Abstract
The mammalian Na+/Ca2+ exchanger, NCX1.1, serves as the main mechanism for Ca2+ efflux across the sarcolemma following cardiac contraction. In addition to transporting Ca2+, NCX1.1 activity is also strongly regulated by Ca2+ binding to two intracellular regulatory domains, CBD1 and CBD2. The structures of both of these domains have been solved by NMR spectroscopy and x-ray crystallography, greatly enhancing our understanding of Ca2+ regulation. Nevertheless, the mechanisms by which Ca2+ regulates the exchanger remain incompletely understood. The initial NMR study showed that the first regulatory domain, CBD1, unfolds in the absence of regulatory Ca2+. It was further demonstrated that a mutation of an acidic residue involved in Ca2+ binding, E454K, prevents this structural unfolding. A contradictory result was recently obtained in a second NMR study in which Ca2+ removal merely triggered local rearrangements of CBD1. To address this issue, we solved the crystal structure of the E454K-CBD1 mutant and performed electrophysiological analyses of the full-length exchanger with mutations at position 454. We show that the lysine substitution replaces the Ca2+ ion at position 1 of the CBD1 Ca2+ binding site and participates in a charge compensation mechanism. Electrophysiological analyses show that mutations of residue Glu-454 have no impact on Ca2+ regulation of NCX1.1. Together, structural and mutational analyses indicate that only two of the four Ca2+ ions that bind to CBD1 are important for regulating exchanger activity.
Cardiac contraction/relaxation relies upon Ca2+ fluxes across the plasma membrane (sarcolemma) of cardiomyocytes. Rapid Ca2+ influx (primarily through L-type Ca2+ channels) triggers the release of additional Ca2+ from the sarcoplasmic reticulum (SR),4 resulting in cardiomyocyte contraction. Removal of cytosolic Ca2+ by reuptake into the SR (through the SR Ca2+-ATPase) and expulsion from the cell (primarily through the Na+/Ca2+ exchanger, NCX1.1) results in relaxation (1). Altered Ca2+ cycling is observed in a number of pathophysiological situations including ischemia, hypertrophy, and heart failure (2). Understanding the function and regulation of NCX1.1 is thus of fundamental importance to understand cardiac physiology.
NCX1.1 utilizes the electrochemical potential of the Na+ gradient to extrude Ca2+ in a ratio of three Na+ ions to one Ca2+ ion (3). In addition to transporting both Na+ and Ca2+, NCX1.1 is also strongly regulated by these two ions. Intracellular Na+ can induce NCX1.1 to enter an inactivated state, whereas Ca2+ bound to regulatory sites removes Na+-dependent inactivation and also activates Na+/Ca2+ exchange (3). These regulatory sites are located on a large cytoplasmic loop (∼500 residues located between transmembrane helices V and VI) containing two calcium binding domains (CBD1 and CBD2), which sense cytosolic Ca2+ levels. We have previously shown that Ca2+ binding to the primary site in CBD2 is required for full exchange regulation (4); CBD1, however, is a site of higher affinity and appears to dominate the activation of exchange activity by Ca2+.
Both CBDs have an immunoglobulin fold formed from two antiparallel β sheets generating a β sandwich with a differing number of Ca2+ ions coordinated at the tip of the domain (4, 5). CBD1 binds four Ca2+ ions, whereas CBD2 binds only two Ca2+ ions. An initial NMR study revealed a local unfolding of the upper portion of CBD1 upon release of Ca2+ (6). In contrast, CBD2 did not display an unfolding response upon Ca2+ removal. A comparative analysis between CBDs revealed a difference in charge at residues in equivalent positions near the Ca2+ coordination site; Glu-454 in CBD1 is replaced by Lys-585 in CBD2. The unstructuring of CBD1 upon Ca2+ removal was alleviated by reversing the charge of the acidic residue (E454K) involved in Ca2+ coordination (6). Previously, we solved the structures of the Ca2+-bound and -free conformations of CBD2 and revealed a charge compensation mechanism involving Lys-585 (4). The positively charged lysine residue assumes the position of one of the Ca2+ ions upon Ca2+ depletion, permitting CBD2 to retain its overall fold (4). A similar phenomenon is predicted to take place in E454K-CBD1 mutant. In addition, Hilge et al. (6) showed that the E454K mutation of CBD1 decreases Ca2+ affinity to a level similar to that of CBD2 and suggested that the E454K mutation would cause the loss of primary regulation of NCX1.1 by CBD1.
