-H., Khang, C.H., Park, S.-Y., Kang, S., Valent, B., and Lee, Y.-H. (February 27, 2009). The ER chaperone MoLHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Mehrabi, R., and Xu, J.-R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.