Abstract
Plant epidermal cells dedicate more than half of their lipid metabolism to the synthesis of cuticular lipids, which seal and protect the plant shoot. The cuticle is made up of a cutin polymer and waxes, diverse hydrophobic compounds including very-long-chain fatty acids and their derivatives. How such hydrophobic compounds are exported to the cuticle, especially through the hydrophilic plant cell wall, is not known. By performing a reverse genetic screen, we have identified LTPG, a glycosylphosphatidylinositol-anchored lipid transfer protein that is highly expressed in the epidermis during cuticle biosynthesis in Arabidopsis thaliana inflorescence stems. Mutant plant lines with decreased LTPG expression had reduced wax load on the stem surface, showing that LTPG is involved either directly or indirectly in cuticular lipid deposition. In vitro 2-p-toluidinonaphthalene-6-sulfonate assays showed that recombinant LTPG has the capacity to bind to this lipid probe. LTPG was primarily localized to the plasma membrane on all faces of stem epidermal cells in the growing regions of inflorescence stems where wax is actively secreted. These data suggest that LTPG may function as a component of the cuticular lipid export machinery.
INTRODUCTION
A common feature of all terrestrial plants is the cuticle, a hydrophobic structure that covers aerial surfaces, prevents nonstomatal water loss, and provides protection against pathogens (Riederer, 2006). The cuticle consists of wax embedded in and covering a polyester matrix called cutin (Jeffree, 2006). Cuticular wax contains very-long-chain fatty acid derivatives, such as alkanes and alcohols, with chain lengths >20 carbons, and wax composition varies according to species, organ, and developmental state (Samuels et al., 2008). Wax components are synthesized by epidermal cells, especially during rapid elongative growth (Suh et al., 2005).
Export of cuticular wax to the cuticle is not fully understood. In Arabidopsis thaliana, two ATP binding cassette (ABC) transporters, ABCG12/CER5 and ABCG11/WBC11, have been shown to be involved in wax transport (Pighin et al., 2004; Bird et al., 2007), but the mechanism of export from the plasma membrane (PM) through the surrounding aqueous cell wall to the plant surface is unknown. This remains an interesting problem because once a wax molecule is extruded from an epidermal cell, it must traverse a hydrophilic and charged extracellular environment (Samuels et al., 2008). The polysaccharide components of the cell wall underlying the cuticle, such as pectin (Jeffree, 2006), may also represent a physical impediment to cuticular wax transport.
Lipid transfer proteins (LTPs) were proposed as candidates that played a role in the delivery of wax components during the assembly of the cuticle (Sterk et al., 1991; Yeats and Rose, 2008). Originally described as proteins that facilitate lipid transfer between liposomes in vitro (Kader et al., 1984), LTPs possess several characteristics that make them suitable for delivering wax to the cuticle: they have a hydrophobic cavity (Kader, 1996); they are capable of binding fatty acids in vitro, and binding is impaired upon disruption of the hydrophobic pocket (Zachowski et al., 1998); and they are extracellular proteins and at <25 kD (Beisson et al., 2003) are likely small enough to fit through the pores of the plant cell wall (Baron-Epel et al., 1988). Nuclear magnetic resonance data have demonstrated that wheat (Triticum aestivum) LTP forms a complex with prostaglandin B2, suggesting that LTP structures can accommodate larger hydrophobic compounds, even those with large, planar functional groups (Tassin-Moindrot et al., 2000). LTPs have been localized to the cell wall (Thoma et al., 1993; Pyee et al., 1994), and they have been extracted from plant surfaces with nonpolar solvents (Pyee et al., 1994). Genes encoding LTPs are upregulated during drought-induced cuticular wax deposition (Cameron et al., 2006). While there has been much speculation based on the above, indirect evidence, a direct relationship between LTPs and cuticle deposition has not yet been demonstrated experimentally.
The objective of this study was to test whether LTPs are required for cuticular wax deposition in the stems of the model plant Arabidopsis. The Arabidopsis genome contains >70 annotated LTP genes (Beisson et al., 2003). To narrow down the number of candidate genes encoding LTPs that could act in cuticular wax export, we decided to focus on those LTPs that were highly expressed in the epidermis during cuticle biosynthesis (Suh et al., 2005). This approach reduced the list of candidate genes to five, and subsequent analyses of T-DNA insertional mutants in the LTP genes of interest resulted in the identification of LTPG, a gene encoding a glycosylphosphatidylinositol (GPI)-anchored protein. Here, we report the functional characterization of LTPG, using T-DNA mutant and RNA interference (RNAi) transgenic plants with reduced LTPG transcript levels, in vitro lipid binding assays of recombinant LTPG, and confocal analysis of the subcellular location of yellow fluorescent protein (YFP)-LTPG.
RESULTS
Identification of ltpg-1
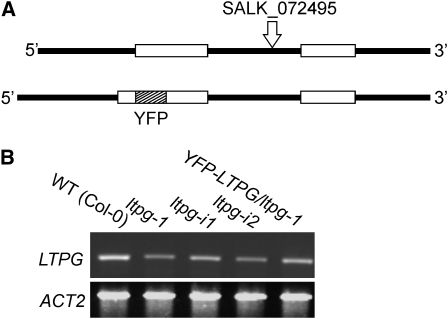
To test if LTPs are involved in cuticular wax deposition in Arabidopsis stems, five candidate LTPs were chosen for further analysis using microarray expression data based on two criteria (Suh et al., 2005): the transcripts corresponding to LTPs were abundant in the epidermis at the top of the inflorescence stem, an area undergoing rapid expansion and cuticle deposition, and there was a high ratio of LTP gene expression in the epidermis compared with the whole stem (see Supplemental Table 1 online). T-DNA insertional lines generated by the Salk Institute were obtained from the ABRC for each LTP (Alonso et al., 2003). Following PCR identification of homozygous individuals, the ltp mutants were screened for the glossy or eceriferum phenotype that may accompany changes in cuticular wax. Since some mutants with altered wax load and/or composition do not display this phenotype, quantitative screening using gas chromatography with flame ionization detection (GC-FID) was also performed. With the exception of At1g27950 (ltpg-1, SALK_072495), a gene experimentally identified as a GPI-anchored protein in global proteomics studies (Borner et al., 2003; Elortza et al., 2006), none of the other mutant lines displayed reductions in stem cuticular wax load (see Supplemental Table 1 online). Therefore, we named this gene LTP-GPI-ANCHORED or LTPG. The T-DNA insertion site was verified by sequencing to be in the only intron, 889 bp downstream from the predicted translation start site (Figure 1A, top). RT-PCR analysis demonstrated that this mutation resulted in reduced LTPG transcript abundance in ltpg-1 stems (Figure 1B).
