Abstract
Hsp70 molecular chaperones play a variety of functions in every organism, cell type and organelle, and their activities have been implicated in a number of human pathologies, ranging from cancer to neurodegenerative diseases. The functions, regulations and structure of Hsp70s were intensively studied for about three decades, yet much still remains to be learned about these essential folding enzymes. Genome sequencing efforts revealed that most genomes contain multiple members of the Hsp70 family, some of which co-exist in the same cellular compartment. For example, the human cytosol and nucleus contain six highly homologous Hsp70 proteins while the yeast Saccharomyces cerevisiae contains four canonical Hsp70s and three fungal-specific ribosome-associated and specialized Hsp70s. The reasons and significance of the requirement for multiple Hsp70s is still a subject of debate. It has been postulated for a long time that these Hsp70 isoforms are functionally redundant and differ only by their spatio-temporal expression patterns. However, several studies in yeast and higher eukaryotic organisms challenged this widely accepted idea by demonstrating functional specificity among Hsp70 isoforms. Another element of complexity is brought about by specific cofactors, such as Hsp40s or nucleotide exchange factors that modulate the activity of Hsp70s and their binding to client proteins. Hence, a dynamic network of chaperone/co-chaperone interactions has evolved in each organism to efficiently take advantage of the multiple cellular roles Hsp70s can play. We summarize here our current knowledge of the functions and regulations of these molecular chaperones, and shed light on the known functional specificities among isoforms.
Key Words: Hsp70, Ssa1, chaperone network, functional specificity.
INTRODUCTION
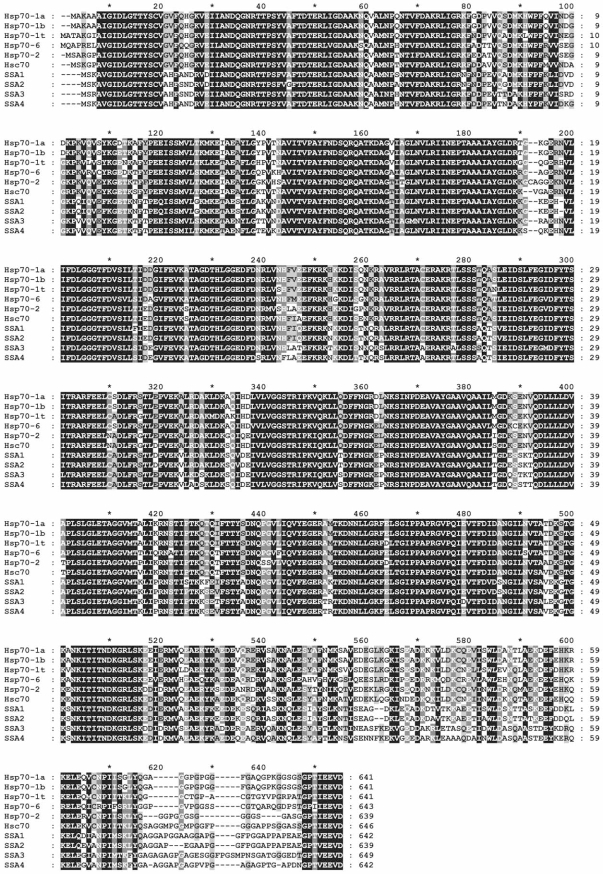
The 70-kDa heat-shock proteins (Hsp70s) are a ubiquitous family of molecular chaperones found in all organisms and sub-cellular compartments where they play essential housekeeping functions in protein folding, synthesis, assembly, transport across biological membranes and degradation. They are also involved in quality control processes, such as protein refolding after a stress injury, and control the activity of regulatory proteins in signal transduction pathways [1]. Hsp70s constitute one of the most conserved protein families in evolution (Fig. 1), and members of this family may be constitutively expressed or stress-inducible (hence their classification as heat-shock proteins) [2-6].
Fig. (1).
The alignment of yeast and human cytosolic Hsp70s shows their remarkable evolutionary conservation and highlights the variability of the C-terminal lid domain (this figure was made using clustalX 1.83 and GeneDoc v2.6).
The functional pleiotropy of Hsp70s is achieved through the evolutionary amplification and diversification of HSP70 genes, cofactors that recruit and regulate Hsp70s for specific cellular functions, and cooperation of Hsp70s with other chaperone systems such as TRiC/CCT or Hsp90 [1]. All of these cellular activities depend on the ability of Hsp70s to interact with hydrophobic stretches of proteins in an ATP-dependent manner. Hsp70s are highly conserved and are composed of an N-terminal 44-kDa ATPase domain (also named adenine nucleotide-binding domain or NBD), an 18-kDa peptide-binding domain (PBD) and a C-terminal 10-kDa “lid” domain. The NBD has a bilobal structure and each globular lobe (I and II) is further conventionally divided into two subdomains (A and B) [7]. The interdomain linker connecting the NBD to the PBD is highly conserved and plays a critical role in the allosteric regulation of Hsp70s [8, 9]. Cytosolic Hsp70s also contain a G/P-rich C-terminal region containing an EEVD-motif that mediates their binding to Tetra-Trico-Peptide repeat (TPR)-domain containing co-chaperones such as the C-terminus-Hsp70-Interacting Protein CHIP. The EEVD motif is absent from specialized Hsp70s such as the ribosome-bound Ssb1/2p in yeast (see below).
Hsp70s cycle between two stable conformations with different affinities for substrates: in the ATP-bound state, Hsp70s display fast on-and-off rates of peptide binding, whereas in the ADP-bound state these constants are slowed, resulting in tighter association of the substrate within the PBD [10, 11]. The modulation of the affinity for polypeptide substrates is triggered by a conformational change in the lid that, upon ATP hydrolysis, closes on the substrate that is then trapped within the PBD [12]. It appears that ADP-ATP exchange is critical for substrate release and for the recycling of the molecular chaperone. This allosteric regulation of Hsp70s activity is tightly controlled by specific co-chaperones (Fig. 1A).
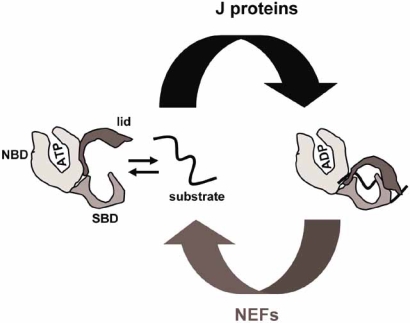
The well characterized Hsp40/DnaJ family members share a common ~70-amino-acid J-domain that mediates binding to Hsp70s and activation of their weak intrinsic ATPase activity (Fig. 2). These cofactors, by virtue of their cellular localization and/or association to specific complexes, recruit Hsp70s to specific sites of the cell and therefore greatly contribute to the wide range of functions assigned to Hsp70s (reviewed in [13-17]).
Fig. (2).
The Hsp70 ATPase cycle. In the ATP-bound state, Hsp70 have low affinity for substrates. Upon J-protein-stimulated ATP hydrolysis, a conformational switch of the lid tightly locks the substrate within the substrate binding domain. NEF catalyzed-ADP release and subsequent ATP re-binding trigger substrate release.
