Abstract
Here, we report our analysis of a mutant of Volvox carteri, InvB, whose embryos fail to execute inversion, the process in which each Volvox embryo normally turns itself inside-out at the end of embryogenesis, thereby achieving the adult configuration. The invB gene encodes a nucleotide-sugar transporter that exhibits GDP-mannose transport activity when expressed in yeast. In wild-type embryos, the invB transcript is maximally abundant before and during inversion. A mannoside probe (fluorescent concanavalin A) stains the glycoprotein-rich gonidial vesicle (GV) surrounding wild-type embryos much more strongly than it stains the GV surrounding InvB embryos. Direct measurements revealed that throughout embryogenesis the GV surrounding a wild-type embryo increases in size much more than the GV surrounding an InvB embryo does, and the fully cleaved InvB embryo is much more tightly packed within its GV than a wild-type embryo is. To test the hypothesis that the restraint imposed by a smaller than normal GV directly causes the inversion defect in the mutant, we released InvB embryos from their GVs microsurgically. The resulting embryos inverted normally, demonstrating that controlled enlargement of the GV, by a process in which requires the InvB nucleotide-sugar transporter, is essential to provide the embryo sufficient space to complete inversion.
INTRODUCTION
In many kinds of developing embryos, a cytoskeletal rearrangement that results in a change in the curvature of a multicellular sheet is often an important early step in generating tissues, organs, or organisms of characteristic shapes. In plants, for example, formation of a root hair is a subcellular process in which a cytoskeletal rearrangement generates a bulge on a single epidermal cell, followed by polarized outgrowth in the bulged region (Hepler et al., 2001; Mathur, 2004). In animal embryos, by contrast, deformation of an epithelial sheet, as is seen at the beginning of many important developmental processes, such as gastrulation, neurulation, and gland morphogenesis, generally is a supercellular process, involving coordinated changes in the shapes and adhesive properties of a group of neighboring cells that act in concert to curl the cell sheet a predictable manner (Schock and Perrimon, 2002; Leptin, 2005; Pilot and Lecuit, 2005). Regulation of such shape-generating events is generally not well understood but appears to involve rather complex interactions among various cell-intrinsic factors as well as extrinsic influences arising from interactions of the cells with one another and with the adjacent cell walls or extracellular matrix (Keller, 1981; Schoenwolf and Franks, 1984; Hay, 1989; Kam et al., 1991; Mathur, 2004).
Previous studies have shown that the green alga Volvox carteri provides an exceptionally attractive model system for studying the cellular and molecular mechanisms and the genetic foundations of an extreme example of morphogenetic curling of a cellular monolayer: namely, inversion, the process by which each Volvox embryo turns itself completely inside-out at the end of embryogenesis (Kirk et al., 1982; Kirk, 1998; Kirk and Nishii, 2001; Nishii et al., 2003; Ueki and Nishii, 2008).
A V. carteri adult (called a spheroid) consists of ∼2000 biflagellate somatic cells arranged in a monolayer at the surface of a transparent sphere of extracellular matrix and ∼16 asexual reproductive cells (called gonidia) just internal to the somatic cells (Figure 1A). Each mature gonidium initiates a series of 11 rounds of rapid, synchronous cleavage divisions. A predictable subset of these divisions is visibly asymmetric, producing large and small sister cells that serve as gonidial precursors and somatic-cell precursors, respectively. At the end of cleavage, each V. carteri embryo contains all of the cells of both types that will be present in an adult of the next generation. However, at that stage the orientation of the cells is the opposite of what it is in a wild-type adult: the somatic cells all have their flagellar ends pointing inward, and the gonidia are protruding from the outer surface of the embryo. This inside-out arrangement is corrected by inversion, in which the cell sheet turns itself right-side-out through an opening called the phialopore at one pole of the embryo (Starr, 1968, 1969). In the context of this study, it is important to note that cleavage and inversion of a Volvox embryo both occur inside a glycoprotein gonidial vesicle (GV) that is attached tightly to the plasmalemma of a gonidium during its development, but then separates from the gonidial plasmalemma just before cleavage begins (Kirk, 1998).
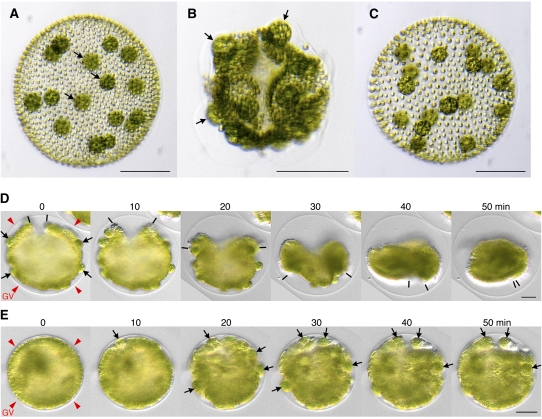
Figure 1.
The Inversionless Phenotype of the InvB Mutant.
(A) to (C) Adults of three related strains of V. carteri: (A) the wild type (CRH22), (B) an InvB mutant derived from CRH22, and (C) a stable revertant derived from InvB. In the wild-type adult, the gonidia (asexual reproductive cells, arrows) are completely enclosed within a spherical monolayer of small somatic cells. However, in the InvB mutant, the gonidia (arrows) are exposed on the surface of the somatic cell monolayer because inversion of the embryo did not occur in the mutant. Bars = 100 μm.
(D) and (E) Individual frames taken from time-lapse video movies of wild-type (D) and InvB (E) embryos during the time period when inversion occurs in the wild-type embryo. Numbers above images indicate time in minutes after the first inversion movements were seen. Red arrowheads mark the GV surrounding each embryo; note how much more space there is between the embryo and the GV at the beginning of inversion in the wild type than in InvB. In (D), the black lines track the progress of inversion by marking the cells that were closest to the phialopore before inversion and that are at the leading edges of the inverting cell sheet. In (E), the InvB phialopore is difficult to recognize in these individual frames, but in the movie from which they were taken, the phialopore is recognizable as the place where cellular movements begin and then propagate to adjacent regions (see Supplemental Movie 1 online). Arrows in (D) (0) mark some of the gonidia that protrude into the space between the embryo and the GV at the beginning of inversion; by the end of inversion, they have all been moved into the interior. In (E) (0), the gonidia are less distinct because they are pressed into the somatic cell layer by the closely appressed GV, and by the end of the inversion period ([E]; 40 to 50 min) the gonidia (arrows) remain on the surface of the embryo. Bars = 20 μm.
It has been known for decades that V. carteri mutants with a variety of different inversion defects can be isolated readily (Sessoms and Huskey, 1973; Kirk et al., 1982), raising the tantalizing possibility that the genetic programming of this dramatic morphogenetic process might be analyzed in some detail. However, all efforts to analyze the primary defects in such mutants were unsuccessful for many years, until a gene-tagging system based on an endogenous transposon called Jordan was developed (Miller et al., 1993), making tagging, recovery, and sequencing of such genes possible. Jordan was used to tag, recover, and characterize an inversionless gene called invA, which encodes a kinesin that provides the motile force to turn the cell sheet inside-out during inversion (Nishii et al., 2003). More recently, we reported the discovery of a second gene-tagging system, based on the Idaten family of endogenous transposons, and used it to tag and recover a second inversionless gene, invC (Ueki and Nishii, 2008), which encodes a glycosyltransferase that belongs to the LARGE family (Grewal et al., 2005). In discussing the mechanism by which a mutant glycosyltransferase might cause an inversion block, we noted that the glycoprotein-rich vesicle that surrounds all V. carteri embryos appears to be much smaller and tighter in the InvC mutant than it is in the wild type, and its compactness might conceivably interfere with inversion mechanically (Ueki and Nishii, 2008).
Here, we characterize another Idaten-tagged inversionless mutant, InvB, which, like InvC, has embryos that appear to be compressed at the end of cleavage by a GV that is too small and that fail to invert. The tagged gene, invB, encodes a nucleotide-sugar transporter, a protein that transfers sugar nucleotides, such as GDP-mannose, from the cytoplasm to the Golgi lumen to be used for synthesis of glycosylated proteins such as those that are found in the Volvox GV. We established that the InvB GV enlarges less during cleavage and inversion than the wild-type GV does, and we then demonstrated that if the InvB GV is removed mechanically, InvB embryos invert normally. This indicates that during normal development, the size of the GV is actively controlled, in a process that requires the activity of the invB product to provide enough room for the embryo to invert.
RESULTS
InvB Is a Transposon-Tagged Inversionless Mutant of V. carteri
InvB, the mutant that is the subject of this study, was isolated in an earlier screen for mutants with inversion defects (Ueki and Nishii, 2008). Whereas wild-type V. carteri adults have a smooth, spheroidal conformation, with all of their gonidia on the interior of the sphere (Figure 1A), adults of the InvB mutant are irregular in shape, extensively wrinkled, and with all of their gonidia on the outside, protruding from the surface (Figure 1B). The inversionless phenotype of InvB is similar to that previously reported for InvC (Ueki and Nishii, 2008) but quite different from that of InvA (Nishii et al., 2003), in which embryos begin to invert, but then arrest, producing adults that resemble Bowler hats, similar to a transitional phase of the wild-type phenotype (Figure 1D, 20 min). Strain InvB is stable at the standard culture temperature of 32°C, but it repeatedly yields wild-type revertants (Figure 1C) when it is cultured at 24°C, which is the temperature at which Idaten transposition is activated.
We used time-lapse video microscopy to compare the behavior of wild-type and InvB embryos during the period when wild-type inversion occurs (Figures 1D and 1E). Before inversion, both types of embryos are hollow spheres, with gonidia protruding from their outer surfaces. But whereas the wild-type gonidia are clearly visible between the outer surface of the somatic cell layer and the inner surface of the GV (Figure 1D, 0 min, black arrows), the InvB gonidia are difficult to see at this stage (Figure 1E, 0 min) because the embryos are so tightly packed within their GVs that the gonidia are pressed into the somatic cell layer, instead of protruding from it as they do in the wild type.