Figure 1.
LTPG Gene Structure and Expression of LTPG in T-DNA and Transgenic RNAi Lines.
(A) Schematics illustrating the intron/exon pattern of LTPG and the position of the T-DNA insertion in ltpg-1 (top). YFP was inserted into LTPG (bottom) immediately following the predicted N-terminal signal sequence cleavage site. Open boxes represent exons and solid lines represent introns and untranslated regions.
(B) LTPG expression analysis by RT-PCR in primary inflorescence stems of Arabidopsis, comparing the wild type (Col-0) with the mutant (ltpg-1) and transgenic lines harboring RNAi constructs (ltpg-i1 and ltpg-i2) and the YFP-LTPG fusion. ACTIN2 was used as an amplification and loading control.
ltpg Mutants Have Reduced Stem Cuticular Wax
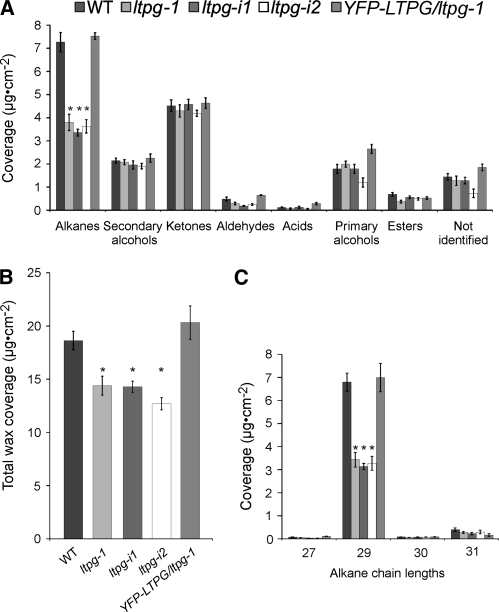
To determine the chemical phenotype of ltpg-1 mutant in detail, GC-FID analyses were performed. ltpg-1 stems showed a 50% reduction in alkanes (Figure 2A), the major class of components of Arabidopsis stem wax, which led to a 25% reduction in total wax load when compared with the wild type (Columbia-0 [Col-0]) (Figure 2B). No other wax components differed significantly from the wild type. Examination of alkane chain lengths showed that nonacosane (C29-alkane) levels were reduced by 50% compared with the wild type (Figure 2C). Alkanes of other chain lengths were not significantly different from the wild type. Besides this cuticular wax phenotype, ltpg-1 mutants were indistinguishable from the wild type and displayed no organ fusions, unlike some other mutants with cuticle defects (Kurdyukov et al., 2006; Bird et al., 2007).
Figure 2.
Reduction of Stem Cuticular Wax Load in ltpg Mutants Detected by Gas Chromatography.
(A) ltpg-1 mutants and lines expressing RNAi construct (ltpg-i1 and ltpg-i2) have lower amounts of alkanes, the most abundant compound class in wild-type wax. Alkane coverage was significantly different by analysis of variance (ANOVA), F (4, 31.106) = 29.1112, P < 0.0001.
(B) Total stem wax coverage is reduced. ANOVA indicated significant differences among genotypes, F (4, 32.127) = 10.9559, P < 0.0001.
(C) Examination of alkane chain lengths shows that only the C29 alkane (nonacosane) is reduced in the mutants. ANOVA indicated significant differences among genotypes, F (4, 31.106) = 29.1112, P < 0.0001. In the lines expressing YFP-LTPG driven by the native promoter in the ltpg-1 mutant background, wax levels are not different from the wild type. Asterisks indicate difference from the wild type by Games-Howell test (P < 0.05). Values are means with se for at least n = 10.
To compare the phenotypes of additional mutant alleles, two independent transgenic plant lines expressing an RNAi construct directed against LTPG were obtained from the Arabidopsis Genomic RNAi Knockout Line Analysis project (Hilson et al., 2004). Neither RNAi plant line, designated ltpg-i1 and ltpg-i2, was a complete transcriptional knockout (Figure 1B), but their wax coverage phenocopied the T-DNA mutant by displaying reduced total cuticular wax loads (68% to 76% of wild-type levels; Figure 2B) as a result of reduced C29-alkane (50 to 54% of wild-type levels; Figure 2C). The amounts of other wax components were statistically indistinguishable among ltpg-i1, ltpg-i2, and the wild type.
Native Promoter–Driven LTPG and YFP-LTPG Expression Rescue the Cuticular Wax Phenotype
To confirm that differences in wax load and composition were a result of a mutation at the LTPG locus, and to determine the subcellular location of LTPG, a genomic copy of the gene with an internal fusion of citrine YFP (Griesbeck et al., 2001) was generated for complementation of the ltpg-1 mutant. It was necessary to clone the YFP in frame, downstream from the predicted N-terminal signal sequence cleavage site (Bendtsen et al., 2004), so as not to disturb endoplasmic reticulum (ER) targeting and signal peptidase activity nor the C-terminal ω-site for the GPI-anchor attachment in the ER (Figure 1A, bottom). Twelve independent lines expressing YFP-LTPG in the ltpg-1 mutant background were recovered, and 10 restored the total wax coverage to wild-type levels (Figure 2B), rescuing the C29-alkane phenotype (Figure 2C) and showing that the fusion protein was biologically active. YFP-LTPG was as effective in complementing the ltpg-1 mutant as the wild-type gene. This was confirmed by analyses of 12 independent transgenic lines transformed with the LTPG gene (see Supplemental Figure 1 online). These results indicate that the detected wax phenotype was caused by a mutation in the LTPG gene.