Whereas ATP hydrolysis is stimulated by members of a common Hsp40/DnaJ family, ADP-ATP exchange is catalyzed by several evolutionary unrelated classes of nucleotide exchange factors (NEFs) (Fig. 2); these include GrpE homologs in prokaryotes and the Bag1, HspBP1/Fes1p and Hsp110 families in eukaryotes (reviewed in [1, 18]). It should be noted that Hsp110s are molecular chaperones homologous in sequence and structure to Hsp70s and together these proteins form the super-family of Hsp70s [19, 20]. The domain architecture of Hsp110 is therefore very similar to that of Hsp70s; the main noticeable difference lies in large (~100-140 amino acids) acidic insertions between the PBD and the lid and also at the C-terminus [20]. Hsp110s are not protein folding chaperones, yet they act as efficient “holdases” by binding to and preventing aggregation of denatured proteins [21, 22]. Mammalian Hsp110 and yeast Sse1p were shown to functionally and physically interact with counterpart Hsp70s [23-25], and to be potent nucleotide-activated NEFs [26-28].
THE EUKARYOTIC CYTOSOL CONTAINS MULTIPLE MEMBERS OF THE HSP70 SUPER-FAMILY
Genome sequencing efforts confirmed early observations that eukaryotes have several Hsp70 encoding genes [4]. Evidently, some of these Hsp70s are compartment specific and fulfil unique and essential functions in the endoplasmic reticulum, mitochondria or plastids. These organelle-specific Hsp70s are generally encoded by a single gene in most organisms. Strikingly, multiple Hsp70 isoforms very often coexist in the same cytosol and are encoded by different homologous genes. In Table (1) we describe the cytosolic Hsp70 machinery of various fully sequenced eukaryotic organisms, ranging from yeast to human. For most of these selected species, the Hsp70 system has been recently described in details at the genomic and/or functional levels [6, 29-34].
Table 1.
The Cytosolic Hsp70s of Various Eukaryotes are Shown Together with Some Important Co-Chaperones (Because of their High Number, J-Proteins are not Shown). UniProt Accession Numbers are Provided in Most Cases, Along with the Alternative Acronyms by which these Proteins were Described when Appropriate
| Yeasts | Nematode | Green Algae | Ascidian | Plant | Fruit fly | Human | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | S. pombe | C. albicans | Y. lipolytica | C. elegans | C. reinhardtii | C. intestinalis | A. thaliana | D. melanogaster | H. sapiens | |
| Canonical Hsp70's | Ssa1p (P10591) | Sp.Ssa1p (Q10265) | Ca.Ssa1p (P41797) | Ssa5p (Q6C0E9) | Hsp70-1 (P09446) | Cr.Hsp70-3 (P25840) | Ci.HSPA1/6/7-like (P91874) | At.Hsp70-1 (P22953) | Hsp70Aa (P82910) | Hsp70-1a (Hsp70, Hsp72, Hsp70-1) (P08107) |
| Ssa2p (P10592) | Sp.Ssa2p (O59855) | Ca.Ssa2p (P46587) | Ssa6p (Q6C3G5) | Hsp70-7 (Q9XTL8) | Ci.HSPA2/8 | At.Hsp70-2 (P22954) | Hsp70Ab (P02825) | Hsp70-1b (Hsp70, Hsp72, Hsp70-1) (P08107) | ||
| Ssa3p (P09435) | Ssa7p (Q6C9V0) | Hsp70-8 (Q9XTL8) | At.Hsp70-3 (O65719) | Hsp70Ba (Q8INI8) | Hsp70-1t (Hsp70-hom) (P34931) | |||||
| Ssa4p (P22202) | Ssa8p (Q6C864) | Hsp70-9 (O45246) | At.Hsp70-4 (Q9LHA8) | Hsp70Bbb (Q9VG58) | Hsp70-2 (Hsp70-3) (P54652) | |||||
| At.Hsp70-5 (Q959N1) | Hsp70Bb (Q9BIS2) | Hsp70-6 (Hsp70B') (P17066) | ||||||||
| Hsp70Bc (Q9BIR7) | Hsc70 (Hsp70-8, Hsp73) (P11142) | |||||||||
| Hsc70-1 (P29843) | ||||||||||
| Hsc70-2 (P11146) | ||||||||||
| Hsc70-4 (P11147) | ||||||||||
| Hsp68 (097125) | ||||||||||
| Other Hsp70's | Ssb1p (P11484) | Sp.Ssb1p (Sks2) (Q10284) | Ca.Ssb1p (P87222) | Yl.Ssb1p (Q6CIA7) | Hsp70-10 (Q9TW52) | - | - | - | - | - |
| Ssb2p (P40150) | ||||||||||
| Ssz1p (P38788) | Sp.Ssz1p(P87142) | Ca.Ssz1p (Q5A678) | Yl.Ssz1p (Q6CEW0) | - | - | - | - | - | Hsp70-14 (Hsp70L1) (Q0VDF9) | |
| Hsp110's | Sse1p (P32589) | Sp.Sse1p (Pss1) (O59838) | Ca.Sse1p (Q96VB9) | Yl.Sse1p (Q6C618) | Hsp110-1 (Q05036) | - | Ci.HSPA4/4L/HSPH1 | At.Hsp70-14 (Q957C0) | Hsc70Cb (Q9XZT5) | Hsp110 (Hsp70-4) (P34932) |
| Sse2p (P32590) | At.Hsp70-15 (Q9CA95) | Hsp110 (Hsp70-4L) (O95757) | ||||||||
| At.Hsp70-16 (A8MRM9) | Hsp105 (Q92598) | |||||||||
| HspBP1 | Fes1p (P38260) | Sp.Fes1p (O43030) | Ca.Fes1p (Q59NN8) | Yl.Fes1p (Q6C239) | - | - | - | At.HspBP1-1 (Q9M346) | ? | HspBP1 (Q9NZL4) |
| At.HspBP1-2 (Q84J81) | ||||||||||
| At.HspBP1-3 (Q9LZL7) | ||||||||||
| Bag proteins | Snl1p (P40548) | Bag-1A(O59739) | Ca.Snl1p (Q59NB3) | Yl.Bag1 (Q6C9J3) | Ce.Bag-1 (044739) | - | CI.BAG1 | At.BaBag-1 (Q0WUQ1) | starvin, isoform A (Q9VU81) | Bag-1L, Bag-1M, Bag-1S (Q99933) |
| Bag-1B (O59739) | Unc-23a (O61980) | CI.BAG3 | At.Bag-2 (Q8LEP8) | starvin, isoform B (Q9VU83) | Bag-2 (O95816) | |||||
| Unc-23b (Q86S24) | Ci.BAT3 | At.Bag-3 (Q9LYP4) | starvin, isoform C (Q9VU82) | Bag-3 (O95817) | ||||||
| Unc-23c (Q5TKA9) | At.Bag-4 (O65021) | Bag-4 (O95429) | ||||||||
| At.Bag-5 (O65373) | Bag-5 (Q9UL15) | |||||||||
| At.Bag-6 (O82345) | Bag-6, BAT3 (P46379) | |||||||||
| At.Bag-7 (Q9LVA0) | ||||||||||
| CHIP | - | - | - | Yl.Chn1p (Q6C1F4) | Chn-1 (Q9BMU2) | Cr.CHIP (A8J756) | - | At.CHIP (Q9SRS9) | Dm.CHIP (Q9XYW6) | CHIP (Q9UNE7) |
With the exception of ribosome-associated specialized Hsp70s (that do not contain the C-terminal EEVD motif), canonical Hsp70s are generally believed to be functionally redundant, the main differences between isoforms lying in their spatio-temporal expression. This is exemplified by studies in yeast that showed that only one member of the Ssa group of Hsp70s can support yeast growth if expressed at sufficient levels [29]. Additionally, heterologously expressed Hsp70s were shown to protect mammalian cells and transgenic animals from various stresses, demonstrating that the protective role of these molecular chaperones is highly conserved [35-38].