The significance of some of these observations is unclear as a recent NMR study (7) of CBD1 under more physiologically relevant conditions revealed no significant alteration in tertiary structure in the absence of Ca2+. It was hypothesized that Ca2+ binding induces localized conformational and dynamic changes involving several of the binding site residues. To clarify this issue, we solved the crystal structure of the E454K-CBD1 mutant and examined the functional effects of different CBD1 mutations in the full-length NCX1.1. The results indicate that charge compensation is indeed provided by the residue Lys-454 to replace one Ca2+, whereas the overall E454K-CBD1 structure is only slightly perturbed. The charge compensation, however, has no impact on Ca2+ regulation of NCX1.1.
EXPERIMENTAL PROCEDURES
Expression and Purification of E454K-CBD1—The mutation E454K was introduced into CBD1 of canine NCX1.1 (residues 370–509) by QuikChange mutagenesis (Stratagene) using a pET47b(+) plasmid (Novagen, San Diego, CA) and was confirmed by DNA sequencing. Escherichia coli (HMS174(DE3), Novagen) were transformed with the plasmid and grown in LB to an A of 0.6 before protein expression was induced using 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 2 h. Cells were solubilized in a 1× BugBuster (Novagen) solution in wash buffer 1 (300 mm NaCl, 1% Triton X-100, 20 mm Tris-Cl, pH 8) supplemented with 5 mm 2-mercaptoethanol, 0.25 units/ml DNase, 40 units/ml egg lysozyme, and EDTA-free protease inhibitors (Roche Applied Science). The solution was cleared by centrifugation, and the soluble fraction was applied to 10 ml of nickel-nitrilotriacetic acid-Sepharose (Qiagen, Valencia, CA), washed with wash buffer 2 (25 mm MES, pH 6.3, 300 mm NaCl, 10% glycerol, 0.1 mm CaCl2), and eluted with wash buffer 2 supplemented with 250 mm imidazole, pH 7.4. Eluted E454K-CBD1 was digested with HRV-3C (80 units/6 mmol CBD1-E454K; Novagen) overnight at 4 °C and dialyzed against protease buffer (150 mm NaCl, 50 mm Tris-Cl, pH 7.5) to remove the N-terminal His tag. Cleaved protein that did not bind to nickel-nitrilotriacetic acid-Sepharose was concentrated with a Centricon 30 (Millipore, Bedford, MA) and run on a HiPrep 16/60 Sephacryl S-100 column (Amersham Biosciences) in 20 mm Tris-Cl, pH 8, 300 mm NaCl, 0.1 mm CaCl2. Fractions containing CBD1-E454K were dialyzed against 10 mm Tris-Cl, pH 7.5, and concentrated with a Centricon 30–60 mg/ml.
Crystal Growth and Structure Determination—Purified E454K-CBD1 was spun at 400,000 × g for 30 min at 4 °C prior to crystallization trials. Initial trials were carried out at 20 °C with the mosquito crystallization robot (TTP Labtech) at a protein concentration of 20 mg/ml using the hanging drop vapor diffusion technique. Initial screens yielded crystal that grew in 3.5 m ammonium chloride, 0.1 m sodium acetate, pH 4.6. The crystal was cryoprotected by transferring to a solution containing 10% glycerol in addition to the mother liquor solution and was flash-frozen in liquid nitrogen.