As inversion begins in wild-type embryos, the phialopore (the cross-shaped opening at one pole of the embryo) becomes clearly visible, as the opening widens dramatically and the lips of cells flanking the opening (marked by bars in Figure 1D) bend outward. In the InvB embryo, however, even though cellular movements become visible at the time inversion would begin in a wild-type embryo, the phialopore never opens as it does in the wild type (see Supplemental Movie 1 online). Instead of turning the embryo right-side out, the cellular activities of the InvB embryo merely cause a sequence of uncoordinated folding movements of the cell sheet in various regions, at which time some of the gonidia in those fold regions become visible as they are moved away from the GV (Figure 1E, arrows). Such movements stop after ∼50 min, which corresponds to the period required for wild-type inversion. Thereafter, the InvB embryos grow while retaining their irregular shapes, becoming wrinkled adults that are characteristic of this mutant strain (Figure 1B).
The invB Gene Encodes a GDP-Mannose Transporter
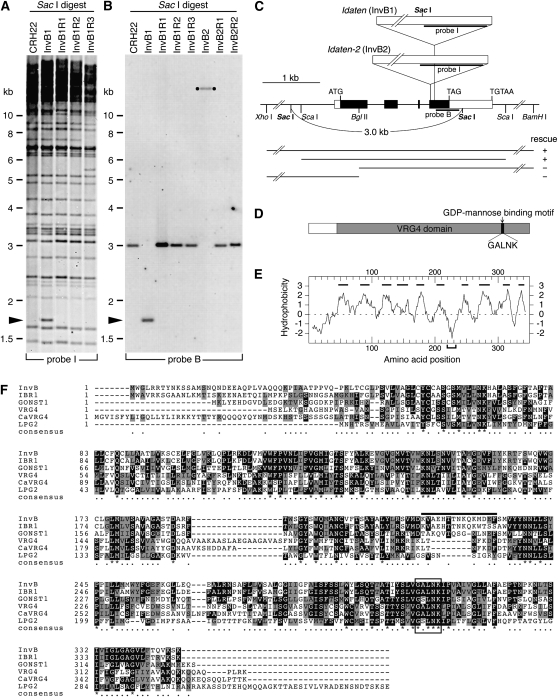
DNA gel blot analysis revealed a restriction fragment length polymorphism that was clearly linked to the InvB phenotype: an Idaten probe (probe I) detected a SacI fragment at 1.7 kb (Figure 2A, arrowhead) that is present in the InvB lane but missing in the lanes containing DNAs from both the wild-type progenitor of InvB (CRH22) and three independent revertants of InvB. Part of this InvB-specific DNA fragment was isolated using inverse PCR (Ueki and Nishii, 2008). When we then used a portion of this fragment that lacked any Idaten sequences as a probe (probe B, Figure 2C), it detected the same InvB-specific 1.7-kb band as had been detected with the Idaten probe, but it also detected a 3-kb fragment that was present in CRH22 (the progenitor of InvB) and all three InvB revertants, but not in InvB itself (Figure 2B). When we used probe B to screen other inversionless mutants we had isolated previously (Ueki and Nishii, 2008), we found a second mutant with a phenotype similar to InvB that had a different Idaten insertion in the invB gene (Figure 2B, lane InvB2). We renamed the original mutant InvB1 and named the second one InvB2. InvB1 has an insertion of Idaten, which contains a SacI site, whereas InvB2 has an insertion of Idaten-2 (Ueki and Nishii, 2008), which lacks a SacI site (Figure 2C). This difference accounts for the fact that the probe B–positive fragment in strain InvB2 is so much larger than the one in InvB1 (Figure 2B). The fact that all phenotypic revertants of both InvB1 and InvB2 that we tested had lost their Idaten or Idaten-2 insertions proved conclusively that those insertions were responsible for the inversionless phenotypes of both mutants.
Figure 2.
Cloning and Characterization of the invB Gene.
(A) DNA gel blot analysis of SacI-digested DNA of CRH22, InvB1, and three revertants that were independently isolated from InvB1; the blot was probed with a part of the Idaten transposon (probe I). A 1.7-kb fragment (indicated by arrowhead) is specifically detected in InvB1.
(B) A non-Idaten part of the 1.7-kb fragment (probe B) was used to probe a similar blot. The 1.7-kb fragment was again detected in InvB1 DNA, whereas a 3.0-kb fragment was detected in CRH22 and all three InvB1 revertants. A different restriction fragment length polymorphism is detected in mutant InvB2, but its revertants have the same 3.0-kb fragment seen in CRH22 and the InvB1 revertants.
(C) Structure of the invB gene and the insertion sites of Idaten and Idaten-2 in InvB1 and InvB2, respectively. The DNA regions corresponding to probes I and B are marked. The 1896-bp coding region of invB consists of four exons. A 7.1-kb genomic fragment covering the invB gene could be used to rescue InvB mutants by transformation, but shorter fragments could not, as indicated by bars below the map.
(D) The invB gene encodes a protein with a VRG4 domain that is conserved in nucleotide-sugar transporters. The predicted amino acid sequence of the InvB protein is 348 residues long with a theoretical molecular mass of 38 kD and a pI of 9.07.
(E) A hydrophobicity plot of InvB amino acid sequence predicts 10 transmembrane regions indicated by bars. A less hydrophobic region used as antigen is shown by square bracket.
(F) Aligned amino acid sequences of the V. carteri InvB, C. reinhardtii IBR1 (the InvB ortholog), Arabidopsis GONST1, S. cerevisiae VRG4, Candida globrata CaVRG4, and Leishmania donovani LPG2, shaded with BoxShade 3.21 software. Black boxes indicate residues that are identical in at least half of the proteins. The gray boxes indicate conservative substitutions, as defined by the BoxShade 3.21 default parameters. The conserved GDP-mannose binding motif is denoted by an open rectangle. The anti-InvB antibody was prepared in rabbits using as the immunogen a chemically synthesized peptide corresponding to the portion of the sequence denoted by an over-bar.
An invB cDNA clone was retrieved from our presequenced full-length cDNA library (I. Nishii, unpublished data), and the plasmid containing this clone was named pfcinvB. Probe B was used to screen V. carteri BAC filter (from the Arizona Genome Institute), and seven positive clones were identified. The invB gene was subcloned from one of those BAC clones and then sequenced. Comparison of the cDNA and genomic sequences revealed that the invB gene was 1.9 kb long, with four exons and three introns (Figure 2C). Two fragments that each covered the full length of the invB gene could be used to rescue the inversionless phenotype of InvB by transformation, but shorter fragments could not (Figure 2C). This was further proof that the InvB phenotype was caused by the Idaten family insertions in this genomic region in strains InvB1 and InvB2.
The deduced InvB amino acid sequence has a conserved nucleotide-sugar transporter domain (the VRG4 domain, Figure 2D) that contains 10 predicted transmembrane regions (Figure 2E). Aligned sequences of InvB and five other proteins containing the VRG4 domain (Figure 2F) revealed many conserved residues, including the GDP-mannose binding motif, GXLNK (Gao et al., 2001), which is boxed in Figure 2F. GDP-mannose transporters are reported to be localized in the Golgi membrane, where they function to transport cytoplasmic GDP-mannose into the Golgi lumen, where glycoconjugates are synthesized (reviewed in Kawakita et al., 1998).
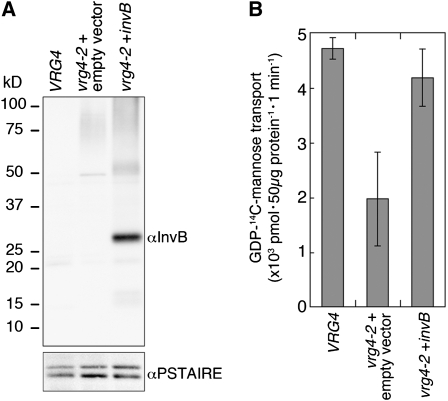
We tested whether wild-type InvB protein could function as a GDP-mannose transporter by expressing it in the yeast vrg4-2 mutant (Dean et al., 1997; Gao et al., 2001) (Figure 3A). Figure 3B shows that GDP-mannose transport activity was much lower in the P100 fraction of the vrg4-2 mutant than it is in the corresponding fraction of wild-type cells, but was almost back to normal in the yeast strain expressing the invB transgene product. We take this as evidence that the InvB protein almost certainly functions as a nucleotide-sugar transporter in Volvox also. (We note that the InvB protein expressed in yeast exhibits anomalous gel mobility and so does the InvB protein that is expressed in Volvox, as will be discussed in the next section.)
Figure 3.
GDP-Mannose Uptake by Vesicles from Yeast Cells Expressing InvB Protein.
(A) Proteins extracted from control yeast (VRG4), mutant vrg4-2 transformed with empty vector, and vrg4-2 transformed with invB were resolved by SDS-PAGE and subjected to immunoblot analysis using the anti-InvB antiserum. The anti PSTAIRE-domain antibody was used for a loading control.
(B) GDP-14C-mannose transport activity of Golgi-rich vesicles prepared from the same types of yeast cells as in (A). Averages of triplicate assays with standard error bars are shown.