However, many studies also raised the possibility of functional specificity among Hsp70 isoforms, meaning that a particular Hsp70 will for instance preferentially bind to a subset of client proteins and/or co-chaperones to perform a unique task. Such a specialization among Hsp70s may explain the need for several Hsp70-encoding genes and their different tissue-expression patterns in complex multicellular organisms. In this review, we will discuss examples of functional specificities among Hsp70s in yeast and humans, hypothesize on the molecular basis that may govern this specificity, and suggest directions for future research in this field.
REDUNDANT AND SPECIALIZED FUNCTIONS AMONG HSP70S IN YEAST
The Cytosolic Hsp70 System in Yeast
The yeast Saccharomyces cerevisiae contains seven cytosolic Hsp70s that fall into two major groups: 1) the canonical Ssa1, Ssa2p, Ssa3p and Ssa4p proteins and 2) the ribosome-associated Ssb1p, Ssb2p and Ssz1p proteins (Table 1) [4].
The highly homologous Ssa proteins differ by their expression pattern: the SSA2 gene is constitutively expressed at high levels; the SSA1 gene is constitutively expressed, yet at lower levels than SSA2, and is also induced by stress; the SSA3 and SSA4 genes are not expressed during normal vegetative growth, but their expression is dramatically induced upon stress; additionally, Ssa3p levels are increased upon entry into stationary phase [29, 39-42]. The simultaneous deletion of these four genes is lethal, but can be complemented by the over-expression of either one of them, suggesting redundant functions for these molecular chaperones [29]. It should be noted that a Δssa1Δssa2 mutant is viable and constitutively thermotolerant because of the upregulation of SSA3, SSA4 and other components of the general stress response [29, 39, 40]. However, the Δssa1Δssa2 mutant forms small colonies at 23°C and is thermosensitive for growth at 37°C, suggesting that the functions carried by Ssa1p and/or Ssa2p cannot be fully complemented by the induction of SSA3 and SSA4 [29, 39, 40].
The nearly identical Ssb1p and Ssb2p proteins interact with nascent chains at the yeast ribosome and provide a dispensable function in the process of translation and in early folding events [43, 44]. These Hsp70s are unable to complement the lack of Ssa proteins, indicating that these two groups of Hsp70s clearly have distinct non overlapping functions [4]. Ssb1p and Ssb2p are recruited to the ribosome through their interaction with the stable heterodimeric Ribosome-Associated Complex (RAC) that is composed of the J-domain protein Zuo1p (or zuotin) and the atypical Hsp70 Ssz1p [45, 46]. The simultaneous deletion of SSB1 and SSB2 results in slow growth, cold sensitivity and hypersensitivity to translation-inhibiting drugs such as hygromycin B or paromomycin. Identical phenotypes are observed upon deletion of SSZ1 or ZUO1, suggesting that Ssb1/2p and RAC form a functional triad in vivo [45-48]. The Ssb1/2p proteins are not stimulated by other J-proteins than Zuo1p and require their PBD for function [49, 50]. The Ssz1p protein diverges markedly from other Hsp70s: it contains an unusually short PBD and lacks the C-terminal lid domain; in fact, truncated Ssz1p lacking nearly the entire PBD is fully functional in vivo [51]; Ssz1p is not an ATPase in vitro and its nucleotide binding ability is not strictly required in vivo [52]. Ssz1p has been proposed to serve as a structural scaffold between Ssb1/2p and Zuo1p [53], although this may not be its only function [52]. While Ssb proteins are generally considered fungal-specific, we have identified an uncharacterized Ssb homolog in C. elegans (Table 1). Moreover, a mammalian RAC complex has been recently described, and is comprised of the Zuo1 homolog MPP11 and the Ssz1 homolog Hsp70L1 (or Hsp70-14; Table 1) [54].
In S. cerevisiae, the Ssa proteins are recruited and activated by 11 cytosolic J-proteins [55], and both Ssa and Ssb proteins can be regulated by the Fes1p and Sse1/2p NEFs [23, 24, 26, 27, 56, 57]. Whether the different Ssa isoforms have preferences and/or different affinities for distinct J-proteins or NEFs has not been systematically tested, but remains an attractive possibility that could partly explain the functional specificities observed among Hsp70 isoforms. Indeed, a functional distinction between cytosolic, ER or mitochondrial Hsp70s has been greatly documented and was attributed to specific interactions between these Hsp70s and their partner J-proteins. In particular, cytosolic and ER lumenal Hsp70s could not replace one another in in vitro reconstitution assays of protein translocation across the ER membrane [58, 59]. Moreover, mitochondrial Hsp70 was unable to replace the ER lumenal Hsp70 in similar assays [60]. In both cases, this has been attributed to impaired interactions of the “extraneous” Hsp70 with the “endogenous” compartment-specific J-proteins, suggesting convergent evolution of chaperone and co-chaperones for proper interaction [16, 58, 60].
As shown in Table (1), Ssa, Ssb and Ssz homologues are present in most yeasts that mainly differ with respect to the number of Ssa-encoding genes: only two in Schizosaccharomyces pombe and Candida albicans, and four in Y. lipolytica. We are currently investigating the regulations and functions of the four Y. lipolytica Ssa proteins that we have named Ssa5-8 to avoid confusion with the S. cerevisiae Hsp70 system (Martineau C.N. and Kabani M., unpublished data). A comparison of the Hsp70 systems in these two distantly related yeasts will provide useful information on the plasticity and dynamics of Hsp70-containing chaperone networks.