LTPG Displays Lipid Binding Activity
The feature of protein sequences that leads to their annotation as LTPs is the conserved eight-Cys motif, but this alone is not a sufficient predictor of lipid binding activity. The conserved eight-Cys motif and α-helix structure is not unique to LTPs: it occurs in several plant protease and amylase inhibitors as well as in proteins of unknown function (José-Estanyol et al., 2004). Other than the conservation of this motif, the percentage of sequence identity of LTPG and structurally characterized LTPs is quite low. When the 75 amino acids comprising the LTP-like domain of LTPG are compared with the sequence of LTP from a maize (Zea mays) seedling (PDB 1MZL), which is the closest solved x-ray crystal structure (Han et al., 2001), identity is ∼20%. Therefore, the predicted protein structure of LTPG was modeled. Both the four amphipathic α-helices (Jpred; Cole et al., 2008) and eight Cys residues capable of forming four disulfide bonds (DISULFIND; Ceroni et al., 2006) were predicted (see Supplemental Figure 2 online), leading to the question of whether the structure of LTPG supports lipid binding.
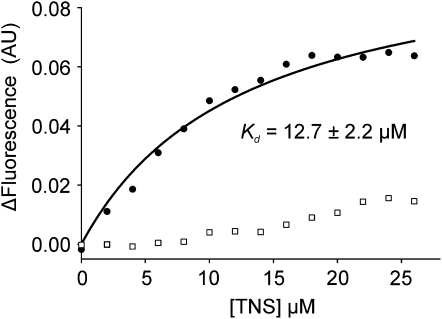
To answer this question, an in vitro assay was performed using purified LTPG and a fluorescent lipid substrate. To produce the processed, soluble version of LTPG, which we named ΔLTPG, the cDNA lacking both the N-terminal signal sequence and the C-terminus, including the GPI-anchor attachment ω-site was expressed in Escherichia coli. The overexpressed protein was purified by affinity, ion exchange and then size exclusion chromatography (see Supplemental Figure 3 online). To assess potential lipid binding activity, ΔLTPG was titrated with the fluorescent lipophilic probe 2-p-toluidinonaphthalene-6-sulfonate (TNS) that fluoresces intensely when bound in a hydrophobic environment, but weakly in aqueous solution. The binding isotherm of ΔLTPG with TNS indicates a Kd of 12.6 ± 2.2 μM (Figure 3, circles).
Figure 3.
Recombinant Purified ΔLTPG Binds the Lipophilic Probe TNS.
Recombinant LTPG (ΔLTPG) was engineered to resemble the mature processed protein by omitting both the N-terminal signal sequence and GPI-anchor domain and then purified following overexpression in E. coli. ΔLTPG, incubated with increasing concentrations of TNS, exhibited saturation binding defined by a Kd of 12.7 ± 2.2 μM (circles). No saturation binding was observed (squares) if the protein was denatured by boiling in a solution of 6 M urea and 200 mM DTT.
When ΔLTPG was denatured and reduced by boiling in DTT prior to the TNS assay, the resulting TNS fluorescence was decreased to close to background levels (Figure 3, squares), suggesting that the intact protein structure is required for lipid binding. While LTPG annotation as a putative LTP was based on its conserved eight-Cys motif (Beisson et al., 2003), this result provides experimental evidence supporting the hypothesis that LTPG is a lipid binding protein.
LTPG Resides Primarily in the PM
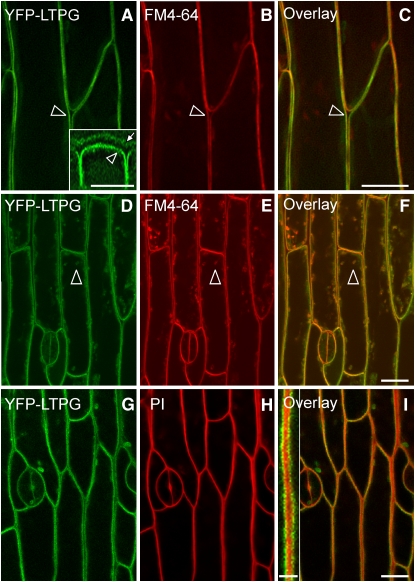
Initial screening of the 10 lines complemented by expression of the YFP-LTPG showed that the YFP fluorescence signal originated from the periphery of the cell. Since GPI-anchored proteins are generally found on the extracellular leaflet of the PM, transgenic lines were stained with either propidium iodide or the styryl dye FM4-64 (Bolte et al., 2004), markers for the cell wall and PM, respectively. The YFP-LTPG signal colocalized with FM4-64 in the PM of epidermal cells (Figures 4A to 4C). In some cells, YFP-LTPG fluorescence was also detected in the cytoplasm as small puncta that colocalized with endocytosed FM4-64 from the PM (Figures 4D to 4F, arrowheads). YFP-LTPG did not localize to anticlinal cell walls as determined by staining with propidium iodide (Figures 4G and 4I, inset). Epidermal cells stained with the ER counterstain hexyl rhodamine B showed that the strong peripheral YFP-LTPG signal did not colocalize with the ER (see Supplemental Figures 4A to 4C online).
Figure 4.
PM Localization of LTPG Visualized by Confocal Microscopy.
(A) YFP-LTPG signal at the PM (arrowhead). Inset: Three-dimensional reconstruction of the signal indicates an additional YFP-LTPG signal is present in the cell wall near the cuticle (arrow). Inset bar = 8 μm.
(B) PM of epidermal cells stained with FM4-64.
(C) YFP-LTPG colocalized with PMs stained with FM4-64.
(D) During late cell expansion, YFP-LTPG is found in puncta throughout the cytoplasm (arrowhead).
(E) Epidermal cells stained with FM4-64 showed internal puncta, indicating that endocytosis of the FM4-64 occurred.
(F) Some of the YFP-LTPG puncta colocalized with FM4-64.
(G) YFP-LTPG signal.
(H) Cell walls stained with propidium iodide.