The RNA and Protein Products of invB Both Peak around the Time of Inversion
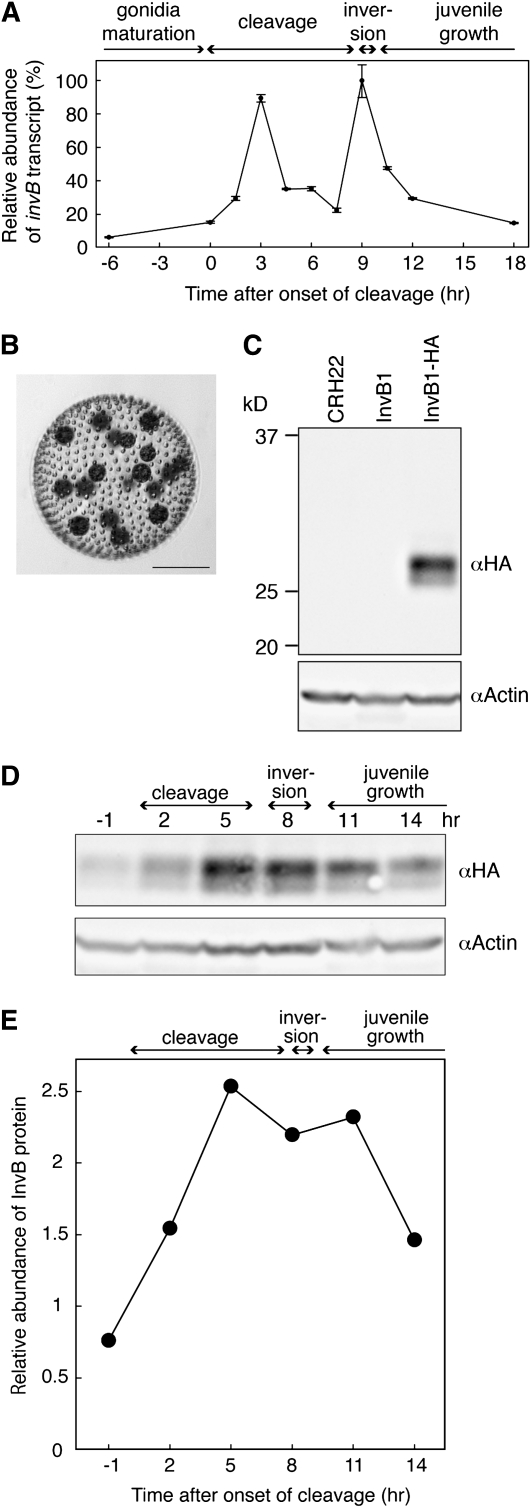
The abundance of invB transcripts at various times during V. carteri embryogenesis was quantified by real-time RT-PCR (Figure 4A). The invB transcript level is low in maturing gonidia, but increases after the onset of cleavage, subsequently reaching one abundance peak during mid-cleavage and a second peak during inversion. After inversion, the transcript level gradually returned to the basal level.
Figure 4.
Stage-Specific Abundance of invB Transcript and InvB Protein.
(A) Real-time RT-PCR analysis of the invB transcript. Stage-specific RNAs prepared from isolated embryos were reverse transcribed and analyzed by the Applied Biosystems 7500 real-time PCR system using the comparative Ct method, with 18S rRNA as a calibrator. Levels of invB transcript were normalized by setting the 9-h stage to 100%. Stages indicated on the top of graph were determined by observation. Bars indicate standard errors of the mean from technical replicates (n = 3).
(B) An adult of strain InvB1-HA, a morphologically wild-type strain produced by transforming the InvB1 mutant strain with an epitope-tagged invB construct. Bar = 100 μm.
(C) The transgene product expressed in strain InvB1-HA was detected with an anti-HA antibody. Actin protein levels are shown as a loading control.
(D) The HA-InvB protein was abundant from late cleavage through postinversion. The numbers above the blot indicate hours after the onset of cleavage.
(E) HA-InvB signals on the immunoblot detected by a Bio-Rad VersaDoc imager were quantified using NIH ImageJ software and normalized using actin as an internal standard. This experiment has been repeated with similar results.
To study expression of the InvB protein during development, we transformed strain InvB1 with a construct that encodes a hemagglutinin-tagged InvB (InvB-HA) and recovered a morphologically wild-type transformant (strain InvB1-HA, Figure 4B) that produces HA-tagged InvB. Anti-HA antibody detected the tagged transgene product on immunoblots in the 28-kD region, which is significantly lower than the ∼38-kD molecular mass predicted on the basis of the deduced amino acid sequence. We presume that this discrepancy is because the 10 hydrophobic transmembrane regions in the InvB amino acid sequence cause the protein to migrate anomalously in SDS-PAGE. In accord with the pattern of transcript abundance described above, we found the InvB protein to be present in high abundance from late cleavage to early postinversion, whereafter it declined (Figures 4D and 4E), suggesting a function for the InvB protein during inversion.
InvB Increases the Abundance of α-Mannosyl Groups in the GV at the Time of Inversion
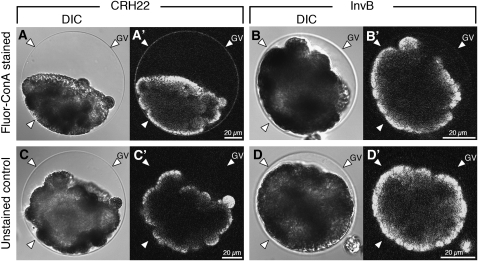
Because we had shown that InvB can act as a GDP-mannose transporter (Figure 3), we used fluorescent-labeled concanavalin A (Fluor-ConA), a lectin that specifically binds to sugar chains that include α-mannosyl groups, to determine where mannose-rich molecules may be localized during inversion and how different the localization pattern may be between the wild type and InvB mutants. A strong fluorescent ConA signal was observed on the wild-type GV (Figure 5A′), whereas the signal from the Fluor-ConA–stained InvB GV (Figure 5B′) was not significantly stronger than the signal from the unstained control (Figure 5D′). This marked difference in the fluorescence of the wild-type and mutant GVs (Figures 5A′ and 5B′) was in contrast with the very similar levels of chlorophyll autofluorescence exhibited by the wild-type and mutant embryos themselves (Figures 5C′ and 5D′).
Figure 5.
Differential Interference Contrast and Fluorescence Images of Fluorescent Con A–Stained Wild-Type and InvB Embryos.
Top row: Live wild-type ([A] and [A′]) and InvB ([B] and [B′]) embryos were collected during the time when wild-type embryos were inverting and were stained using Alexa 488–conjugated concanavalin A (Fluor-ConA). Bottom row: Unstained control embryos of similar genotypes and developmental stages as above. In each pair of images, the one on the left ([A], [B], [C], or [D]) is a differential interference contrast (DIC) micrograph, and the one on the right ([A′], [B′], [C′], or [D′]) is a confocal laser micrograph of the same embryo. Note that in the DIC images the GVs (arrowheads) are similar in appearance in wild-type and mutant embryos ([A] versus [B]; [C] versus [D]) and are also similar in stained and unstained embryos ([A] versus [C]; [B] versus [D]). In marked contrast, in the fluorescence images, the signal coming from the GV of the stained, wild-type embryo (A′) is much brighter than the signals coming from the stained InvB embryo (B′) or either of the unstained images ([C′] or [D′]). Note also that the fluorescence intensity of the embryos themselves is very similar in stained and unstained embryos and thus can be attributed to chlorophyll autofluorescence. The fact that the embryos are all in contact with one spot on the GV is an artifact caused by the fact that the embryos were centrifuged during the staining and washing process. Note the difference in the diameter of the vesicles between wild-type and InvB embryos.
These results lead us to conclude that a major function of the InvB protein is to transport GDP-mannose to the site of glycosylation of proteins that are destined to become constituents of the GV.
A Smaller than Normal GV Prevents Inversion in InvB
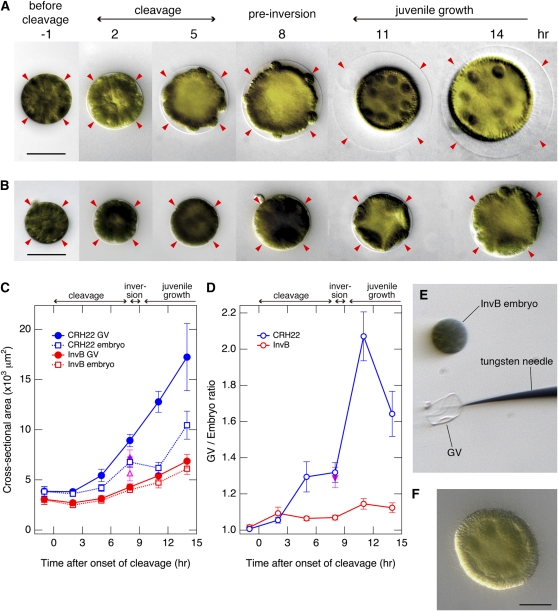
The crowded appearance of the InvB embryo within its GV at the time when inversion should occur, but does not (Figure 1E; see Supplemental Movie 1 online), and the extremely low signal from the InvB GV after staining with Fluor-ConA (Figure 5B′) focused our attention on a smaller than normal GV as the probable cause of the inversionless phenotype of InvB. So we measured the cross-sectional areas of wild-type and InvB embryos and their corresponding GVs from the one-cell stage until after the wild-type embryo had completed inversion and had become a growing juvenile spheroid (Figures 6A to 6C).
Figure 6.
A Slow-Growing GV in InvB Interferes with Inversion.
(A) and (B) DIC micrographs of wild-type (A) and InvB (B) embryos at various stages of embryogenesis from the one-cell stage through postinversion. Images with computer-enhanced contrast are shown to facilitate visualization of the GVs. Note that the area enclosed by the GV (arrowheads) increases gradually during cleavage and then more rapidly during and after inversion in the wild type, but increases much more slowly in the InvB mutant. Bars = 50 μm.
(C) The cross-sectional areas of the embryos and their GVs were quantified. The closed and open magenta triangles indicate the sizes that were attained by the CRH22 GV and embryo, respectively, when embryos were isolated from the parental spheroid before cleavage and then allowed to develop in isolation. Bars indicate standard errors (n = 20).
(D) The GV:embryo ratios were calculated by dividing the cross-sectional area enclosed by each GV by the cross-sectional area of the embryo that it enclosed. The inverted magenta triangle indicates the ratio for CRH22 embryos cultured in isolation from the parent. Bars indicate standard errors (n = 20).
(E) The GV of an InvB embryo was removed with a fine tungsten needle at the one-cell stage, and then the embryo was allowed to develop.
(F) In contrast with InvB embryos with intact GVs (which never inverted), GV-free InvB embryos inverted normally; the embryo shown here was photographed immediately after inversion, showing gonidia on the inside and flagella on the outside of the juvenile spheroid. Bar = 20 μm.
The wild-type embryo increases in diameter during cleavage as its internal lumen expands. However, the GV, which is initially in contact with the surface of the embryo, expands faster than the embryo does, so that by the end of cleavage there is a visible space between the somatic cell layer of the embryo and the GV, and the gonidia protrude into that space, making contact with the overlying GV (Figures 6A, 5 to 8 h, and 6C). Following inversion there is an even greater difference in the rates at which a wild-type embryo and its GV expand, so that the space surrounding the embryo increases in relative size during this period (Figure 6A, 8 to 14 h). To more clearly illustrate these relationships, we calculated the ratio of the area enclosed by each GV to the cross-sectional area of the embryo that it surrounds, thereby obtaining the GV:embryo ratios that are plotted in Figure 6D. This approach makes it obvious that the wild-type GV is ∼30% larger than the embryo at the time of inversion. After (and as a result of) inversion, the diameter of the embryo (now a juvenile) decreases temporarily, while the GV continues to increase in size (Figure 6C), so that the GV:embryo ratio rises sharply at this time (Figures 6A, 8 to 14 h, and 6D).