Examples of Functional Specificity Among the Ssa Group of Hsp70s
Early evidence for functional specificity among Ssa proteins was brought by a study from Eisenberg and collaborators that investigated the ability of yeast cytoplasmic Hsp70s to dissociate clathrin from bovine brain coated vesicles [61]. Using purified Hsp70 preparations from wild-type yeast, they first showed that the in vitro clathrin uncoating activity is associated with Ssa but not Ssb proteins. They then showed that higher uncoating activities could be achieved using Hsp70 preparations from a Δssa1 mutant than from a Δssa2 mutant. Because Ssa3p and Ssa4p are nearly undetectable in these preparations, they concluded that Ssa2p has a markedly higher uncoating activity than Ssa1p. Moreover, they showed that Ssa1p inhibits the uncoating activity of Ssa2p, probably by blocking Hsp70-binding sites on clathrin baskets [61]. The molecular basis of this functional difference in clathrin uncoating between these highly homologous Hsp70s remains to be directly addressed. One possible explanation is that Ssa2p and Ssa1p could have markedly different affinities for clathrin and/or the J-protein auxilin (Swa2p) which is specifically required to activate Hsc70 during endocytosis [62]. In humans and fruit fly, clathrin uncoating was similarly shown to depend on Hsc70 and Hsc70-4, respectively, which are the most abundant and constitutive Hsp70s in these organisms [63, 64]. This is not surprising given that Hsp70 acts stoichiometrically in clathrin uncoating, and therefore fair amounts of the molecular chaperone are required to fulfil this vital cellular process [65].
Another functional distinction between Ssa1p and Ssa2p has been reported with respect to their roles in prion propagation. The yeast epigenetic factors [PSI+], [URE3] and [PIN+] are the prion forms of the Sup35p, Ure2p and Rnq1p proteins, respectively [66-68]. These infectious proteins self-propagate as amyloids [66, 67, 69-72] and therefore constitute a powerful and safe model to investigate mammalian prion-related diseases such as sheep scrapie or transmissible spongiform encephalopathies [73]. Molecular chaperones of the Hsp104, Hsp70 and Hsp40 families play critical roles in prion formation and propagation [74, 75]. The fungal specific Hsp104 molecular chaperone promotes the ATP-dependent solubilization of aggregated misfolded proteins with the help of Hsp70 and Hsp40 [76]. Hsp104 is strictly required for yeast prion propagation, as the deletion of the HSP104 gene or the inactivation of Hsp104 by growth in the presence of 5 mM guanidium chloride cure cells from [PSI+], [URE3] and [PIN+]. In contrast, the overexpression of Hsp104 cures [PSI+] but not [URE3] [67, 77-80]. Hsp104 was proposed to mediate protein-only inheritance by remodelling large prion aggregates into new self-replicating particles (or seeds) [69, 81-83]. The effects of Hsp70s and Hsp40s on prion propagation are complex and prion-dependent [75]. Mutation of the cytosolic Hsp70 Ssa1p impairs [PSI+] propagation while its overexpression was shown to inhibit [PSI+] curing by overexpressed Hsp104 [84, 85]. The overproduction of Ssa1p, but not Ssa2p, cures [URE3] [86], whereas mutations in SSA2, but not SSA1, impair [URE3] propagation [87]. The overexpression of Ydj1p, the most abundant yeast Hsp40, cures cells from [URE3] [78], whereas mutations in Sis1p cure [PIN+] [88]. Similarly, NEFs have been shown to influence prion propagation. Daniel Masison and collaborators have isolated an SSA1-21 mutation that weakens [PSI+] propagation [84]. They then showed that in the SSA1-21 background the depletion of Fes1p further destabilized [PSI+], whereas its overexpression restored almost normal [PSI+] propagation [89]. However, contrasting results have been reported for [PSI+] propagation when FES1 is deleted in an otherwise wild-type background [89, 90]. The deletion of SSE1 and FES1 completely blocked [URE3] propagation, but only the overexpression of Sse1p cured it [90-92]. In addition, the deletion of SSE1 severely inhibited [PSI+] formation while the overexpression of Sse1p improved it [92]. The Hsp70 machinery appears as an important modulator of amyloid formation and protein-only inheritance, and the complex phenotypes obtained upon mutations in its components indicates that a dynamic network of specific chaperone/co-chaperone/substrate interactions occur in vivo. A better understanding of the role of the Hsp70 system in the process of amyloidogenesis will benefit from in vitro approaches where the effects of purified chaperones and co-chaperones on the assembly of amyloid fibrils can be precisely tested [93, 94].
We have recently showed that mutations in components of the cytosolic Hsp70 machinery (e.g. FES1, SSE1, YDJ1, SSA1) dramatically impair biofilm (or ‘mat’) formation in yeast [95]. We showed that the overexpression of FES1 rescues the defect in biofilm formation of a Δsse1 strain, but the opposite is not true: the Δfes1 mutant is unable to form ‘mats’ on semi-solid medium with or without SSE1/2 overexpression [95]. Thus, Fes1p and Sse1/2p play both overlapping and distinct functions as suggested by earlier studies [27, 96]. Remarkably, we showed that a Δssa1 mutant, in an otherwise wild-type background for the other SSA genes, is severely affected in biofilm formation [95]. The deletion of SSA2 had more subtle effects suggesting that Ssa1p is specifically required for ‘mat’ formation [95]. Moreover, we showed that the additional deletion of SSA3 and SSA4 aggravated the phenotypes of the Δssa1 and Δssa2 mutants, indicating a possible functional cooperation between the constitutive and inducible Hsp70s [95].
THE HUMAN HSP70 SYTEM
The Cytosolic Hsp70 System in Humans
Several reviews and studies attempted to provide an accurate description of the human Hsp70 family, a task that has been complicated by confusing and conflicting nomenclature in the literature [6, 34, 97]. However, extensive literature and database search, facilitated by the availability of the human genome, recently allowed Alberto Macario and collaborators to provide a detailed description of human Hsp70 encoding genes [34]. They identified up to 17 Hsp70-encoding genes that fall into evolutionary and functionally distinct groups, as well as 30 pseudogenes [34]. For each of these genes, an impressively high number of mRNA variants and isoforms were identified through EST analysis, although their functional relevance is still unknown [34]. In Table (1), we chose to present only the bona fide human cytosolic Hsp70s for which expression has been proved and that correspond to Ssa or Sse-like proteins [6, 34, 97].
The human cytosolic Hsp70s are comprised of six canonical members, named Hsp70-1a, Hsp70-1b, Hsp70-1t, Hsp70-2, Hsp70-6 and Hsc70, and of an Ssz-like protein named Hsp70-14 (or Hsp70L1) [6, 34, 97] (Table 1). These proteins were also described with different acronyms that are indicated in Table (1). While the expression of Hsp70-1a, Hsp70-1b and Hsp70-6 is strongly induced by stress [6, 34], all these cytosolic Hsp70s differ considerably with respect to their expression patterns in the different tissues and developmental stages, as predicted by assessing the relative number of ESTs per tissue type [34].