Data were collected at beamline 8.2.1 at the Advanced Light Source (Berkeley, CA) to a resolution of 2.4 Å (supplemental Table 1). The crystal belongs to space group P41 21 2 with cell dimensions of a = b = 91.858 Å and c = 65.59 Å. Image data were processed using the programs DENZO and SCALEPACK (8). The structure of E454K-CBD1 was solved by molecular replacement using modified coordinates from the crystal structure of CBD1 (PDB accession code 2DPK) by the program PHASER (9). The structure was manually built using the program COOT (10) and refined in REFMAC (11) with a final R and Rfree of 19.3 and 25.8%, respectively. The final model encompasses residues 370–465 and 482–501 and binds three Ca2+ ions in the Ca2+ binding site (position 2: 75% occupancy). Despite acidic crystallization conditions, glutamates and aspartates are coordinating Ca2+; thus the structure is likely an accurate representation under physiological condition. Coordinates were deposited at the PDB under accession code 3GIN.
Functional and Mutational Analysis—Inside-out giant patch recordings of outward Na+/Ca2+ exchanger currents were performed as described previously (12, 13). Xenopus laevis oocytes were injected with RNAs coding for WT or mutant NCX1.1 and kept at 18 °C for 4–7 days. Borosilicate glass pipettes of ∼20–30 μm were used. After excision of a membrane patch, intracellular solutions were rapidly changed by using a computer-controlled 20-channel solution switcher. Measurements were obtained by using the following solutions. The pipette solution was: 100 mm N-methylglucamine, 10 mm Hepes, 20 mm tetraethyl-ammonium hydroxide, 0.2 mm niflumic acid, 0.2 mm ouabain, 8 mm Ca(OH)2 (pH 7) (by using MES). The bath solution was: 100 mm CsOH or 100 mm NaOH, 20 mm tetraethyl-ammonium hydroxide, 10 mm Hepes, 10 mm EGTA, or HEDTA and different Ca(OH)2 concentrations to obtain the desired final Ca2+-free concentration (pH 7; adjusted with MES). Free Ca2+ concentrations were calculated according to the MAXc program (14) and confirmed with a Ca2+ electrode.
Data were normalized to maximal values and fitted to a Hill function. Values are mean ± S.E. pCLAMP (Axon Instruments, Burlingame, CA) software was used for acquisition and analysis. Data were acquired on line at 4 ms/point and fitted at 50 Hz by using an eight-pole Bessel filter. Experiments were performed at 35 °C at a holding potential of 0 mV.
RESULTS
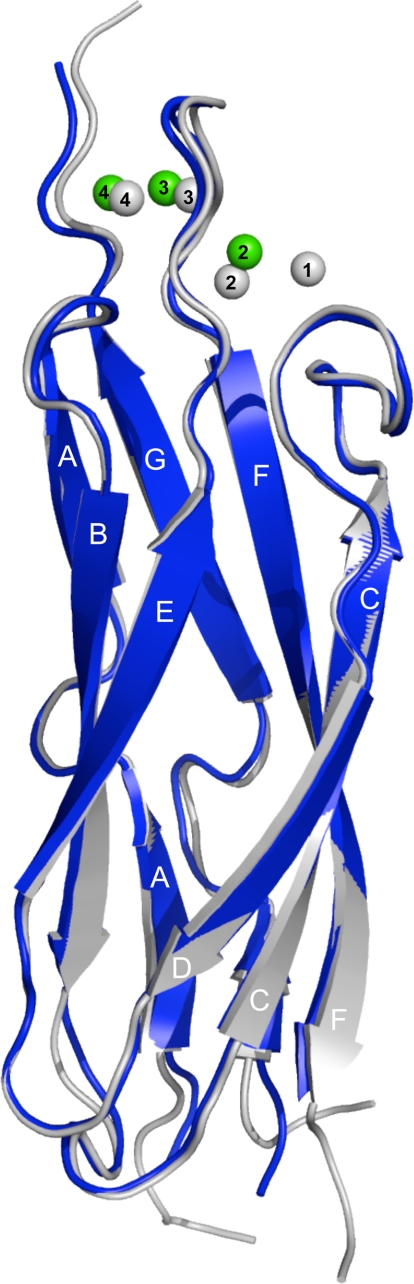
Structure Overview—The overall structure of the E454K-CBD1 mutant is virtually identical to the structure of the WT protein with an r.m.s.d. of 0.5 Å for 115 Cα atoms. As observed in both the x-ray and the NMR structures of the wild type protein, the 17-amino-acid loop between strands F and G was not observed, presumably due to inherent disorder within this region (5, 6). Deletion studies removing the FG loop had no significant influence on NCX function (not shown). The asymmetric unit is composed of two virtually identical parallel molecules with an r.m.s.d. of 0.39 Å for 115 Cα atoms; only one molecule will be discussed further. A structural overlay between E454K-CBD1 and wild type CBD1 reveals modest structural displacements with the most significant alterations located in the Ca2+ binding site around position 454 (Figs. 1 and 2).