The studies just described involved embryos that had remained within intact parental spheroids, under standard culture conditions, until they reached the developmental stage of interest, whereupon they were isolated and measured. To determine whether the preinversion increase in GV size that has just been described required the presence of the cells and/or extracellular matrix of the parental spheroid, a supplemental set of measurements were made on embryos that were removed from parental spheroids before they had begun to cleave, cultured in isolation, and then measured when they had reached the inversion stage. When cultured in isolation, both the embryo and its surrounding vesicle increased in size slightly less than the embryos in intact spheroids did (Figure 6C, triangles). Importantly, however, the GV:embryo ratio attained by the onset of inversion in these isolated embryos was indistinguishable from the GV:embryo ratio attained by embryos in intact parental spheroids (Figure 6D), and embryos developing in isolation this way regularly invert normally.
The behavior of the GV in the InvB mutant is very different from the behavior of the wild-type GV that has just been described. At the beginning of cleavage, the InvB GV is already smaller than the wild-type GV in diameter, and the InvB embryo itself appears considerably smaller and denser, as if it is being compressed (Figure 6A versus 6B, 2 to 5 h). In addition to starting out smaller, the InvB vesicle grows much more slowly than the wild-type vesicle (Figure 6C) so that the visible space that is seen between the wild-type embryo and its vesicle never appears in the mutant (Figure 6A versus 6B, 5 to 14 h; Figure 6D), and (as described earlier) while the wild-type embryo is inverting, the InvB embryo exhibits vigorous cell movements inside its GV, but those movements are wholly ineffectual and do not result in inversion (see Supplemental Movie 1 online).
These findings led us to postulate that if the InvB embryo were released from the constraint of its smaller than normal GV, it might be able to invert. To test this hypothesis, we removed the vesicles from three InvB embryos at the one-cell stage, using a tungsten needle (Figure 6E), and then followed their subsequent development. They all inverted normally at the appropriate time and became essentially indistinguishable from wild-type juveniles, with their gonidia on the inside and their elongating flagella on the outside (Figure 6F). We conclude from this observation that normal inversion of the Volvox embryo requires an invB-dependent enlargement of the GV.
DISCUSSION
The Size of the GV Plays a Controlling Role in Volvox Morphogenesis
Most previous studies of inversion in Volvox have focused on the way in which changes in the cells of the embryo, changes in their shapes, their movements, and their connections to one another, contribute to the dynamic process of inversion (Pocock, 1933; Kelland 1977; Viamontes and Kirk, 1977; Viamontes et al., 1979; Green et al., 1981; Kirk et al., 1982; Nishii and Ogihara, 1999; Kirk and Nishii, 2001; Nishii et al., 2003). However, the fact that the GV plays a role in the inversion process has not been overlooked entirely. Viamontes et al. (1979) measured the circumference of the GV and the perimeter of the fully cleaved embryo and concluded that at this stage the embryo is “too big for its vesicle,” and that this was the reason that the gonidia of the fully cleaved embryo are pressed against the GV (causing some distortion of the somatic cell layer in that region) and that the lips of cells flanking the phialopore are curled inward. They noted that it was only when the embryo contracted (as a result of a decrease in the diameter of each of its cells) that it moved away from the GV far enough to make room for its phialopore to open and for the cells flanking the phialopore to curl outward instead of inward. They then performed two experiments that supported their hypothesis that the phialopore would open immediately (mimicking the first step of inversion) if they could artificially relieve the compressive stress that the GV was exerting on the preinversion embryo. Throughout the discussion of this topic by Viamontes et al. (1979), however, there clearly was an implication that the GV is a passive player in inversion, merely providing a physical barrier that prevents premature opening of the phialopore. They did not recognize that the only reason that there normally is room inside the GV for the embryo to invert is because the GV increases in diameter during cleavage. Likewise, we failed to recognize the importance of the normal GV enlargement process in our previous study, in which we stated that the glycoprotein-rich vesicle that surrounds all V. carteri embryos appears to be much smaller and tighter in the InvC mutant than it is in wild-type V. carteri, and its compactness might conceivably interfere with inversion mechanically (Ueki and Nishii, 2008).
Here, however, we have documented the fact that during late cleavage and early inversion the GV increases in diameter faster than the embryo does, so that the space between the embryo and the GV is increased enough to make room for the embryo to invert (Figures 6A, 6C, and 6D, 2 to 8 h). By contrast, because the InvB vesicle does not increase in diameter any faster than the embryo does during this same interval, the InvB embryo lacks the space to invert (Figures 6B to D, 2 to 8 h). Our demonstration that the InvB embryo can invert normally if it is freed of its GV proves conclusively that the reason it normally fails to invert is because of the impediment imposed by a smaller than normal GV, rather than because of any deficiency in the inversion ability of the embryo itself.
The Size of the GV Is Controlled by the Embryo in a Process That Requires the GDP-Mannose Transport Activity of the InvB Protein
The size increase of the wild-type GV during embryogenesis is so reproducible in magnitude (Figure 6C) that it clearly must be a closely controlled process. The fact that this controlled enlargement process does not occur when the invB gene has been inactivated by a transposon insertion, but is fully restored when the transposon excises from the invB gene (Figure 1), shows that activity of InvB, the GDP-mannose transporter encoded by invB, is required for normal GV enlargement.
This requirement for GDP-mannose transporter activity to modify the extracellular matrix (ECM) is not without precedent. GDP-mannose transporter mutants that have defects in their cell walls or other extracellular materials have been reported in several other organisms. As just one example, such mutants of a fungal pathogen, Cryptococcus neoformans, make it unable to form its usual large capsules (Cottrell et al., 2007).
Both wild-type embryos that are isolated from the parent before the beginning of cleavage and forced to develop in isolation, as well as those that are allowed to develop within intact parental spheroids, develop a GV that encloses a space 30% larger than the embryo by the end of cleavage (Figure 6D). Under both conditions, the embryos are then able to invert normally. This demonstrates conclusively that the embryo itself is able to provide the InvB mannose transporter activity that is required for vesicle enlargement. Stated differently, each embryo is responsible for enlarging its own GV to make enough room for its subsequent inversion. How does it do this? Because there is a gap of increasing size between the embryo and its GV as the GV enlarges (Figure 6D), it is obvious that embryos do not cause their GVs to enlarge by simply pushing them outward physically. They presumably push on their GVs indirectly, with the help of molecules that they synthesize and secrete.
Expansion of the GV Requires a Structural Component That Is Mannosylated in Wild-Type Embryos but Not in InvB Mutant Embryos
It is well established that the major structural components of the Volvox ECM are Hyp-rich, fibrous glycoproteins, a substantial number of which have been characterized (Miller et al., 1974; Sumper and Hallmann, 1998; Hallmann, 2003). In their modular construction and peptide sequences, these glycoproteins bear a certain amount of resemblance to higher-plant cell wall proteins, and they play a variety of roles in such functions as mating, wound healing, and pathogen defense, in addition to their more obvious roles in structuring and supporting the organism (Sumper and Hallmann, 1998; Hallmann, 2003). In electron micrographs, the GV is seen to consist of densely packed fibers (Kirk et al., 1986), resembling in its appearance ECM elements elsewhere in the spheroid that are known to be glycoprotein rich; here, we have shown that when the wild-type GV increases in diameter during embryogenesis, it becomes enriched in mannose. These observations lead us to believe that the developing wild-type embryo produces a mannosylated glycoprotein that contributes directly to both the increase in the circumference of the GV and the increase in its fluorescent-ConA staining (as shown in Figure 5). The phenotype of the InvB mutant indicates clearly that the production of this ECM component requires the activity of the GDP-mannose transporter encoded by the invB gene. A diagram illustrating the manner in which we believe that the InvB protein participates in this process is provided in Figure 7.
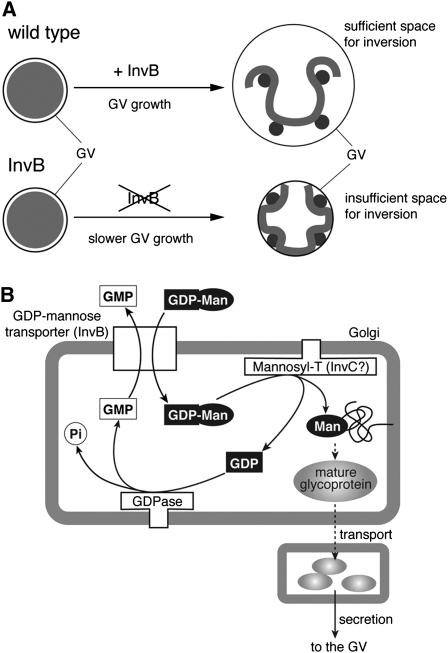
Figure 7.
Model Illustrating the Role That the InvB Protein Plays in Preparation for Inversion of a Volvox Embryo.
(A) Diagram illustrating why embryos lacking normal InvB protein fail to invert. In wild-type Volvox, there is a normal amount of InvB protein, so the GV grows enough during cleavage to provide the embryo with sufficient space for inversion. However, in the InvB mutant, InvB protein is lacking, and as a result the GV does not grow enough to provide the space the embryo needs to turn itself inside-out. A smaller than normal GV simply forms a physical barrier to the inversion movements.
(B) Predicted mechanism of glycosylation in the Volvox Golgi apparatus, based in part on the work of Hirschberg et al. (1998) and Kawakita et al. (1998). The GDP-mannose transporter encoded by invB transports GDP-mannose from the cytosol to the lumen of the Golgi apparatus, where it is used to add mannose residues to the side chains of glycoproteins in reactions catalyzed by specific mannosyltransferases (one of which probably is the InvC protein; Ueki and Nishii, 2008). Such a mannose-rich glycoprotein is then secreted and involved in some critical way in the normal enlargement of the GV surrounding the embryo.