The heat-shock cognate Hsc70 is by far the most expressed Hsp70 in all tissues with particularly high levels in the vascular tissue [34], and is believed to play essential house-keeping functions in protein folding, transport across biological membranes, prevention of protein aggregation and uncoating of clathrin vesicles [6, 98]. The intron-less genes encoding Hsp70-1A, Hsp70-1B and Hsp70-1t are all closely linked and located within the same MHC class III region on chromosome 6 [99, 100], yet their expression patterns differ [6, 34, 101]. Hsp70-1a and Hsp70-1b are expressed in most tissues, and in most cases Hsp70-1a levels are much higher than Hsp70-1b levels [6, 34]. Moreover, very high expression levels were observed for Hsp70-1a in the spleen [34]. On the other hand, Hsp70-1t is expressed at almost undetectable levels in most tissues, except in testis, adipose tissue and pituitary glands where its levels are much higher [6, 34, 101]. In contrast to Hsp70-1a and Hsp70-1b, the levels of Hsp70-1t are not induced by heat stress [101]. Hsp70-2 is expressed at low to undetectable levels in most tissues, and at high levels in testis, brain and umbilical cord [6, 34]. The expression of Hsp70-6 is only induced after severe stress insults [102, 103], although moderate levels of this protein have been reported in some blood cells as well as in the bladder, trachea and esophagus [6, 34].
Thus, all cytosolic Hsp70s are constitutively expressed at varying levels making each tissue and developmental stage unique in its Hsp70 content [6, 34]. Important efforts will be needed to define the specific roles of each Hsp70 in each cell and tissue type, taking into account the possibility that these Hsp70s may functionally interact with one another, either positively or negatively, depending on their respective abundance. Whether the Hsp70 co-chaperones content also varies among cell and tissue types has yet to be determined, but could also be a major determinant of the functional specialization of Hsp70s.
Examples of Functional Specificity Among the Human Cytosolic Hsp70s
Understanding the need for up to six Hsp70 family members in human cells and sorting out their redundant from their specialized functions have proven difficult and technically challenging. Most of the published data on human Hsp70s were obtained by studying the constitutive Hsc70 or the major heat-inducible Hsp70 (which in fact refers to both Hsp70-1a and Hsp70-1b) that were often considered as equivalent and functionally interchangeable. [6, 104]. However, a clear distinction between constitutive and inducible Hsp70 isoforms emerged from several studies, including the characterization of mouse knock-out models [6]. It appeared very logically that constitutive Hsp70s mostly play important housekeeping functions while inducible Hsp70s are required to cope with stress situations [6].
Hence, Hsc70 was shown to be essential for viability in vertebrates, including fruit fly [105], mouse [106] and human cell lines [107, 108], whereas mice deficient for the homologues of human Hsp70-1a and Hsp70-1b (Hsp70.1 and Hsp70.3) develop normally in standard conditions but show significantly decreased resistance to stress situations such as heat shock, cardiac ischemia or radiation [6, 109-117].
Additionally, Hsp70 and Hsc70 were shown to have differential and antagonistic effects with regard to the intracellular trafficking of ENaC, an epithelial sodium chloride channel: Hsp70 promoted the maturation and functional cell surface expression of ENaC in Xenopus oocytes, whereas Hsc70 disfavoured it [104]. Similarly, both the overexpression of Hsp70 and the decrease of Hsc70 levels via the use of sodium 4-phenylbutyrate were shown to promote the intracellular trafficking of the ΔF508 mutant of the cystic fibrosis transmembrane conductance regulator (CFTR) [118-120].
Hsp70 levels are abnormally high in a wide variety of tumor cell types and contribute to tumorigenesis and resistance to chemotherapy by directly interfering with several key components of the apoptotic signalling pathway [121]. Similarly, an up-regulation of the HspBP1 NEF was observed in various tumor cell types [122, 123]. HspBP1 has been shown to modulate Hsp70s activity either negatively [124, 125] (at high HspBP1/Hsp70 molar ratios) or positively (at low HspBP1/Hsp70 molar ratios) [126]. Interestingly, HspBP1 was shown in vivo to bind with a much higher affinity to the stress-inducible Hsp70 than to the abundant and constitutively expressed Hsc70. Hence, HspBP1 is present in great excess (~10 fold) compared to its preferential Hsp70 partner in normal cells. However, the HspBP1/Hsp70 molar ratio may be lowered considerably in tumor cells (from ~10 to ~2) because the up-regulation of Hsp70 is greater than that of HspBP1 in these cells [123]. This may contribute to the Hsp70-mediated resistance to chemotherapy, as tumor cells with high HspBP1/Hsp70 molar ratios were much more susceptible to anticancer drugs than were those with a low ratio [123]. In addition, anticancer drugs up-regulated HspBP1 in various cancer cells, while no effect was observed on Hsp70 levels [123]. These findings suggest that HspBP1 antagonizes the prosurvival function of Hsp70 [123]. In support of this hypothesis, RNAi-mediated depletion of HspBP1 markedly reduced the susceptibility of tumor cells to anticancer drugs [123]. Conversely, transient ectopic expression of HspBP1 in tumor cells enhanced the cathepsin-mediated cell death induced by anticancer drugs [123]. This pro-apoptotic function of HspBP1 appeared to be dependent on its ability to bind to Hsp70 and was inhibited by heat-shock induced up-regulation of Hsp70 [123]. These findings exemplify that part of the functional distinction among Hsp70 isoforms may lie in differential interaction with their co-chaperones, thereby forming specific functional networks in each cell or tissue type. Remarkably, both primate and plant Hsc70 were able to support growth of a Δssa1-4 yeast mutant, whereas the corresponding inducible Hsp70s did not [127], suggesting an evolutionary conserved functional distinction among these molecular chaperones.
The Hsp70-2 protein appears to play an important role in spermatogenesis in humans and mice via its essential chaperoning functions for the cyclinB/cdc2 complex during meiosis, and for spermatid DNA-packaging proteins involved in post-meiotic genome reorganization [6, 128-132]. Additionally, Hsp70-2 is upregulated in some primary and metastatic breast cancers, and was shown to be required for the growth and survival of several human cancer cells [6, 107, 108, 133]. The depletion of Hsp70 or Hsp70-2 from cancer cells resulted in markedly different phenotypes and gene expression profiles, whereas their concomitant depletion resulted in synergistic antiproliferative effects, suggesting separate non-overlapping functions for these Hsp70s [108].
By virtue of their cellular protective role, Hsp70s are important candidates in gene-longevity association studies [134, 135]. Indeed, the ability to cope with cellular stress by the induction of Hsp70 was shown to decline with age in numerous in vivo and in vitro models [134, 136-139]. Moreover, the presence of extra-copies of a heat-inducible Hsp70-encoding gene increased the lifespan of transgenic fly models after a transient mild heat-shock [140]. Specifically, polymorphisms positively or negatively associated with longevity were identified in the genes encoding the MHC-III-linked Hsp70-1a, Hsp70-1b and Hsp70-1t proteins, suggesting that unique anti-ageing functions are fulfilled by these isoforms (reviewed in [134, 135]).