(I) Overlay of (G) and (H) shows that YFP-LTPG does not colocalize to anticlinal cell walls. Inset: YFP-LTPG is exclusive from propidium iodide. Inset bar = 2.5 μm.
Bars = 10 μm.
To determine whether LTPG is also localized in the cell wall adjacent to the cuticle, we collected a z-series of images through the depth of the epidermal cell layer using spinning-disk confocal microscopy and assembled it into a three-dimensional data set, which could be sampled in the x, y, and z dimensions. In addition to the YFP signal observed at the PM as shown above, a weaker signal at the periclinal cell wall was seen, giving the appearance of two parallel fluorescent lines (Figure 4A, inset). When the signal from the YFP-LTPG was merged with the propidium iodide cell wall label, the fluorescence signals colocalized (see Supplemental Figure 5 online, arrowheads). The YFP-LTPG signal detected in the cell wall could not be found in the absence of the PM signal, such as under conditions of strong plasmolysis.
LTPG Is Localized to All Faces of Epidermal Cells
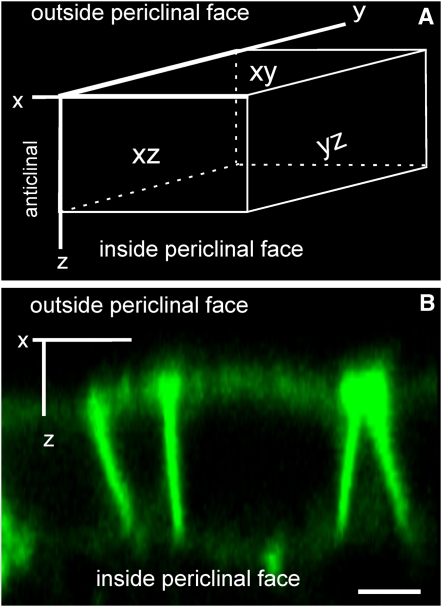
Because GPI-anchored proteins can have polar distribution (Lisanti et al., 1989; Fischer et al., 2004), it was important to determine if the YFP-LTPG was present in the PM on all faces of epidermal cells or only on the outer periclinal cell face near the cuticle (Figure 5A). Examination of three-dimensional reconstructions from z-series images generated by two-photon imaging showed that YFP-LTPG is found on all sides of the epidermal cells (Figure 5B). Anticlinal faces appeared brighter than the periclinal faces due to the presence of appressed PMs from adjacent cells doubling the signal and the configuration of the pinhole in two-photon imaging, which leads to overestimation of intensity in the z-dimension. Nevertheless, these results clearly demonstrate that LTPG is not limited to the PM facing the cuticle.
Figure 5.
LTPG Is Localized to All Faces of Primary Inflorescence Stem Epidermal Cells.
(A) A three-dimensional representation of an epidermal cell identifies the anticlinal (xz and yz) and periclinal (xy) faces.
(B) Two-photon microscopy shows that YFP-LTPG does not display polar localization. Bar = 5 μm.
LTPG Is Found in Epidermal Cells of Expanding Regions of Inflorescence Stems
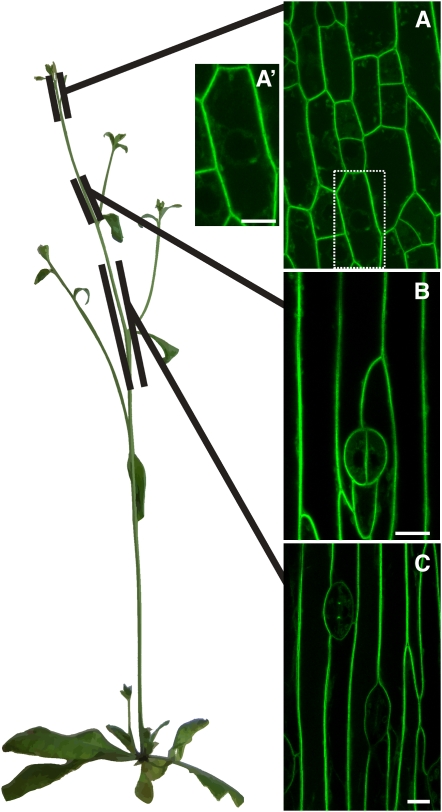
Our early observations of YFP-LTPG localization in the stem epidermis revealed the presence of the YFP-LTPG fluorescence signal in puncta, presumably in the secretory system, in addition to the PM. This YFP fluorescence in puncta was only detectable in young epidermal cells, but not in mature cells, suggesting that there might be developmental differences in the subcellular localization of LTPG. Therefore, we more closely examined the distribution of YFP-LTPG along the expanding stem of Arabidopsis. We defined three areas of study along the stem as follows: early, 0 to 1 cm (from the shoot apex) (Figure 6A); expanding, 2 to 3 cm (Figure 6B); and mature, 4 to 7cm (Figure 6C). As before, YFP-LTPG was found at the PM in all of the above stages, but the frequency of organellar localization changed (Figures 6A to 6C). In young meristematic cells, YFP-LTPG was observed in a pattern consistent with the ER (e.g., in transvacuolar strands and surrounding the nucleus) and in puncta resembling the Golgi (Figure 6A′). In expanding or recently matured cells that have undergone anisotropic expansion so the large central vacuole dominates in the cell, the signal was only associated with the PM and was not observed in transvacuolar strands, around the laterally displaced nucleus, or in puncta (Figure 6C). Beyond 8 cm from the apical meristem, in the most mature region of the stem epidermis, YFP-LTPG PM signal diminished and could not be detected. These results suggest that LTPG expression is concomitant with expansion of inflorescence stems.
Figure 6.
Localization of YFP-LTPG Over the Length of the Arabidopsis Stem Shows High Expression in Regions of Rapid Expansion.
(A) At 0 to 1cm from the shoot apical meristem, fluorescence was found at the PM with some internal signal.
(A′) YFP-LTPG signal in the interior of immature cells, in perinuclear distribution and puncta resembling endomembranes. Bar = 14 μm.
(B) At 2 to 3 cm from the shoot apex, YFP-LTPG signal was localized to the PM.