Another mutant, InvC, which we recovered from the same mutant screen as InvB, is also informative. InvC exhibits a more severe noninverter phenotype than InvB; it cannot initiate inversion at all, and it has an even smaller GV than InvB at the stage when inversion should occur (Ueki and Nishii, 2008). Importantly, a GV-free InvC embryo also inverts successfully, just as InvB does (our unpublished data), indicating that in this mutant also, the reason for the inversion defect resides in the smaller than usual GV. Because invC encodes a putative glycosyltransferase belonging to the LARGE family (Grewal et al., 2005), it is likely that InvC is involved in glycosylation together with InvB. In Figure 7B, we propose that InvB transports GDP-mannose into the Golgi lumen, and InvC then uses the GDP-mannose to add mannose to one or more glycoproteins that are secreted and used to make the GV larger. (The more severe phenotype of InvC might be because InvC catalyzes other types of glycosylation reactions that are also required for GV growth.)
Our supposition is that the mannosylated glycoprotein(s) produced by the sequential actions of InvB and InvC function by inserting into the preexisting fibrous network of the GV, thereby permitting the GV to expand a bit. This would be far from the first example of self-assembly and growth of a structural element of the Volvox ECM at some distance from the nearest cells. The V. carteri ECM has a highly complex and regular morphology, involving a diverse array of fibrous elements that surround the entire spheroid, form individual compartments around each cell, separate different regions of the spheroid, etc. (Kirk et al., 1986). The first step in the formation of this complex ECM has been shown to be the self-assembly of a single secreted glycoprotein, inversion-specific glycoprotein (ISG), into a fibrous network that surrounds each newly inverted embryo, forming the inner layer of the boundary zone of ECM that will eventually surround the adult and hold its cells firmly in place. Once assembled, the ISG network serves as a scaffold upon which additional fibrous elements then self-assemble on both its inner and outer surfaces (Hallmann and Kirk, 2000). If, for any reason, assembly of the ISG layer fails to occur during the first few hours after inversion has been completed, none of the rest of the ECM is assembled, and the postinversion embryo falls apart into a single-cell suspension (Ertl et al., 1992; Hallmann and Kirk, 2000). ECM self-assembly does not end with the embryonic period, however, because during the 2 d following inversion, the Volvox spheroid increases in volume by ∼10,000-fold, with all of its component ECM structures increasing in size proportionately, and most of this ECM expansion occurs at a considerable distance from the nearest cell (just as the expansion of the GV does during embryogenesis).
Throughout the period of spheroid expansion, glycoprotein components of the ECM are synthesized within the somatic cells via a conventional rough endoplasmic reticulum, Golgi apparatus, secretory vesicle pathway, and distinct fibrous elements of significant size can already be visualized within the secretory vesicles before they reach the cell surface (Dauwalder et al., 1980; Sumper and Hallmann, 1998).
A Second Type of Secreted Product Is Almost Certainly Required for GV Expansion
In all likelihood, there is at least one additional type of secretory product of the embryo that is involved in GV expansion: a less fibrous, highly hydrophilic polymer that attracts water from the medium and forms a mucilaginous sol that inflates the space internal to the GV (Figures 5 and 6). Several considerations lead us to propose the existence of such a mucilaginous component: First, formation of these two distinctive types of ECM components, fibrous and mucilaginous, is a regular and characteristic feature of most members of the order Volvocales, including Volvox (reviewed in Kirk, 1998), and multiple investigators over many years have inferred that all of the compartments of the Volvox ECM that are bounded by fibrous elements are filled with mucilaginous material (also reviewed in Kirk et al., 1986). Second, in her ground-breaking mutational analysis of Volvox development, Sessoms (1974) found that when a mutation knocked out the production of a mucilaginous product that normally would have filled a particular ECM compartment, the fibrous component that would normally have formed a smooth border around that compartment accumulated in a shapeless heap of randomly folded sheets, like the skin of an collapsed balloon. But instead of that sort of disheveled appearance, the V. carteri GV maintains a nearly perfect spherical shape during all stages of its enlargement, like a fully inflated balloon (Figures 5 and 6). Third, when we were performing microsurgery to remove the GVs from one-cell embryos (Figure 6E), we often observed that as soon as we nicked the GV with the tungsten needle, the embryo popped part way out through the open hole, as if the contents of the GV had been under pressure.
We propose that it is hydrostatic pressure exerted by one or more hydrophilic, mucilaginous ECM components that is secreted by the embryo and accumulated in the space enclosed by the GV that is responsible for keeping the GV in an inflated, spherical configuration and for causing it to expand as additional fibrous, mannosylated glycoprotein molecules are added to it.
What the chemical nature of this postulated mucilaginous substance might be is uncertain. In contrast with the ECM glycoproteins, which have received considerable attention from investigators, the chemistry of the mucilaginous components of the Volvox ECM is essentially unexplored territory. However, preliminary studies of such materials in two close relatives of Volvox (Eudorina/Pleodorina californica, Tautvydis, 1978; and Platydorina caudata, Crayton, 1980) suggest that such mucilage may be a rather complex mixture of highly sulfated polysaccharides and proteoglycans.
Other GDP-Mannose Transporter Proteins May Be Involved in Synthesis of Other Important ECM Components
The primary sequence and function of InvB was deduced from the sequences of invB cDNA and genomic clones. However, we now know that in the V. carteri genome database (http://genome.jgi-psf.org/Volca1/Volca1.home.html) there are seven putative genes in addition to invB that encode nucleotide-sugar transporters related to VRG4 of yeast and GONST1 of Arabidopsis thaliana (see Supplemental Table 1 online). A phylogenetic analysis indicates that two of these genes are much more closely related to invB than the other five are and that each of these three closely related homologs has an apparent ortholog in Chlamydomonas reinhardtii (see Supplemental Table 1 and Supplemental Figure 1 online). The mutant phenotype of the InvB mutant clearly indicates, however, that none of those other GDP-mannose transporters is able to substitute for InvB in the production of the mannosylated ECM component that is required for GV expansion. The concept that invB is expressed at a much higher level than its two most closely related paralogues is reinforced by the observation that the Joint Genome Institute homepage identifies nine EST clones corresponding to invB, but none corresponding to Vca_89221 or Vca_66734 (http://genomeportal.jgi-psf.org/Volca1/Volca1.home.html). Thus, although those two proteins might be similar enough to InvB to have some overlapping functions, the fact that they are unable to support normal GV expansion may be due to their very low expression levels.
The InvB GV is not constant in size during embryogenesis, but enlarges slightly (Figure 6C). This might be the result of a small contribution from the redundant function of one or both of the closely related proteins discussed above. Alternatively, because the InvB embryo is pressed tightly against the GV during cleavage, it is conceivable that it exerts enough outward pressure on the GV to cause it to expand slightly.
Multiple GDP-mannose transporters that execute different functions have been described in land plants. For example, it has been shown that the five GONST family GDP-mannose transporters of Arabidopsis exhibit different tissue-specific expression patterns (Baldwin et al., 2001; Handford et al., 2004). Therefore, it is possible that InvB and the seven other GDP-mannose transporters in Volvox may play different specialized roles in production of various parts of the complex ECM of Volvox. It would be of interest to determine the stage and cell type specificity of expression of the genes encoding the various GDP-mannose transporters. It would be equally interesting to screen for transposon-induced ECM mutants of the sort recovered by Sessoms following chemical mutagenesis of Volvox (i.e., expansionless and dissolver mutants; Sessoms and Huskey, 1973; Sessoms, 1974) and to determine whether any of them are the result of mutations in the genes encoding nucleotide-sugar transporters.
The invB Function May Have Evolved before V. carteri Did
Extensive similarities between the glycoproteins of the V. carteri ECM and the cell walls of C. reinhardtii (the closest unicellular relative of V. carteri) in modular construction patterns, amino acid compositions, sequence motifs, glycosylation patterns, and morphological patterns at the electron microscopic level have been described repeatedly (Adair et al., 1987; Goodenough and Heuser, 1988; Woessner and Goodenough, 1994; Sumper and Hallmann, 1998). Indeed, glycoproteins of the crystalline layer of the Chlamydomonas cell wall and the V. carteri boundary zone are so similar that they can substitute for one another in an in vitro reassembly system (Adair et al., 1987). Now we find that these two species are similar with respect to genes encoding nucleotide-sugar transporters of the VRG4/GONST1/InvB family also: There are eight such genes in V. carteri, and seven of them in C. reinhardtii (Merchant et al., 2007; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), including one, IBR1 (invB related gene 1) that appears to be orthologous to invB (Figure 2F; see Supplemental Figure 1 online). These findings suggest strongly that such genes and the proteins that they encode must have been present ∼200 million years ago in the last common ancestor of unicellular C. reinhardtii and multicellular V. carteri (Herron et al., 2009).
It would be interesting to know whether the functions of any of these nucleotide-sugar transporters have been conserved as long as their sequences have. For example, is it possible that the protein encoded by IBR1 plays an essential role in the enlargement of the cell wall that occurs during the C. reinhardtii cell cycle? (During its characteristic cell cycle, C. reinhardtii increases in volume as much as 32-fold before undergoing up to five rapid cleavage divisions inside the mother cell wall, and during that enlargement period, there is evidence of considerable turnover of cell wall glycoproteins [Voigt, 1985].) Each new finding leads to many interesting new questions.
METHODS
Volvox carteri Strains, Cultivation Conditions, and Isolation of the InvB Mutant
The standard wild-type strain of V. carteri that is currently used for many studies in our laboratory and others is EVE (Adams et al., 1990), which is a subclone of HK10, the female strain originally isolated by Richard Starr (Starr, 1968, 1969). However, the morphologically wild-type strain that was used as the progenitor of the InvB mutants in this investigation was CRH22, which was derived from EVE by mutagenesis (Miller et al., 1993). CRH22 and all of its progeny carry a partial deletion of nitA, the nitrate reductase–encoding locus, which makes them suitable for cotransformation studies, using the nitA gene as the selectable marker. All cultures were maintained in standard Volvox medium (SVM), and all other culture conditions and procedures, and the methods used for isolation of the mutants, were as previously described (Ueki and Nishii, 2008). Two independent InvB mutants (subsequently named InvB1 and InvB2) were isolated from CRH22 cultures maintained at 24°C, the temperature at which transposition of both the Jordan and Idaten transposons is activated. Both of these strains were genetically stable when maintained at 32°C, but produced wild-type revertants with a moderate frequency when cultured at 24°C for ∼10 d, indicating that their mutations were very likely due to insertions of cold-inducible transposons.