CONCLUSIONS AND FUTURE DIRECTIONS: MOLECULAR BASIS OF HSP70 SPECIALIZATION
Despite a high degree of sequence conservation and overlapping chaperoning functions, members of the Hsp70 superfamily appear to have evolved specialized functions for which they can not replace each other. This functional specificity is most apparent and widely accepted for compartment-specific and ribosome-associated Hsp70s, and also for the Hsp110 family members that evolved to act as Hsp70 NEFs [6, 98]. However, less attention has been paid to the possibility that a similar specialization might occur among the highly conserved cytosolic Hsp70s, although it may provide an answer to why some organisms such as humans or Drosophila melanogaster need up to 6 or 10 cytosolic Hsp70s, respectively (Table 1). A recent study demonstrated functional differences among highly conserved ribosomal protein paralogs in yeast, indicating that the co-existence of multiple members of a given protein family is not only the result of gene duplication and mere redundancy, but also significantly contributes to cellular homeostasis [141].
Future efforts will have to identify and characterize the unique functions played by individual Hsp70s in particular cell or tissue types. Several non-mutually exclusive hypotheses can be made, according to our current knowledge of Hsp70 function, to understand the molecular basis of such specialization. A first model would be that Hsp70 isoforms differentially interact with the various cytosolic Hsp70 co-chaperones, as observed for the preferred interaction of HspBP1 with Hsp70 and not with Hsc70 [123]. A second model would be that Hsp70 isoforms bind to different sets of client proteins or to different types of substrates (e. g. native client proteins, nascent proteins, misfolded proteins, protein aggregates, etc.). In support of this hypothesis, different peptide binding specificities were described for bacterial DnaK, mammalian Hsc70 and BiP [142, 143]. Similarly, the DnaK protein from the archea Methanosarcina mazei failed to fully complement a dnak mutant in Escherichia coli, and was shown to have markedly different peptide binding properties [144, 145]. Importantly, the 10-kDa C-terminal domain of Hsp70s is the less conserved region and could play an important role in modulating Hsp70-substrate interactions (Fig. 1). Additionally, different Hsp70 isoforms could cooperate in the process of protein folding by binding to different exposed sites on a given substrate. To our knowledge, the side-by-side and thorough biochemical characterization of Hsp70 isoforms from a single model organism has not been reported but would certainly help clarifying the functions and regulations of these essential folding enzymes. Because Hsp70s are relevant targets for drug-based treatments for cancers or protein folding disorders, the identification of the molecular determinants that govern their functional specificities in particular cell or tissue types will greatly improve the efficacy of these approaches without unwillingly affecting vital cellular processes.
ACKNOWLEDGEMENTS
This work was supported by the Centre National de la Recherche Scientifique (CNRS). CNM is the beneficiary of a doctoral grant from the French Ministry of Research.
REFERENCES
- 1.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt C, Morimoto RI. Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70. Proc. Natl. Acad. Sci. USA. 1985;82:6455–6459. doi: 10.1073/pnas.82.19.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 4.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RS, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 1994;4:1104–1114. doi: 10.1016/s0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 6.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein [see comments] Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 8.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 10.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 11.McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 15.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabani M, Beckerich JM, Brodsky JL. The yeast Sls1p and Fes1p proteins define a new family of Hsp70 nucleotide exchange factors. Curr. Genomics. 2003;4:465–473. [Google Scholar]
- 19.Liu Q, Hendrickson WA. Insights into hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner L, Morano KA. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones. 2007;12:1–8. doi: 10.1379/CSC-245R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J. Biol. Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 22.Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J. Biol. Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- 23.Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 24.Yam AY, Albanese V, Lin HT, Frydman J. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
- 25.Yamagishi N, Ishihara K, Hatayama T. Hsp105alpha suppresses Hsc70 chaperone activity by inhibiting Hsc70 ATPase activity. J. Biol. Chem. 2004;279:41727–41733. doi: 10.1074/jbc.M407947200. [DOI] [PubMed] [Google Scholar]
- 26.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 Is a Nucleotide-activated Exchange Factor for Hsp70. J. Biol. Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 29.Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolaidis N, Nei M. Concerted and nonconcerted evolution of the Hsp70 gene superfamily in two sibling species of nematodes. Mol. Biol. Evol. 2004;21:498–505. doi: 10.1093/molbev/msh041. [DOI] [PubMed] [Google Scholar]
- 32.Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada S, Hamada M, Satoh N. A genomewide analysis of genes for the heat shock protein 70 chaperone system in the ascidian Ciona intestinalis. Cell Stress Chaperones. 2006;11:23–33. doi: 10.1379/CSC-137R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brocchieri L, Conway de Macario E, Macario AJ. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008;8:19–0. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelham HR. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li GC, Li L, Liu RY, Rehman M, Lee WM. Heat shock protein hsp70 protects cells from thermal stress even after deletion of its ATP-binding domain. Proc. Natl. Acad. Sci. USA. 1992;89:2036–2040. doi: 10.1073/pnas.89.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plumier JC, Ross BM, Currie RW, Angelidis CE, Kazlaris H, Kollias G, Pagoulatos GN. Transgenic mice expressing the human heat shock protein 70 have improved post-ischemic myocardial recovery. J. Clin. Invest. 1995;95:1854–1860. doi: 10.1172/JCI117865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radford NB, Fina M, Benjamin IJ, Moreadith RW, Graves KH, Zhao P, Gavva S, Wiethoff A, Sherry AD, Malloy CR, Williams RS. Cardioprotective effects of 70-kDa heat shock protein in transgenic mice. Proc. Natl. Acad. Sci. USA. 1996;93:2339–2342. doi: 10.1073/pnas.93.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boorstein WR, Craig EA. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:18912–18921. [PubMed] [Google Scholar]
- 40.Boorstein WR, Craig EA. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:3262–3267. doi: 10.1128/mcb.10.6.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner-Washburne M, Becker J, Kosic-Smithers J, Craig EA. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werner-Washburne M, Craig EA. Expression of members of the Saccharomyces cerevisiae hsp70 multigene family. Genome. 1989;31:684–689. doi: 10.1139/g89-125. [DOI] [PubMed] [Google Scholar]