FIGURE 1.
Structural comparison between WT-CBD1 (gray, PDB code: 2DPK) and E454K-CBD1 (blue). The three Ca2+ ions bound to E454K-CBD1 are displayed as green spheres and numbered according to WT-CBD1 numbering (5). For clarity, the β strands are labeled A–G.
FIGURE 2.
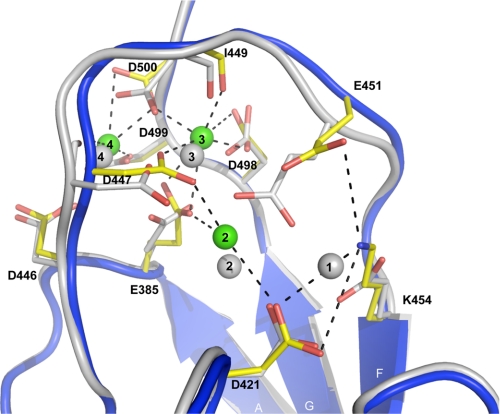
Overlay of the Ca2+ binding sites of WT-CBD1 and E454K-CBD1. E454K-CBD1 is shown as a blue drawing, and Ca2+ ions are depicted as green spheres. Ca2+-coordinating residues are displayed as sticks colored by atom type (Cα is yellow). Relevant metal ion coordinations and charge pairs of E454K-CBD1 are shown as black dotted lines. WT-CBD1 is shown as a gray drawing, and equivalent residues are displayed as sticks colored by atom type. Ca2+ ions are numbered according to Ref. 5.
Ca2+ Binding Site—WT-CBD1 binds four Ca2+ ions clustered in two pairs, numbered 1 and 2, and 3 and 4 (Fig. 2) (5). In contrast, the E454K-CBD1 mutant coordinates only three Ca2+ ions, which were assigned without ambiguity in the electron density (supplemental Fig. 1). Lys-454 replaces the Ca2+ ion at position 1 and forms intramolecular charge pairs with Asp-421 and Glu-451 (coordinating distances ∼3.6 Å) (Fig. 2). As a consequence, residue Glu-451 is shifted 2.3 Å away from its original position, where it no longer coordinates Ca2+ at positions 1 and 2. The latter Ca2+ is loosely coordinated by Glu-385 (2.61 Å), Asp-421 (3.19 Å), forms a new interaction with Asp-447 (2.37 Å), and displays partial occupancy (∼75%). In contrast, Ca2+ ions at positions 3 and 4 are present at 100% occupancy, and all coordinating residues, together with their coordination pattern, display strong similarity to the WT-CBD1 structure.
An additional, nonphysiological, Ca2+ ion is observed at the crystal-packing interface between symmetrical molecules. This Ca2+ is loosely coordinated by five water molecules (supplemental Fig. 2), where four of them are oriented via bidentate interactions from Asp-453 and its corresponding residue on the crystallographic symmetric molecule. This location and coordination pattern indicates that this additional Ca2+ ion has no biological relevance but is likely present due to crystallization restraints.
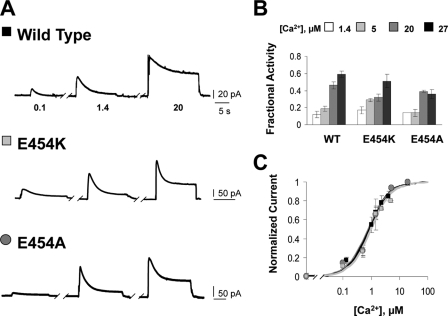
Mutational Analysis—To understand the impact of these observations on NCX1.1 function, effects of mutations at position 454 were investigated by electrophysiology. WT and mutant exchanger cRNAs were injected into Xenopus oocytes, and biophysical properties were examined using the giant excised patch technique (15). Outward currents were generated by fast application of 100 mm cytoplasmic Na+ to patches maintaining 8 mm Ca2+ within the pipette at the extracellular surface. Various concentrations of regulatory Ca2+ were present within the bath solution. Fig. 3A shows representative outward currents recorded from WT and mutant exchangers in the presence of the indicated levels of regulatory Ca2+.