Most of the microscopy studies to measure the size of wild-type and mutant embryos were performed using synchronized 2-d-old cultures. Spheroids containing embryos at each desired stage were broken by a Dounce homogenizer, and the released embryos were quickly collected and photographed. For a few supplemental studies discussed in the text, we isolated embryos at the beginning of cleavage and allowed them to develop in isolation under standard culture condition until the beginning of inversion.
DNA Gel Blot Analysis
Preparation of genomic DNAs, electrophoresis, blotting, and detection of Idaten-positive bands were all performed as in the previous article (Ueki and Nishii, 2008), except that in this case the genomic DNAs were digested with SacI (TOYOBO) at 37°C for at least 8 h.
Identification and Cloning of the invB Locus
DNA enriched in the InvB1-specific 1.7-kb SacI fragment (Figure 2A, arrowhead) was prepared as follows: 50 μg of InvB1 genomic DNA was digested with 250 units of SacI at 37°C overnight; the digest was then electrophoresed overnight in a Mupid-ex preparative gel apparatus (Advance) using 1.2% SeaPlaque GTG Agarose (Cambrex Bio Science) in TAE buffer with circulation at 4°C at 25 V with a GeneRuler 1-kb ladder (Fermentas) in the adjacent lane. The region of the gel in the ∼1.4- to 3-kb fragment range was fractionated into 3-mm sections with a razor blade, and the section containing the 1.7-kb SacI fragment was then identified by DNA gel blotting, and its DNA was purified with a QIAEXII gel extraction kit (Qiagen). Self-ligation of one-half of the resulting DNA was performed using 2 units of T4 ligase (TOYOBO) at 37°C for 1 h in a total volume of 450 μL; then 10 μL of this ligation mixture was used directly for inverse PCR in a total volume of 50 μL. The PCR reaction program was 94°C for 5 min followed by 32 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min; the final incubation was at 68°C for 6 min. The primers used were IF02 (5′-TGCTGAATCTGCCAAGTAGGCTCT-3′) and IR02 (5′-TGGCTGTTATGCCACGTTGACAAC-3′) as previously reported for the cloning of invC (Ueki and Nishii, 2008). The amplified fragment, which had an estimated length of 0.7 kb, was purified from an agarose gel and sequenced directly using IF02 and IR02 primers.
In an effort to confirm that this inverse PCR product was derived from the 1.7-kb genomic fragment, we used it as a PCR template with the primer pair BF01 (5′-TGACACGCCCGCAATTCCGTTAT-3′) and BR01 (5′-TTTCTGCTCTCGGCG GCATCATT-3′) and amplified a 0.4-kb fragment. That product, subsequently called probe B (Figure 2C), consistently detected a 3-kb SacI fragment in DNAs from CRH22 and the InvB1 revertants, but only a 1.7-kb fragment in InvB1 DNA. Probe B was also used for analyzing InvB2 and its revertants (Figure 2B).
A single invB cDNA clone was retrieved from our presequenced full-length cDNA library (I. Nishii, unpublished data) and cloned into a plasmid that we named pfcinvB. In the Joint Genome Initiative Volvox database (http://genome.jgi-psf.org/Volca1/Volca1.home.html), it is consistent with a single gene model (protein ID: 105037, scaffold_22: 1764313-1767060), although a cDNA corresponding to this model would have seven extra bases at the 5′ untranslated region, as has been found in a single EST clone (JGI_CBGZ14932).
A genomic clone covering the invB locus was generated as follows: A V. carteri BAC filter (VCNEBb) from the Arizona Genomics Institute (http://www.genome.arizona.edu) was hybridized with probe B (using the methods of hybridization and detection previously described for DNA gel blot analyses; Ueki and Nishii, 2008), and seven positive clones were detected. DNA from one of those positive clones (ID: 14M17) was prepared with the Plasmid Midi Kit (Qiagen) and digested with BamHI and XhoI. The resulting digest was electrophoresed, and an ∼7-kb band including the invB gene (Figure 2C) was gel purified and subcloned into pBluescript KS− (KS-inv423BX-7kb).
Construction of an Epitope-Tagged Version of the invB Gene
An HA epitope tag (YPYDVPDYA; Jarvik and Telmer, 1998) was inserted near the N-terminal of invB using a PCR-oriented method (Sambrook et al., 1989). The oligonucleotide encoding the HA tag (5′-TACCCGTACGACGTCCCGGACTACGCC-3′) with 12 additional nucleotides (5′-GGTTGGGGT-3′) was inserted between the 4th and 5th codons of the InvB-coding region of invB using KS-inv423BX-7kb that contains 2.1 kb upstream of the invB transcript (Figure 2C, XhoI → BamHI fragment). The resulting plasmid, ks_inv423BX_7kb_N_HA, was sequenced and then used for transformation of the InvB1 mutant.
DNA Sequencing
DNA sequencing was performed on an ABI 3730 DNA Analyzer using BigDye v3.1 (Applied Biosystems) with the M13 forward and M13 reverse primers used for PCR and was extended by primer walking.
Nuclear Transformation
Biolistic cotransformation with the nitA gene carried on plasmid pVcNR15 as the selectable marker (Schiedlmeier et al., 1994; Gruber et al., 1996) was used to test the ability of genomic invB clones and an epitope-tagged invB transgene to rescue InvB mutants. Isolated embryos in a vacuum chamber were bombarded with DNA-coated gold particles using a commercial ballistic device (IDERA GIE-III; Tanaka) and 0.65 MPa of helium gas. Bombarded samples were suspended in selective medium lacking reduced nitrogen, dispensed into multiwell plates, and monitored for the appearance of green, growing (Nit+) survivors, each of which was cultured in fresh selective medium and scored for its inversion phenotype. No more than one wild-type strain was saved per initial culture well. We then checked each such putative cotransformant with wild-type morphology to ensure that it was really a transformant, rather than a revertant, by performing genomic PCR to amplify both the host invB locus containing the Idaten insertion and the invB transgene lacking such an insertion. Primers BF01 and IR02 (5′-TGGCTGTTATGCCACGTTGACAAC-3′) were used to amplify a 535-bp product from the InvB1 gene containing Idaten, and primers BF01 and BR5772 (5′-TGCGTATGGCAAGATTCCGTGGTA-3′) were used to amplify an 866-bp product from the invB transgene lacking Idaten.
Antibody Production and Immunoblotting
Anti-InvB antiserum was raised in rabbits against a synthetic peptide (NH2-CKVAEHTTNKQKMDEF-COOH; the italicized region of this sequence corresponds to residues 219 to 233 of InvB, as indicated by an over-bar in Figure 2F). Volvox extracts for immunoblots were prepared as previously described (Kirk et al., 1999) except for omitting the boiling step before loading (because boiling causes aggregation of hydrophobic proteins, like InvB, that have multiple membrane-spanning motifs; Burke et al., 2000; Baldwin et al., 2001). SuperSep Ace 10% (Wako Pure Chemical Industries) was used for SDS-PAGE; following electrophoresis, the proteins were transferred to Hybond-P PVDF Membrane (GE Healthcare) in a semidry type blotter (NA-1512; Nihon Eido) for 60 min at 2 mA/cm2; the membrane was blocked with TBS containing 0.1% Tween 20 (TTBS) and 5% nonfat dried milk overnight at 4°C and washed with TTBS three times. The membrane was then incubated with a primary antibody diluted in the blocking solution (anti-InvB, 1:2000, or anti PSTAIRE, 1:2000 [Santa Cruz Biotechnology]; anti-HA, 1:5000 [Sigma-Aldrich]; anti-actin, 1:5000 [provided by D. Kirk]) for 60 min. After washing the membrane with TTBS three times, immunoreactive proteins were visualized with an horseradish peroxidase–linked secondary antibody incubated for 60 min at room temperature (1:2000 for InvB, 1:1000 for PSTAIRE, and 1:5000 for HA and actin [Amersham Biosciences] in blocking solution) plus SuperSignal Chemiluminescent Substrate (Pierce Biochemicals). The chemiluminescent image was recorded with VersaDoc Imaging system model 5000 with Quantity One software (Bio-Rad).
Microscopy and Morphometry
Light microscopy images of adult spheroids at lower magnification were acquired on a SMZ1500 dissection microscope with an HR Plan Apo ×1.6 objective (WD24; Nikon) equipped with a ZEISS AxioCam MRc5 CCD camera and Axiovision 4.6.3 Software on PC (Carl Zeiss). DIC images of isolated embryos from synchronized cultures were taken using the same camera and software on a ZEISS Axio Imager Z1 microscope with an EC Plan-Neofluar ×40/0.75 objective. To measure the size of the GV and the embryo in such an image, the inner perimeter of the GV and the outline of the embryo were traced manually, using Adobe Photoshop CS (Adobe Systems), and then the areas bounded by the trace were measured using the NIH ImageJ 1.38 analysis program (http://rsb.info.nih.gov/ij/; Abramoff et al., 2004).
For video microscopy, isolated embryos were attached to 0.01% PEI-coated cover slips prepared as previously described (Nishii and Ogihara, 1999), and DIC images at 30-s intervals were recorded with the same microscope, lens, and software described above. These time-lapse images were processed with the NIH ImageJ program and converted by QuickTime 7 Pro software on PowerMac G5 (Apple) into the movie file included here as Supplement Movie 1 online.
For fluorescent detection of α-mannosyl–rich structures, living embryos were incubated with 100 μg/mL Alexa fluor 488–conjugated ConA (Invitrogen Molecular Probes) in SVM for 30 min at room temperature in darkness; they were then washed three times with SVM (being pelleted centrifugally after each wash) and were resuspended in fresh SVM. Fluorescence was visualized using a Zeiss inverted laser scanning confocal microscope (LSM 510 META system) with excitation at 488 nm and a water immersion objective lens C-Apochromat ×40/1.2W. Images were captured with LSM510 v3.5 software (Carl Zeiss).