- 43.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 44.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michimoto T, Aoki T, Toh-e A, Kikuchi Y. Yeast Pdr13p and Zuo1p molecular chaperones are new functional Hsp70 and Hsp40 partners. Gene. 2000;257:131–137. doi: 10.1016/s0378-1119(00)00381-4. [DOI] [PubMed] [Google Scholar]
- 46.Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Buesa P, Pfund C, Craig EA. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains . Proc. Natl. Acad. Sci. USA. 1998;95:15253–15258. doi: 10.1073/pnas.95.26.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfund C, Huang P, Lopez-Hoyo N, Craig EA. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell. 2001;12:3773–3782. doi: 10.1091/mbc.12.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA. 2002;99:4203–4208. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conz C, Otto H, Peisker K, Gautschi M, Wolfle T, Mayer MP, Rospert S. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J. Biol. Chem. 2007;282:33977–33984. doi: 10.1074/jbc.M706737200. [DOI] [PubMed] [Google Scholar]
- 53.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 54.Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA. 2005;102:10064–10069. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast hsp70 molecular chaperone ssa1p. Mol. Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol. Chem. 2006;387:1593–1600. doi: 10.1515/BC.2006.198. [DOI] [PubMed] [Google Scholar]
- 58.Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J. Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brodsky JL. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem. Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- 60.Brodsky JL, Bauerle M, Horst M, McClellan AJ. Mitochondrial Hsp70 cannot replace BiP in driving protein translocation into the yeast endoplasmic reticulum. FEBS Lett. 1998;435:183–186. doi: 10.1016/s0014-5793(98)01065-5. [DOI] [PubMed] [Google Scholar]
- 61.Gao BC, Biosca J, Craig EA, Greene LE, Eisenberg E. Uncoating of coated vesicles by yeast hsp70 proteins. J. Biol. Chem. 1991;266:19565–19571. [PubMed] [Google Scholar]
- 62.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 63.Chappell TG, Welch WJ, Schlossman DM, Palter KB, Schlesinger MJ, Rothman JE. Uncoating ATPase is a member of the 70 kilodalton family of stress proteins. Cell. 1986;45:3–13. doi: 10.1016/0092-8674(86)90532-5. [DOI] [PubMed] [Google Scholar]
- 64.Chang HC, Newmyer SL, Hull MJ, Ebersold M, Schmid SL, Mellman I. Hsc70 is required for endocytosis and clathrin function in Drosophila. J. Cell Biol. 2002;159:477–487. doi: 10.1083/jcb.200205086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greene LE, Eisenberg E. Dissociation of clathrin from coated vesicles by the uncoating ATPase. J. Biol. Chem. 1990;265:6682–6687. [PubMed] [Google Scholar]
- 66.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 67.Sondheimer N, Lindquist S. Rnq1 an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 68.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 69.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 70.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 71.King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 73.Prusiner SB. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tuite MF, Cox BS. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 75.Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 77.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 78.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 80.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 81.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 82.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roberts BT, Moriyama H, Wickner RB. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107–117. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- 88.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lian HY, Zhang H, Zhang ZR, Loovers HM, Jones GW, Rowling PJ, Itzhaki LS, Zhou JM, Perrett S. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J. Biol. Chem. 2007;282:11931–11940. doi: 10.1074/jbc.M606856200. [DOI] [PubMed] [Google Scholar]
- 92.Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de Novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martineau CN, Beckerich JM, Kabani M. Flo11p-independent control of "mat" formation by hsp70 molecular chaperones and nucleotide exchange factors in yeast. Genetics. 2007;177:1679–1689. doi: 10.1534/genetics.107.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 99.Goate AM, Cooper DN, Hall C, Leung TK, Solomon E, Lim L. Localization of a human heat-shock HSP 70 gene sequence to chromosome 6 and detection of two other loci by somatic-cell hybrid and restriction fragment length polymorphism analysis. Hum. Genet. 1987;75:123–128. doi: 10.1007/BF00591072. [DOI] [PubMed] [Google Scholar]
- 100.Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32:242–251. doi: 10.1007/BF00187095. [DOI] [PubMed] [Google Scholar]
- 101.Fourie AM, Peterson PA, Yang Y. Characterization and regulation of the major histocompatibility complex-encoded proteins Hsp70-Hom and Hsp70-1/2. Cell Stress Chaperones. 2001;6:282–295. doi: 10.1379/1466-1268(2001)006<0282:carotm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leung TK, Rajendran MY, Monfries C, Hall C, Lim L. The human heat-shock protein family Expression of a novel heat-inducible HSP70 (HSP70B') and isolation of its cDNA and genomic DNA. Biochem. J. 1990;267:125–132. doi: 10.1042/bj2670125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parsian AJ, Sheren JE, Tao TY, Goswami PC, Malyapa R, Van Rheeden R, Watson MS, Hunt CR. The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim. Biophys. Acta. 2000;1494:201–205. doi: 10.1016/s0167-4781(00)00203-7. [DOI] [PubMed] [Google Scholar]
- 104.Goldfarb SB, Kashlan OB, Watkins JN, Suaud L, Yan W, Kleyman TR, Rubenstein RC. Differential effects of Hsc70 and Hsp70 on the intracellular trafficking and functional expression of epithelial sodium channels. Proc. Natl. Acad. Sci. USA. 2006;103:5817–5822. doi: 10.1073/pnas.0507903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elefant F, Palter KB. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol. Biol. Cell. 1999;10:2101–2117. doi: 10.1091/mbc.10.7.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Muller M, Streeck RE, Sapp M. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol. 2004;78:5546–5553. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Daugaard M, Jaattela M, Rohde M. Hsp70-2 is required for tumor cell growth and survival. Cell Cycle. 2005;4:877–880. doi: 10.4161/cc.4.7.1838. [DOI] [PubMed] [Google Scholar]
- 108.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee SH, Kim M, Yoon BW, Kim YJ, Ma SJ, Roh JK, Lee JS, Seo JS. Targeted hsp70.1 disruption increases infarction volume after focal cerebral ischemia in mice. Stroke. 2001;32:2905–2912. doi: 10.1161/hs1201.099604. [DOI] [PubMed] [Google Scholar]
- 110.Van Molle W, Wielockx B, Mahieu T, Takada M, Taniguchi T, Sekikawa K, Libert C. HSP70 protects against TNF-induced lethal inflammatory shock. Immunity. 2002;16:685–695. doi: 10.1016/s1074-7613(02)00310-2. [DOI] [PubMed] [Google Scholar]