FIGURE 3.
Mutations at position 454 do not affect Ca2+ regulation of the Na+-Ca2+ exchanger. A, recordings from giant excised patches expressing the indicated construct. Currents were activated by raising the intracellular Na+ concentration from 0 to 100 mm in the presence of 0.1, 1.4, or 20 μm intracellular Ca2+. Similar to WT, high intracellular Ca2+ increased the exchange currents of E454K and E454A and diminished the extent of the Na+-dependent inactivation. B, summary of the fractional activity values for the indicated exchangers. Fractional activity is calculated as the ratio of steady state current to peak current and was measured in the presence of four different cytoplasmic Ca2+ concentrations, as indicated. Each point is the average of 3–8 experiments. C, dependence of exchange current on regulatory Ca2+ for WT and the indicated exchanger mutants. Amplitudes were measured at peak currents. Residual current recorded in the absence of Ca2+ has been subtracted. Data points were averaged from 2–6 individual experiments.
Like the WT exchanger, currents generated by E454K and E454A mutants peaked and then decayed over several seconds due to Na+-dependent inactivation. To quantify the degree of Na+-dependent inactivation, the ratio of the steady state current to the peak current, measured at four different levels of regulatory Ca2+, was determined (fractional activity; Fig. 3B). WT, E454K, and E454A exchangers display similar effects of Ca2+ on the Na+-dependent inactivation.
Increasing intracellular Ca2+ concentrations stimulate exchange activity and rescue the exchanger from the Na+-dependent inactivated state. The apparent affinity for regulatory Ca2+ was determined by measuring the peak current at different intracellular Ca2+ levels (Fig. 3C). WT, E454K-, and E454A-NCX1.1 show a similar dependence of peak current on regulatory Ca2+ with apparent affinities of 0.86, 0.96, and 1.19 μm, respectively.
DISCUSSION
We solved the crystal structure of the E454K-CBD1 mutant and performed electrophysiologic analysis of Glu-454 mutants in the full-length NCX1.1 exchanger to gain insight on Ca2+ regulation. The structure of E454K-CBD1 reveals that Lys-454 replaces the Ca2+ ion at position 1, thereby providing charge compensation through the formation of intramolecular charge pairs with residues previously interacting with Ca2+ at position 1 in WT-CBD1 (Fig. 2). Perhaps the positive charge provided by Lys-454 allows sufficient retention of the protein fold to maintain biophysical properties of NCX1.1 as the E454K-CBD1 mutant has no effect on Ca2+ regulation of NCX1.1 (Fig. 3). To further investigate this possibility, Glu-454 was mutated to the neutral amino acid alanine, which, unlike lysine, would be incapable of coordinating either Asp-421 or Glu-451, resulting in a larger degree of unfolding. Glu-454 is a primary residue in the coordination of Ca2+ ion 1 in WT-CBD1. Mutation to alanine will most likely result in the loss of Ca2+ binding at position 1 and eliminate any positive charge at that location. Electrophysiology indicates that E454A-NCX1.1 has no alteration of Ca2+ regulation and the same apparent affinity for regulatory Ca2+ as the WT exchanger (Fig. 3). We conclude that a positive charge, supplied either by a lysine or by a Ca2+, at position 1 in CBD1 is not needed for Ca2+ regulation of NCX1.1. It also appears that unfolding in this region of CBD1 is not involved in the mechanism controlling Ca2+ regulation of NCX1.1.