(C) At 4 to 7cm, in mature cells, localization was also primarily at the PM.
Bars = 10 μm.
DISCUSSION
In the search for additional components of cuticular lipid export machinery in the plant epidermis, we examined the involvement of LTPs in this process. LTPs are abundantly expressed in epidermal cells, secreted to the extracellular matrix (Thoma et al., 1993), and capable of binding and carrying lipids in vitro (Kader et al., 1984). These properties have long been cited as the rationale for the proposal that LTPs are potential carriers of cutin and wax constituents to the plant surface. In this study, using reverse genetics, we provide evidence that one member of the LTP family in Arabidopsis, LTPG (At1g27950), is indeed required for wax export. Phenotypic analysis of an LTPG T-DNA insertional mutant, and of transgenic plants in which LTPG was downregulated by RNAi, revealed a marked reduction in stem wax alkane levels. Cuticular wax deficiencies in ltpg mutants were complemented by expression of both the wild-type LTPG and the YFP-LTPG fusion protein, an indication that a mutation in the LTPG gene was responsible for this lipid phenotype.
The expression of LTPG without the N-terminal signal sequence and C-terminal GPI-anchor domain in a heterologous E. coli system allowed us to purify LTPG and assess its lipid binding capacity. As reported for several other LTPs (Buhot et al., 2004; Sawano et al., 2008), LTPG did bind to the fluorescent lipid reporter TNS. This lipid binding is not surprising because, although sequence similarity is low, the overall structural features of the protein predicted by the LTPG amino acid sequence resemble characterized LTPs (i.e., structure with four α-helices and four disulfide bonds). The binding of this reporter is interesting because it is consistent with the hydrophobic cavity structural model (Kader, 1996). TNS binding to other, functionally uncharacterized, LTPs could be displaced by C16 and C18 unsaturated fatty acids (Buhot et al., 2004; Sawano et al., 2008), suggesting that aliphatics are capable of competing with TNS for the same hydrophobic binding cavity. In this case, the binding of the biologically relevant very-long-chain lipids, such as nonacosane, would be technically difficult to confirm due to solubility issues.
Examination of transgenic plants expressing the active YFP-tagged LTPG by confocal microscopy revealed the strongest fluorescence signal in the PM of rapidly growing epidermal cells. In these cells, LTPG was also found in the cytoplasm in a pattern resembling the endomembrane system. In elongating epidermal cells, the signal was predominantly PM localized. In regions further down the stem, YFP-LTPG fluorescence was not detected. The identified differences in LTPG expression levels along the developing inflorescence stem match the published microarray data and demonstrate that LTPG expression is correlated with cell expansion in rapidly growing stems (Suh et al., 2005). The presence of YFP-LTPG fluorescence in the endomembrane system of the cells at the shoot tip may represent the need for LTPG biosynthesis to accommodate rapid growth and cuticle deposition. This scenario is supported by the aggregation of YFP-LTPG signal in epidermal cells treated with Brefeldin A (see Supplemental Figure 6 online).
The predominantly PM localization of LTPG suggests that this protein spends at least part of its lifetime at the cell surface. This is in agreement with in silico predictions and experimental evidence (Eisenhaber et al., 2003; Borner et al., 2003; Elortza et al., 2006) that LTPG is a GPI-anchored protein. In contrast with many GPI-anchored proteins that exhibit polar sorting to one domain of the cell (Lisanti et al., 1989), LTPG was localized to all faces of epidermal cells. Although this may be surprising for a process requiring the polar secretion of wax, the same localization pattern was previously reported for ABC transporters ABCG11 and ABCG12 (Pighin et al., 2004; Bird et al., 2007), which are also involved in wax export to the cuticle.
While the YFP-LTPG signal is mostly PM associated, a minor component of the signal was detected in the cell wall near the cuticle. Although this signal was reproducible between instruments and objective lenses, we interpret these data cautiously. If real, these results are consistent with early studies where LTPs were localized to the cell wall (Thoma et al., 1993; Pyee et al., 1994). It is noteworthy that the cell wall signal could not be isolated in the absence of the PM signal, which may indicate that there is rapid movement and/or turnover of LTPG in the cell wall, and without the PM as a source of LTPG, the cell wall signal disappears. Alternatively, the changes of refractive index at the cell wall/cuticle interface could scatter the signal originating from the PM.
Based on this evidence, we cannot definitively conclude that LTPG is directly involved in transport. It may act as a regulatory component of transport or possibly act indirectly in creating the proper conditions for cuticular lipid export. In terms of potential direct roles for LTPG in cuticular wax export, we propose the following two models. The first model predicts that, through its GPI-anchor, LTPG associates with lipid microdomains containing ABC transporters and that the close contact enables ABC transporters to load LTPG with wax cargo. LTPG binds the C29 alkane, thereby temporarily increasing its solubility by shielding it from the hydrophilic extracellular environment. Once loaded, LTPG is free to exchange its cargo with soluble extracellular LTPs that carry wax molecules to the plant surface. The potential problem with this scenario is the fact that ABCG12/CER5 and ABCG11/WBC11 transporters have not been detected in detergent resistant membranes (Borner et al., 2005). However, this may represent a failure of these proteins to meet the researchers' detection criteria (Borner et al., 2005). In the second model, once loaded with cargo, LTPG could be released from the PM to the cell wall through the cleavage of its GPI-anchor by a GPI-specific phospholipase (Metz et al., 1994). LTPG could then move across the cell wall to deliver alkane molecules to the cuticle. In this case, in addition to the PM, a significant fraction of LTPG would be expected to be localized in the cell wall as described for the GPI-anchored cell wall protein COBRA (Roudier et al., 2005). Since the YFP-LTPG fluorescence signal appeared to be predominantly associated with the PM even in the rapidly elongating cells that were depositing cuticle material at high rates, phospholipase release of LTPG and its movement across the cell wall seems less likely. However, it cannot be excluded that the apparent PM localization represents LTPG that has been cleaved from the membrane and is in the extracellular environment directly adjacent to the PM.