Real-Time RT-PCR Analysis
Stage-specific total RNAs were purified from embryos that were isolated from synchronized EVE cultures, frozen in liquid nitrogen, ground with a cold mortar and pestle, and then extracted with the TRI reagent (Sigma-Aldrich) according to the manufacturer′s protocol. Ten micrograms of RNA from each stage was treated with DNase (Ambion turbo DNA-free kit) and then reverse transcribed to generate cDNA using the Takara Prime Script kit and random primer.
Real-time RT-PCR analysis was performed with an Applied Biosystems 7500 Real-time PCR system with software version 1.4.0; we used the software for the 2−ΔΔCt method. As a calibrator, we analyzed a 1:40 diluted template cDNA (1.0 μL) mixed with Universal TaqMan mix (12.5 μL) and Pre-Developed TaqMan Assay Reagents 18S rRNA premix (including primers) (1.25 μL), in a total reaction volume of 25 μl. For invB, we designed a TaqMan probe and primers for amplifying a sequence spanning the 1st and 2nd exons, using Primer Express version 3.0, as a TaqMan probe (invBcDNA-552T; 5′-CAATGTTGTGACAGCAAT-3′) and a primer pair, invBcDNA-488F (5′-GCACCAGCTTCTATGCGCTAA-3′) and invBcDNA-607R (5′-GCCATGAGAATGTCCGTTTGT-3′), and then checked that the amplification efficiency of this probe was optimal. One microliter of the undiluted cDNA mixed with the Universal TaqMan mix (12.5 μL), 2.25 μL of each primer (10 μM), and 1 μL of TaqMan probe (5 μM) in a total volume of 25 μL was analyzed. All analytical PCR reactions used a default program setting (50°C for 2 min, 95°C for 10min, 40 cycles of 95°C for 15 s, and 60°C for 1 min) and were followed by monitoring the dissociation curve of the invB probe to establish that consistent Tm was obtained in all experiments. For each developmental stage, the Ct value of the calibrator was subtracted from the Ct of the invB probe to obtain ΔCt. Then, we set the 9-h stage (where invB is expressed maximally) as the reference and calculated ΔΔCt for each stage with the formula ΔΔCt = ΔCt (that stage) – ΔCt (9 h). Finally, 2−ΔΔCt for each stage was obtained for comparison of the invB expression. For each time point, triplicate PCR amplifications were performed.
Yeast Strains, Growth Conditions, and Transformation
Saccharomyces cerevisiae strains JPY25 6c (MATα ura3-52 his3-Δ200 trp1-Δ901 ade2-101 dpm1) and JPY26 3d (MATα ura3–52 leu2–3 112 ade2–101 vrg4–2 dpm1) were kindly donated by N. Dean (Stony Brook University) and were grown on synthetic complete agar (Sherman, 1991) supplemented with 0.5 M KCl (Poster and Dean, 1996) at 30°C. JPY26 3d transformed with the URA3-containing plasmid was selected on SD-uracil agar. Yeast transformation was performed using the lithium acetate method (Elble, 1992). For immunoblotting, overnight cultured samples were prepared with the glass bead method (Burke et al., 2000). For preparation of membrane vesicles, strains were grown in liquid selective medium until they reached a density of 0.8 to 1.0 OD600.
For the generation of a construct containing the invB gene, the invB cDNA sequence that contains additional EcoRI and XhoI restriction sites was amplified by PCR using pfcinvB as a template and the following primer pair (5′-CTTTTCCTTGTG AATTCATGTGGGGTTTGC-3′ and 5′-ATCCCTCATCCTCGAGCTATTTGCTCTTG-3′). The resultant PCR fragment was digested with EcoRI and XhoI and subcloned into the yeast expression vector p352-GAP, which was also kindly supplied by N. Dean (Gao et al., 1999). The construct was checked by DNA sequencing, and then it and the empty p352-GAP vector were transformed into yeast strain JPY26 3d.
Preparation of Yeast Membrane Vesicles
Preparation of membrane vesicles was performed by the procedure of Ishida et al. (2005), with slight modifications. Yeast cells were washed with ice-cold 10 mM NaN3 and converted into spheroplasts by incubation at 37°C for 90 min in a solution containing 1.4 M sorbitol, 50 mM potassium phosphate, pH 7.5, 10 mM NaN3, 0.25% (v/v) 2-mercaptoethanol, and 2 mg of Zymolyase-100T (Seikagaku-kogyo) per gram of cells. The spheroplasts were pelleted and resuspended in 4 volumes of lysis buffer containing 0.8 M sorbitol, 10 mM HEPES-Tris, pH 7.4, 1 mM EDTA, and a protease inhibitor cocktail (Complete Mini; Roche Diagnostics) and then homogenized by passing 10 times through a 22G needle attached to a 10-mL syringe and homogenized further by passing 10 times through a 26G needle attached to a 10-mL syringe on ice. The lysate was centrifuged at 200g for 5 min at 4°C to remove unlysed cells and debris, whereupon the supernatant was centrifuged at 100,000g for 60 min to yield a pellet of the P100 membrane vesicle fraction that was then resuspended in lysis buffer.
Nucleotide-Sugar Transport Assay
The transport assay was performed essentially as previously described (Sun-Wada et al., 1998; Ishida et al., 2005). The P100 vesicle fractions were incubated at 30°C in a reaction mixture containing 0.8 M sorbitol, 10 mM Tris-HCl, pH7.0, 1 mM MgCl2,0.5 mM dimercaptopropanol, and 10 μM radiolabeled substrate (guanosine diphosphate mannose, [d-mannose-1-14C]; American Radiolabeled Chemicals). After 1 min, reactions were stopped with 1 mL of ice-cold stop buffer (0.8 M sorbitol, 10 mM Tris-HCl, pH 7.0, 1 mM MgCl2, 0.5 mM dimercaptopropanol, and 10 μM nonradiolabeled substrate). Then the vesicles were washed onto 0.45-μm nitrocellulose filters (25 mm diameter; Advantec) by vacuum filtration and rinsed 10 times with 1 mL of ice-cold wash buffer (0.8 M sorbitol, 10 mM Tris-HCl, pH 7.0, 1 mM MgCl2, and 0.5 mM dimercaptopropanol). The vesicles were dried, and the filters were transferred to vials containing 3 mL of scintillation fluid. The radioactivity trapped on the filter was determined by a Beckman LS-6500 liquid scintillation counter (Beckman Coulter).
Microsurgery
The GV was removed from one-cell embryos by the previously described method (Nishii and Ogihara, 1999) with modifications as follows: Isolated gonidia were incubated, with pipetting, in SVM containing 0.1 mg/mL trypsin (Sigma-Aldrich) for 30 s and washed with an excess volume of SVM. Then they were incubated in SVM containing 1% BSA for 2 to 3 min and washed with an excess volume of SVM. The gonidia in ∼500 μL of SVM were allowed to settle on, and attach weakly to, a microscope slide that had been coated with 0.003% polyethylenimine for ∼5 min; then 1 mL of SVM containing 1% BSA was added to coat the gonidia-free regions of the glass surface. The GVs, which had been softened by the trypsin pretreatment, were then removed from the embryos with a fine tungsten needle sharpened by electrolysis. The GV-free embryos were transferred to SVM contained in a plastic six-well plate and allowed to develop at 32°C.
Accession Numbers
The complete sequence of the invB gene was submitted to DDBJ as accession number AB472041. Sequence data from this article can be found in the GeneBank/EMBL data libraries under the following accession numbers: C. reinhardtii IBR1 (XM_001691067), Arabidopsis GONST1 (AJ314836), S. cerevisiae VRG4 (NP_011290), Candida globrata CaVRG4 (AF164627), and Leishmania donovani LPG2 (U26175). As for C. reinhardtii IBR1, we used a modified model by combining with the C-terminal region of a gene model (protein ID: 114747) from C. reinhardtii genome version 3 (http://www.jgi.doe.gov/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phylogenetic Relationships of the invB Homologs to One Another and to Arabidopsis GONST1 and Yeast VRG4.
Supplemental Table 1. The invB Homologs in the V. carteri and C. reinhardtii Genomes.
Supplemental Data Set 1. Text File of Alignment in Figure 2F.
Supplemental Data Set 2. Text File Corresponding to the Phylogenetic Tree in Supplemental Figure 1.
Supplemental Movie 1. Time-Lapse Video of Wild-Type and InvB Embryos during the Time Period when Inversion Occurs in the Wild-Type Embryo.
Supplementary Material
Acknowledgments
We thank D.L. Kirk for insightful suggestions, critical reading of this manuscript, and providing anti-actin antibody; N. Dean for providing plasmids and yeast strains; T. Kishi for anti-PSTAIRE antibody; H. Inadome for helpful suggestions on yeast studies; J. Seino for help with yeast preparation and transport assay; M. Mitsuki for help with ConA fluorescent microscopy; Y. Hashimoto and S. Kitazume-Kawaguchi for their helpful advice on glycobiology; and J. Kadota and A. Nakazawa in our lab for help with the yeast studies and culture maintenance of Volvox, respectively. We are grateful to the Research Resource Center, RIKEN Brain Science Institute, for their support of the DNA sequencing and antibody production. Sequencing of the V. carteri and C. reinhardtii genome has been performed by the Joint Genome Institute in Walnut Creek, CA (http://www.jgi.doe.gov/). A Volvox BAC filter was provided by the Arabidopsis Genome Initiative, Green BAC project. This study was supported by the RIKEN Special Postdoctoral Researchers Program (N.U.) and the RIKEN Initiative Research Unit Program (I.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Noriko Ueki (noriko.ueki@uni-bielefeld.de) and Ichiro Nishii (ichiron@mac.com).