- 111.Huang L, Mivechi NF, Moskophidis D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol. Cell Biol. 2001;21:8575–8591. doi: 10.1128/MCB.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shim EH, Kim JI, Bang ES, Heo JS, Lee JS, Kim EY, Lee JE, Park WY, Kim SH, Kim HS, Smithies O, Jang JJ, Jin DI, Seo JS. Targeted disruption of hsp70.1 sensitizes to osmotic stress. EMBO Rep. 2002;3:857–861. doi: 10.1093/embo-reports/kvf175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwon SB, Young C, Kim DS, Choi HO, Kim KH, Chung JH, Eun HC, Park KC, Oh CK, Seo JS. Impaired repair ability of hsp70.1 KO mouse after UVB irradiation. J. Dermatol. Sci. 2002;28:144–151. doi: 10.1016/s0923-1811(01)00156-6. [DOI] [PubMed] [Google Scholar]
- 114.Hampton CR, Shimamoto A, Rothnie CL, Griscavage-Ennis J, Chong A, Dix DJ, Verrier ED, Pohlman TH. HSP70.1 and -70.3 are required for late-phase protection induced by ischemic preconditioning of mouse hearts. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H866–874. doi: 10.1152/ajpheart.00596.2002. [DOI] [PubMed] [Google Scholar]
- 115.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1 - and Hsp70.3-deficient mice. Mol. Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singleton KD, Wischmeyer PE. Effects of HSP70.1/3 gene knockout on acute respiratory distress syndrome and the inflammatory response following sepsis. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L956–961. doi: 10.1152/ajplung.00466.2005. [DOI] [PubMed] [Google Scholar]
- 117.Dix DJ, Garges JB, Hong RL. Inhibition of hsp70-1 and hsp70-3 expression disrupts preimplantation embryogenesis and heightens embryo sensitivity to arsenic. Mol. Reprod. Dev. 1998;51:373–380. doi: 10.1002/(SICI)1098-2795(199812)51:4<373::AID-MRD3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 118.Rubenstein RC, Zeitlin PL. Sodium 4-phenylbutyrate downregulates Hsc70: implications for intracellular trafficking of DeltaF508-CFTR. Am. J. Physiol. Cell Physiol. 2000;278:C259–267. doi: 10.1152/ajpcell.2000.278.2.C259. [DOI] [PubMed] [Google Scholar]
- 119.Rubenstein RC, Egan ME, Zeitlin PL. In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing delta F508-CFTR. J. Clin. Invest. 1997;100:2457–2465. doi: 10.1172/JCI119788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choo-Kang LR, Zeitlin PL. Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281:L58–68. doi: 10.1152/ajplung.2001.281.1.L58. [DOI] [PubMed] [Google Scholar]
- 121.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- 122.Raynes DA, Graner MW, Bagatell R, McLellan C, Guerriero V. Increased expression of the Hsp70 cochaperone HspBP1 in tumors. Tumour Biol. 2003;24:281–285. doi: 10.1159/000076459. [DOI] [PubMed] [Google Scholar]
- 123.Tanimura S, Hirano AI, Hashizume J, Yasunaga M, Kawabata T, Ozaki K, Kohno M. Anticancer drugs up-regulate HspBP1 and thereby antagonize the prosurvival function of Hsp70 in tumor cells. J. Biol. Chem. 2007;282:35430–35439. doi: 10.1074/jbc.M707547200. [DOI] [PubMed] [Google Scholar]
- 124.McLellan CA, Raynes DA, Guerriero V. HspBP1, an Hsp70 cochaperone, has two structural domains and is capable of altering the conformation of the Hsp70 ATPase domain. J. Biol. Chem. 2003;278:19017–19022. doi: 10.1074/jbc.M301109200. [DOI] [PubMed] [Google Scholar]
- 125.Raynes DA, Guerriero V Jr. Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J. Biol. Chem. 1998;273:32883–32888. doi: 10.1074/jbc.273.49.32883. [DOI] [PubMed] [Google Scholar]
- 126.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 127.Tutar Y, Song Y, Masison DC. Primate Chaperones Hsc70 (Constitutive) and Hsp70 (Induced) Differ Functionally in Supporting Growth and Prion Propagation in Saccharomyces cerevisiae. Genetics. 2006;172:851–861. doi: 10.1534/genetics.105.048926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Son WY, Han CT, Hwang SH, Lee JH, Kim S, Kim YC. Repression of hspA2 messenger RNA in human testes with abnormal spermatogenesis. Fertil. Steril. 2000;73:1138–1144. doi: 10.1016/s0015-0282(00)00496-9. [DOI] [PubMed] [Google Scholar]
- 129.Govin J, Caron C, Escoffier E, Ferro M, Kuhn L, Rousseaux S, Eddy EM, Garin J, Khochbin S. Post-meiotic shifts in HSPA2/HSP70 chaperone activity during mouse spermatogenesis. J. Biol. Chem. 2006;281:37888–37892. doi: 10.1074/jbc.M608147200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, Goulding EH, Eddy EM. Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc. Natl. Acad. Sci. USA. 1996;93:3264–3268. doi: 10.1073/pnas.93.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, Blizard DR, Brown PR, Goulding EH, Strong BD, Eddy EM. HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development. 1997;124:4595–4603. doi: 10.1242/dev.124.22.4595. [DOI] [PubMed] [Google Scholar]
- 132.Zhu D, Dix DJ, Eddy EM. HSP70-2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development. 1997;124:3007–3014. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- 133.Daugaard M, Kirkegaard-Sorensen T, Ostenfeld MS, Aaboe M, Hoyer-Hansen M, Orntoft TF, Rohde M, Jaattela M. Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res. 2007;67:2559–2567. doi: 10.1158/0008-5472.CAN-06-4121. [DOI] [PubMed] [Google Scholar]
- 134.Macario AJ, Conway de Macario E. Sick chaperones cellular stress and disease. N. Engl. J. Med. 2005;353:1489–1501. doi: 10.1056/NEJMra050111. [DOI] [PubMed] [Google Scholar]
- 135.Singh R, Kolvraa S, Rattan SI. Genetics of human longevity with emphasis on the relevance of HSP70 as candidate genes. Front. Biosci. 2007;12:4504–4513. doi: 10.2741/2405. [DOI] [PubMed] [Google Scholar]
- 136.Singh R, Kolvraa S, Bross P, Jensen UB, Gregersen N, Tan Q, Knudsen C, Rattan SI. Reduced heat shock response in human mononuclear cells during aging and its association with polymorphisms in HSP70 genes. Cell Stress Chaperones. 2006;11:208–215. doi: 10.1379/CSC-184R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Njemini R, Abeele MV, Demanet C, Lambert M, Vandebosch S, Mets T. Age-related decrease in the inducibility of heat-shock protein 70 in human peripheral blood mononuclear cells. J. Clin. Immunol. 2002;22:195–205. doi: 10.1023/a:1016036724386. [DOI] [PubMed] [Google Scholar]
- 138.Simar D, Malatesta D, Koechlin C, Cristol JP, Vendrell JP, Caillaud C. Effect of age on Hsp72 expression in leukocytes of healthy active people. Exp. Gerontol. 2004;39:1467–1474. doi: 10.1016/j.exger.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 139.Visala Rao D, Boyle GM, Parsons PG, Watson K, Jones GL. Influence of ageing, heat shock treatment and in vivo total antioxidant status on gene-expression profile and protein synthesis in human peripheral lymphocytes. Mech. Ageing Dev. 2003;124:55–69. doi: 10.1016/s0047-6374(02)00170-7. [DOI] [PubMed] [Google Scholar]
- 140.Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- 141.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fourie AM, Sambrook JF, Gething MJ. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- 143.Gragerov A, Gottesman ME. Different peptide binding specificities of hsp70 family members. J. Mol. Biol. 1994;241:133–135. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- 144.Zmijewski MA, Macario AJ, Lipinska B. Functional similarities and differences of an archaeal Hsp70(DnaK) stress protein compared with its homologue from the bacterium Escherichia coli. J. Mol. Biol. 2004;336:539–549. doi: 10.1016/j.jmb.2003.12.053. [DOI] [PubMed] [Google Scholar]
- 145.Zmijewski MA, Skorko-Glonek J, Tanfani F, Banecki B, Kotlarz A, Macario AJ, Lipinska B. The DnaK chaperones from the archaeon Methanosarcina mazei and the bacterium Escherichia coli have different substrate specificities. Acta Biochim. Pol. 2007;54:509–522. [PubMed] [Google Scholar]