In summary, the demonstration that LTPG is required for normal export of wax to the cuticle and displays lipid binding capacity suggests that, in this instance, the name lipid transfer protein is appropriate. This work generates additional questions about the role of the LTPG GPI-anchor and potential interactions between LTPG and ABCG transporters.
METHODS
Plant Material
Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used for all analyses. Plants were surface sterilized and sown on half-strength Murashige and Skoog medium and then transferred to soil comprised of Sunshine mix 1 and Metro Mix (3:1). Plants were grown in growth chambers under 24 h of light with an intensity of 80 to 100 μE m−2 s−1.
Genotyping
A T-DNA insertional mutant for LTPG (At1g27950), SALK_072495, was obtained from the ABRC. Plants homozygous for the T-DNA insertion in LTPG were analyzed by PCR with primers P1, P2, and LBa1 (all primer sequences are listed in Supplemental Table 2 online). Plant lines (NASC ID N290585) expressing RNAi constructs targeting LTPG were obtained from the Arabidopsis Genomic RNAi Knockout Line Analysis project (www.agrikola.org), and lines were genotyped for the presence of the gene specific target construct with primers AGR 51, 56, 64, and 69.
Wax Analysis
Soluble cuticular waxes were extracted from 10- to 15-cm-long primary inflorescence stems. To determine stem surface areas, digital images of stems were imported into OpenLab 4.0.2 (Improvision) and density sliced/thresholded to generate squared surface area. The squared surface area was multiplied by π to achieve approximate cylindrical stem area for each stem. Derivatization of cuticular wax and data acquisition was performed as described (Greer et al., 2007) except derivatization was performed for 90 min.
Expression Analysis by RT-PCR
RNA was extracted from 10- to 15-cm-long stems as described previously (Zhao et al., 2006). RT-PCR was used to detect LTPG transcript abundance in primary inflorescence stems from wild type (Col-0), ltpg-1, ltpgi-1, ltpgi-2, and YFP-LTPG/ltpg-1. The SuperScript One-Step RT-PCR system (Invitrogen) was used following the manufacturer's instructions with one modification. The cDNA was synthesized at 45°C for 60 min using gene-specific primers P12 and P13 to detect LTPG followed by 35 cycles of PCR. ACTIN2 (At3g18780) was amplified with P14 and P15 as an internal control for 28 cycles of PCR. The number of cycles used for each amplification was empirically determined to be in the logarithmic phase. RT-PCRs presented are representative of three biological replicates.
Gene Cloning
LTPG was amplified from BAC F13K9 (ABRC) using primers P3 and P4 to generate a genomic copy, including 1800 bp of the 5′ promoter region and 5′ untranslated region (UTR) and 800 bp downstream of the annotated 3′UTR. The sequence was introduced into pUC19, verified, and then subcloned into pCambia1300 (Cambia) as an EcoRI-PstI cassette. Citrine YFP (Griesbeck et al., 2001) was used for localization studies. To generate YFP-LTPG, the genomic copy of LTPG was amplified from BAC F13K9 as two fragments: F1 and F2 (see Supplemental Figure 7 online). F1 containing the 5′ promoter region and the 5′UTR as well as the N-terminal signal sequence was amplified with primers P3 and P5 to generate a KpnI-XhoI amplicon. F2 contained the coding region of the gene, including the intron and 800 bp downstream from the annotated 3′UTR. F2 was amplified with P6 and P7 as an XbaI-PstI amplicon. F1 was introduced into pSTBlue1 (Novagen) generating pF1. F2 was introduced into pGEM3z (Promega) generating pF2. Citrine YFP was amplified with P8 and P9 as an XhoI-XbaI amplicon and introduced into pF1 to yield pF1-YFP. The F1-YFP KpnI-XbaI cassette was introduced into pF2 to yield pYFP-LTPG and subcloned into pCambia1300. Each plasmid was maintained and propagated in Escherichia coli strain JM109 (Promega) and verified by sequencing. All constructs were introduced into Arabidopsis by the floral dip method using Agrobacterium tumefaciens GV3101, pMP90 (Clough and Bent, 1998).
Transgenic Lines
Plants were selected on half-strength Murashige and Skoog medium supplemented with 30 μg·mL−1 hygromycin B (Invitrogen). Plants were screened in both the T1 and T2 generations for restoration of wild-type wax on the surface of Arabidopsis stems by GC-FID.
Recombinant Protein Production and Purification
The coding sequence of LTPG without signal peptide and transmembrane domain was amplified by PCR from cDNA (see above) using primers P10 and P11. This amplicon was ligated into the vector pGEX5-1 (GE Healthcare) using EcoRI and XhoI restriction sites yielding the construct pGEX-ΔLTPG and transformed into the E. coli strain XL-10 Gold (Stratagene). The sequence of the coding region of pGEX-ΔLTPG was verified, and pGEX-ΔLTPG was then transformed into the E. coli expression host Rosetta-gami B (Novagen). A 50-mL overnight culture was grown in Luria-Bertani media supplemented with appropriate antibiotics and 0.5% glucose at 37°C. In the morning, the starter culture was added to 1 liter of Luria-Bertani supplemented with 0.5% glucose and antibiotics. When the culture reached OD600 of 0.5, protein expression was induced by addition of 0.1 mM isopropylthio-β-galactoside. After 4 h of growth at 24°C, cells were harvested by centrifugation, flash frozen in liquid nitrogen, and stored at −80°C.
The cell pellet from 1 liter of E. coli culture was thawed on ice and resuspended in 30 mL buffer A (50 mM Tris, 150 mM NaCl, 1 mM DTT, and 0.02% NaN3). Cells were then lysed by two passes through a chilled French press. The lysate was clarified by centrifugation for 15 min at 24,000g followed by filtration through a 0.45-μM membrane filter. Clarified lysate was applied to an equilibrated 1 mL GSTrap FF column (GE Healthcare) at 0.2 mL/min using a peristaltic pump. The column was then washed with 20 column volumes of buffer A followed by 10 column volumes of Factor Xa cleavage buffer (50 mM Tris, 100 mM NaCl, and 5 mM CaCl2). Using a syringe, 1 mL of Factor Xa cleavage buffer containing 80 units of Factor Xa (Novagen) was applied to the column. The column was sealed and incubated at room temperature for 2 d. Cleaved proteins were eluted with 5 mL Factor Xa cleavage buffer. Factor Xa was then removed using X-arrest agarose (Novagen) following the manufacturer's instructions.