Online version contains Web-only data.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image Processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- Adair, W.S., Steinmetz, S.A., Mattson, D.M., Goodenough, U.W., and Heuser, J.E. (1987). Nucleated assembly of Chlamydomonas and Volvox cell walls. J. Cell Biol. 105 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, C.R., Stamer, K.A., Miller, J.K., McNally, J.G., Kirk, M.M., and Kirk, D.L. (1990). Patterns of organellar and nuclear inheritance among progeny of two geographically isolated strains of Volvox carteri. Curr. Genet. 18 141–153. [DOI] [PubMed] [Google Scholar]
- Baldwin, T.C., Handford, M.G., Yuseff, M.I., Orellana, A., and Dupree, P. (2001). Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., and Sterns, T. (2000). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Cottrell, T.R., Griffith, C.L., Liu, H., Nenninger, A.A., and Doering, T.L. (2007). The pathogenic fungus Cryptococcus neoformans expresses two functional GDP-mannose transporters with distinct expression patterns and roles in capsule synthesis. Eukaryot. Cell 6 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crayton, M.A. (1980). Prezence of a sulfated polysaccharide in the extracellular matrix of Platydori caudata (Volvocales, Chlorophyta). J. Phycol. 16 80–87. [Google Scholar]
- Dauwalder, M., Whaley, W.G., and Starr, R.C. (1980). Differentiation and secretion in Volvox. J. Ultrastruct. Res. 70 318–335. [DOI] [PubMed] [Google Scholar]
- Dean, N., Zhang, Y.B., and Poster, J.B. (1997). The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 272 31908–31914. [DOI] [PubMed] [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13 18–20. [PubMed] [Google Scholar]
- Ertl, H., Hallmann, A., Wenzl, S., and Sumper, M. (1992). A novel extensin that may organize extracellular matrix biogenesis in Volvox carteri. EMBO J. 11 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X.D., Kaigorodov, V., and Jigami, Y. (1999). YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J. Biol. Chem. 274 21450–21456. [DOI] [PubMed] [Google Scholar]
- Gao, X.D., Nishikawa, A., and Dean, N. (2001). Identification of a conserved motif in the yeast golgi GDP-mannose transporter required for binding to nucleotide sugar. J. Biol. Chem. 276 4424–4432. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1988). Molecular organization of cell-wall crystals from Chlamydomonas reinhardtii and Volvox carteri. J. Cell Sci. 90 717–733. [Google Scholar]
- Green, K.J., Viamontes, G.I., and Kirk, D.L. (1981). Mechanism of formation, ultrastructure, and function of the cytoplasmic bridge system during morphogenesis in Volvox. J. Cell Biol. 91 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, P.K., McLaughlan, J.M., Moore, C.J., Browning, C.A., and Hewitt, J.E. (2005). Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology 15 912–923. [DOI] [PubMed] [Google Scholar]
- Gruber, H., Kirzinger, S.H., and Schmitt, R. (1996). Expression of the Volvox gene encoding nitrate reductase: Mutation-dependent activation of cryptic splice sites and intron-enhanced gene expression from a cDNA. Plant Mol. Biol. 31 1–12. [DOI] [PubMed] [Google Scholar]
- Hallmann, A. (2003). Extracellular matrix and sex-inducing pheromone in Volvox. Int. Rev. Cytol. 227 131–182. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., and Kirk, D.L. (2000). The developmentally regulated ECM glycoprotein ISG plays an essential role in organizing the ECM and orienting the cells of Volvox. J. Cell Sci. 113 4605–4617. [DOI] [PubMed] [Google Scholar]
- Handford, M.G., Sicilia, F., Brandizzi, F., Chung, J.H., and Dupree, P. (2004). Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol. Genet. Genomics 272 397–410. [DOI] [PubMed] [Google Scholar]
- Hay, E.D. (1989). Extracellular matrix, cell skeletons, and embryonic development. Am. J. Med. Genet. 34 14–29. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Herron, M.D., Hackett, J.D., Aylward, F.O., and Michod, R.E. (2009). Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. USA 106 3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg, C.B., Robbins, P.W., and Abeijon, C. (1998). Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67 49–69. [DOI] [PubMed] [Google Scholar]
- Ishida, N., Kuba, T., Aoki, K., Miyatake, S., Kawakita, M., and Sanai, Y. (2005). Identification and characterization of human Golgi nucleotide sugar transporter SLC35D2, a novel member of the SLC35 nucleotide sugar transporter family. Genomics 85 106–116. [DOI] [PubMed] [Google Scholar]
- Jarvik, J.W., and Telmer, C.A. (1998). Epitope tagging. Annu. Rev. Genet. 32 601–618. [DOI] [PubMed] [Google Scholar]
- Kam, Z., Minden, J.S., Agard, D.A., Sedat, J.W., and Leptin, M. (1991). Drosophila gastrulation: Analysis of cell shape changes in living embryos by three-dimensional fluorescence microscopy. Development 112 365–370. [DOI] [PubMed] [Google Scholar]
- Kawakita, M., Ishida, N., Miura, N., Sun-Wada, G.H., and Yoshioka, S. (1998). Nucleotide sugar transporters: Elucidation of their molecular identity and its implication for future studies. J Biochem 123 777–785. [DOI] [PubMed] [Google Scholar]
- Kelland, J.L. (1977). Inversion in Volvox (Chlorophyceae). J. Phycol. 13 373–378. [Google Scholar]
- Keller, R.E. (1981). An experimental analysis of the role of bottle cells and the deep marginal zone in gastrulation of Xenopus laevis. J. Exp. Zool. 216 81–101. [DOI] [PubMed] [Google Scholar]
- Kirk, D.L. (1998). Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation, Developmental and Cell Biology Series, No. 33. (Cambridge, UK: Cambridge University Press).
- Kirk, D.L., Birchem, R., and King, N. (1986). The extracellular matrix of Volvox: a comparative study and proposed system of nomenclature. J. Cell Sci. 80 207–231. [DOI] [PubMed] [Google Scholar]
- Kirk, D.L., Viamontes, G.I., Green, K.J., and Bryant, J.L. (1982). Integrated morphogenetic behavior of cell sheets: Volvox as a model. In Developmental Order: Its Origin and Regulation, S. Subtelny & P.B. Green, eds (Alan R. Liss, New York), p. 274.
- Kirk, D.L., and Nishii, I. (2001). Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Dev. Growth Differ. 43 621–631. [DOI] [PubMed] [Google Scholar]
- Leptin, M. (2005). Gastrulation movements: The logic and the nuts and bolts. Dev. Cell 8 305–320. [DOI] [PubMed]
- Kirk, M.M., Stark, K., Miller, S.M., Muller, W., Taillon, B.E., Gruber, H., Schmitt, R., and Kirk, D.L. (1999). regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126 639–647. [DOI] [PubMed] [Google Scholar]
- Mathur, J. (2004). Cell shape development in plants. Trends Plant Sci. 9 583–590. [DOI] [PubMed] [Google Scholar]
- Merchant, S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.H., Mellman, I.S., Lamport, D.T., and Miller, M. (1974). The chemical composition of the cell wall of Chlamydomonas gymnogama and the concept of a plant cell wall protein. J. Cell Biol. 63 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.M., Schmitt, R., and Kirk, D.L. (1993). Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii, I., and Ogihara, S. (1999). Actomyosin contraction of the posterior hemisphere is required for inversion of the Volvox embryo. Development 126 2117–2127. [DOI] [PubMed] [Google Scholar]
- Nishii, I., Ogihara, S., and Kirk, D.L. (2003). A kinesin, invA, plays an essential role in Volvox morphogenesis. Cell 113 743–753. [DOI] [PubMed] [Google Scholar]
- Pilot, F., and Lecuit, T. (2005). Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev. Dyn. 232 685–694. [DOI] [PubMed] [Google Scholar]
- Pocock, M.A. (1933). Volvox and associated algae from Kimberly. Annals of the South African Museum. 16 473–521. [Google Scholar]
- Poster, J.B., and Dean, N. (1996). The yeast VRG4 gene is required for normal Golgi functions and defines a new family of related genes. J. Biol. Chem. 271 3837–3845. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiedlmeier, B., Schmitt, R., Muller, W., Kirk, M.M., Gruber, H., Mages, W., and Kirk, D.L. (1994). Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 91 5080–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock, F., and Perrimon, N. (2002). Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol. 18 463–493. [DOI] [PubMed] [Google Scholar]
- Schoenwolf, G.C., and Franks, M.V. (1984). Quantitative analyses of changes in cell shapes during bending of the avian neural plate. Dev. Biol. 105 257–272. [DOI] [PubMed] [Google Scholar]
- Sessoms, A.H. (1974). The Isolation and Characterization of Morphogenetic Mutants of Volvox carteri forma nagariensis Iyengar. PhD dissertation (Charlottesville, VA: University of Virginia).
- Sessoms, A.H., and Huskey, R.J. (1973). Genetic control of development in Volvox: Isolation and characterization of morphogenetic mutants. Proc. Natl. Acad. Sci. USA 70 1335–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194 3–21. [DOI] [PubMed] [Google Scholar]
- Starr, R.C. (1968). Cellular differentiation in Volvox. Proc. Natl. Acad. Sci. USA 59 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, R.C. (1969). Structure, reproduction, and differentiation in Volvox carteri f. nagariensis Iyengar, strains HK 9 and 10. Arch. Protistenkd. 111 204–222. [Google Scholar]
- Sumper, M., and Hallmann, A. (1998). Biochemistry of the extracellular matrix of Volvox. Int. Rev. Cytol. 180 51–85. [DOI] [PubMed] [Google Scholar]
- Sun-Wada, G.H., Yoshioka, S., Ishida, N., and Kawakita, M. (1998). Functional expression of the human UDP-galactose transporters in the yeast Saccharomyces cerevisiae. J Biochem 123 912–917. [DOI] [PubMed] [Google Scholar]
- Tautvydis, K.J. (1978). Isolation and charactrization of an extracellularhydroxyproline-rich glycoprotein and a mannose-rich polysaccharide from Eudorina califoornica (Shaw). Planta 140 213–220. [DOI] [PubMed] [Google Scholar]
- Ueki, N., and Nishii, I. (2008). Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viamontes, G.I., Fochtmann, L.J., and Kirk, D.L. (1979). Morphogenesis in Volvox: Analysis of critical variables. Cell 17 537–550. [DOI] [PubMed] [Google Scholar]
- Viamontes, G.I., and Kirk, D.L. (1977). Cell shape changes and the mechanism of inversion in Volvox. J. Cell Biol. 75 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, J. (1985). Macromolecules released into the culture medium during the vegetative cell cycle of the unicellular green alga Chlamydomonas reinhardii. Biochem. J. 226 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner, J.P., and Goodenough, U.W. (1994). Volvocine cell walls and their constituent glycoproteins: An evolutionary perspective. Protoplasma 181 245–258. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.