A 70-kD contaminant (likely DnaK; Buchberger et al., 1994) was removed by anion exchange chromatography. The protein was exchanged into buffer B (50 mM Tris, 10 mM NaCl, and 0.02% NaN3) using PD-10 desalting columns (GE Healthcare). The sample was then applied to a 1-mL HiTrap Q (GE Healthcare) column and washed with five column volumes of buffer B. The flow-through and wash were collected, pooled, and concentrated with a 3000 MWCO Amicon Ultra-15 ultrafiltration unit (Millipore). The 70-kD contaminant remained bound to the column but could be eluted with buffer B supplemented with 1 M NaCl.
As a final polishing step, the concentrated sample was applied to a Superdex 75 HR 10/30 gel filtration column (GE Healthcare). The column was eluted with buffer A without DTT at 0.5 mL/min, and 0.5-mL fractions were collected and analyzed by SDS-PAGE. Fractions containing the pure ΔLTPG protein were pooled and concentrated as before. The concentration of ΔLTPG was estimated by densitometry of Coomassie Brilliant Blue–stained SDS-PAGE gel and comparison to a BSA standard.
TNS Binding Assays
Fluorescent binding assays were performed essentially as previously described (Buhot et al., 2004). A 1 mM solution of TNS (Research Organics) was added incrementally to a 2-mL stirred cuvette at 25°C containing 50 nM ΔLTPG. This mixture was excited at 320 nm, and after stabilization of the fluorescence signal, the fluorescence emission at 437 nm was recorded using an SLM 8100c spectrofluorometer (SLM Instruments). Fluorescence of TNS in buffer without protein was subtracted to give ΔF. Nonlinear regression fitting of the data to obtain the saturation binding parameters Kd and Bmax was performed using SigmaPlot software (SYSTAT).
Laser Scanning and Spinning-Disk Confocal Microscopy
Unless noted, all images shown are from the area within 1 cm from the floral apical meristem. In all instances, FM4-64 was used at a 10-μM concentration for 15 to 30 min; propidium iodide was used at 1.5 μM for 15 to 30 min; and hexyl rhodamine B was used at 1.6 μM for 10 min. Dyes were purchased from Invitrogen. Brefeldin A (Sigma-Aldrich) solution was made to 125 μM in water. Stem segments were submerged in Brefeldin A for 30 min prior to imaging.
YFP-LTPG localization analyses were performed with a Zeiss Pascal LSM5 (Carl Zeiss). YFP was excited using the 488-nm laser line of an Ar laser and detected with HFT 545 and 505- to 600-nm emission filter. A He-Ne laser was used to detect FM4-64, propidium iodide, and hexyl rhodamine B with the 543-nm laser line and a 560-nm long-pass emission filter. Images were processed with Zeiss AIM software.
Spinning-disk confocal images were acquired using a WaveFX with custom Yokogawa spinning-disk scan head (Quorum Technologies) mounted on a DMI6000 B inverted microscope (Leica Microsystems). YFP was detected using the 491-nm laser diode and 528/38-nm emission filters. FM4-64 and propidium were detected using the 561-nm laser diode and 624/40-nm emission filter. Images were processed and deconvolved with Volocity 4.3.2 (Improvision). Point spread functions were created using the midpoints of excitation and emission spectra.
Two-Photon Microscopy
Two-photon microscopy was performed using a Mai-Tai titanium sapphire laser (Spectra-Physics Lasers) on a Zeiss Meta 510 using an excitation spectrum of 960 nm (Zipfel et al., 2003). Fluorescence was detected with band-pass filter of 500 to 550 nm.
Statistical Analysis
One-way Welch ANOVA was performed with JMP 7 and post-hoc tests were performed with SPSS 16 (SPSS) to compare ltpg-1, ltpg-i1, ltpg-i2, and YFP-LTPG with the wild type (Col-0). The Welch ANOVA with Games-Howell post-hoc test was employed to identify statistical difference from the wild type while accounting for heterogeneity in variance.
Accession Number
The Arabidopsis Genome Initiative locus identifier for LTPG is At1g27950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Native Promoter-Driven, Non-Epitope-Tagged LTPG Rescues the ltpg-1 Wax Phenotype.
Supplemental Figure 2. Predicted LTPG Secondary Structure and Disulfide Bond Modeling
Supplemental Figure 3. SDS-PAGE of of ΔLTPG Fraction Used for Binding Studies.
Supplemental Figure 4. YFP-LTPG Does Not Localize to the Endoplasmic Reticulum.
Supplemental Figure 5. Spinning-Disk Confocal Micrographs of YFP-LTPG Signal in the Cell Wall Proximal to the Cuticle.
Supplemental Figure 6. Spinning-Disk Confocal Images of Brefeldin A–Treated Primary Inflorescence Stem Epidermal Cells.
Supplemental Figure 7. Schematic Showing the Cloning of YFP-LTPG.
Supplemental Table 1. LTPs Enriched in the Epidermis and Corresponding T-DNA Insertional Mutants.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We acknowledge the Salk Institute Genomic Analysis Laboratory for ltpg-1 and the ABRC for providing seed stocks and BAC clone. We thank the AGRIKOLA project for RNAi transgenic plants. We thank Colin MacLeod and Angel Shan for technical assistance, Kevin Hodgson of the University of British Columbia Bioimaging facility for advice and assistance with microscopy, and David Holowka and Barbara Baird of Cornell University Chemistry and Chemical Biology for assistance with spectrofluorometry. We thank Mi Chung Suh for sharing prepublication data. This work was funded by a Canadian Natural Sciences and Engineering Research Council (NSERC) Special Research Opportunity grant (L.K., A.L.S., and R.J.) and NSERC Discovery grant (L.S.), the Canada Foundation for Innovation (RJ), and research support provided to J.K.C.R. by the National Research Initiative of the USDA Copperative State Research, Education, and Extension Service, Grant 2006-35304-17323. T.Y. is supported in part by a National Institutes of Health chemistry/biology interface training grant (T32 GM008500).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lacey Samuels (lsamuels@interchange.ubc.ca).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Baron-Epel, O., Paramjit, K., Gharyal, P.K., and Schindler, M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta 175 389–395. [DOI] [PubMed] [Google Scholar]
- Beisson, F., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Bird, D., Beisson, F., Brigham, A., Shin, J., Greer, S., Jetter, R., Kunst, L., Wu, X., Yephremov, A., and Samuels, L. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52 485–498. [DOI] [PubMed] [Google Scholar]
- Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N.D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214 159–173. [DOI] [PubMed] [Google Scholar]
- Borner, G.H.H., Lilley, K.S., Stevens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored protein in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger, A., Valencia, A., McMacken, R., Sander, C., and Bukau, B. (1994). The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 13 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhot, N., Gomes, E., Milat, M.L., Ponchet, M., Marion, D., Lequeu, J., Delrot, S., Coutos-Thevenot, P., and Blein, J.P. (2004). Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol. Biol. Cell 15 5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, K.D., Teece, M.A., and Smart, L.B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni, A., Passerini, A., Vullo, A., and Frasconi, P. (2006). DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 34 W177–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cole, C., Barber, J.D., and Barton, G.J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36 W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber, B., Wildpaner, M., Schultz, C.J., Borner, G.H., Dupree, P., and Eisenhaber, F. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 133 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elortza, F., Mohammed, S., Bunkenborg, J., Foster, L.J., Nuhse, T.S., Brodbeck, U., Peck, S.C., and Jensen, O.N. (2006). Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J. Proteome Res. 5 935–943. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Men, S., and Grebe, M. (2004). Lipid function in plant cell polarity. Curr. Opin. Plant Biol. 7 670–676. [DOI] [PubMed] [Google Scholar]
- Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., and Jetter, R. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A., and Tsien, R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. J. Biol. Chem. 276 29188–29194. [DOI] [PubMed] [Google Scholar]
- Han, G.W., et al. (2001). Structural basis of non-specific lipid biding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308 263–278. [DOI] [PubMed] [Google Scholar]
- Hilson, P., Allemeersch, J., Altmann, T., Aubourg, S., Avon, A., Beynon, J., Bhalerao, R.P., Bitton, F., Caboche, M., and Cannoot, B. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree, C.E. (2006). Structure and ontogeny of plant cuticles. In Biology of the Plant Cuticle, M. Riederer and C. Muller eds (Oxford, UK: Blackwell), pp. 11–125.
- José-Estanyol, M., Gomis-Rüth, F.X., and Puigdomenech, P. (2004). The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 42 355–365. [DOI] [PubMed] [Google Scholar]
- Kader, J.-C. (1996). Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 627–654. [DOI] [PubMed] [Google Scholar]
- Kader, J.-C., Julienne, M., and Vergnolle, C. (1984). Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur. J. Biochem. 139 411–416. [DOI] [PubMed] [Google Scholar]
- Kurdyukov, S., Faust, A., Nawrath, C., Bar, S., Voisin, D., Efremova, N., Franke, R., Schreiber, L., Saedler, H., Metraux, J.P., and Yephremov, A. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti, M.P., Caras, I.W., Davitz, M.A., and Rodriguez-Boulan, E. (1989). A glycophospholipid membrane anchor acts as an apical targeting signal in polarized cells. J. Cell Biol. 109 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, C.N., Brunner, G., Choi-Muira, N.H., Nguyen, H., Gabrilove, J., Caras, I.W., Altszuler, N., Rifkin, D.B., Wilson, E.L., and Davitz, M.A. (1994). Release of GPI-anchored membrane proteins by a cell-associated GPI-specific phospholipase D. EMBO J. 13 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin, J.A., Zheng, H., Balakshin, L.J., Goodman, I.P., Western, T.L., Jetter, R., Kunst, L., and Samuels, A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306 702–704. [DOI] [PubMed] [Google Scholar]
- Pyee, J., Hongshi, Y., and Kolattukudy, P.E. (1994). Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 311 460–468. [DOI] [PubMed] [Google Scholar]
- Riederer, M. (2006). Introduction: Biology of the plant cuticle. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell), pp. 1–11.
- Roudier, F., Fernandez, A.G., Fujita, M., Himmelspach, R., Borner, G.H.H., Schindelman, G., Song, S., Baskin, T.I., Dupree, P., Wasteneys, G.O., and Benfey, P.N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidylinositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59 683–707. [DOI] [PubMed] [Google Scholar]
- Sawano, Y., Hatano, K., Miyakawa, T., Komagata, H., Miyauchi, Y., Yamazaki, H., and Tanokura, M. (2008). Proteinase inhibitor from Ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol. 146 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, P., Booij, H., Schellekens, G.A., Van Kammen, A., and De Vries, S.C. (1991). Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M.C., Samuels, A.L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., and Beisson, F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin-Moindrot, S., Caille, A., Douilez, J.-P., Marion, D., and Vovelle, F. (2000). The wide binding properties of a wheat nonspecific lipid transfer protein solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 267 1117–1124. [DOI] [PubMed] [Google Scholar]
- Thoma, S.L., Kaneko, Y., and Somerville, C. (1993). An Arabidopsis lipid transfer protein is a cell wall protein. Plant J. 3 427–437. [DOI] [PubMed] [Google Scholar]
- Yeats, T.H., and Rose, J.K.C. (2008). The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 17 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski, A., Guerbette, F., Grosbois, M., Jolliot-Croquin, A., and Kader, J.-C. (1998). Characterisation of acyl binding by plant lipid-transfer protein. Eur. J. Biochem. 257 443–448. [DOI] [PubMed] [Google Scholar]
- Zhao, L., Gross, B.L., Zou, Y., Andrew, J., and Rieseberg, L.H. (2006). Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 15 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, W.R., Williams, R.M., and Webb, W.W. (2003). Non-linear magic: Multiphoton microscopy in the biosciences. Nat. Biotechnol. 21 1369–1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.