Analysis of the E454K-CBD1 structure reveals that the Ca2+ ion at position 2 is only partially occupied and exhibits a weaker coordination environment than that of Ca2+ ions 3 and 4 (Fig. 2). Likewise, the coordination of Ca2+ ion 2 in the WT-CBD1 has weaker interactions (5). Biophysical analyses of Glu-454 mutants show no differences when compared with WT-NCX1.1, indicating that a partial loss of Ca2+ at position 2 has no effect on Ca2+ regulation. Overall, our data indicate that Ca2+ ions at positions 3 and 4 are solely responsible for triggering Ca2+ regulation. Consistent with this assertion, a previous study (16) indicated that mutation of residues involved in the binding of Ca2+ ions 3 and 4 affect the apparent affinity for Ca2+ regulation.
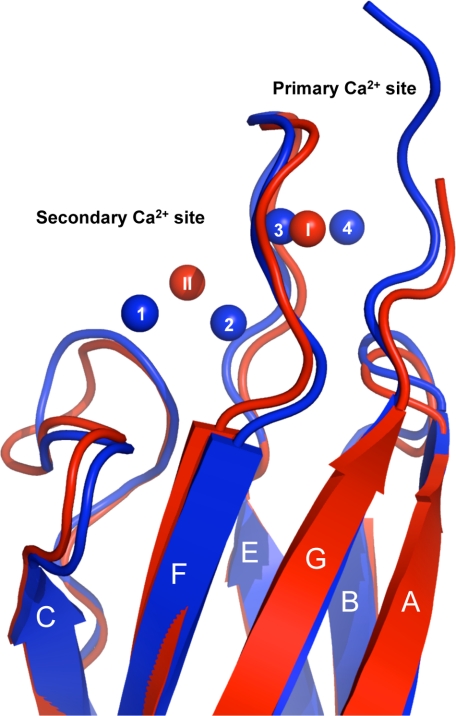
Interestingly, a similar observation has been made for CBD2 where only two Ca2+ ions are present in what we refer to as the primary and secondary Ca2+ binding sites. In our previous study, only the primary Ca2+ binding site was determined to be important in regulation (4). The primary Ca2+ binding site of CBD2 resides in a homologous position as Ca2+ ions 3 and 4 of CBD1 (Fig. 4). A recent study measured Ca2+ binding to purified CBD1 or CBD2 using a fluorescence-based assay and by 45Ca2+ binding (17). This study concluded that between 1.8 and 2.4 Ca2+ ions bind to CBD1, whereas between 1.6 and 2.0 bind to CBD2, suggesting that the binding sites of the CBDs were only partially occupied. Taken together, the findings imply that NCX1.1 activity may be regulated by only a portion of the Ca2+ binding sites in its regulatory domains, via Ca2+ binding at positions 3 and 4 in CBD1 and at the primary Ca2+ binding site in CBD2.
FIGURE 4.
Overlay of WT-CBD1 and CBD2 demonstrating the conserved Ca2+ binding sites. WT-CBD1 is shown as a blue drawing with Ca2+ ions depicted as blue spheres. CBD2 (PDB code: 2QVM) is represented as a red drawing with the primary (I) and secondary (II)Ca2+ ions depicted as red spheres (r.m.s.d. = 1.1 Å over 112 Cα). The CBD2 primary Ca2+ binding site is at a position homologous to Ca2+ ions 3 and 4 of CBD1. For clarity, the strands are labeled A–G.
It appears that CBD1 and CBD2 have arisen from a gene duplication event (18), which is supported by the structural identity observed between the two domains (Fig. 4). Our study further reveals that the sensory domains maintain a conserved global position for Ca2+ binding within both CBDs involved in NCX1.1 regulation.
Supplementary Material
Acknowledgments
We are grateful to Corie Ralston and all personnel of beamline 8.2.1 of the Advanced Light Source (Berkeley, California); the UCLA Department of Physiology Protein Expression Core; and the UCLA-Department of Energy X-ray Crystallography Core Facility (DE-FC02-02ER63421).
This work was supported, in whole or in part, by National Institutes of Health Grant GM07844 (to J. A.) and Grant HL49101 (to K. D. P.). This work was also supported by American Heart Association Grant 0630258N and Human Frontier Science Program (HFSP) Grant RGY0069 (to J. A.).
The atomic coordinates and structure factors (code 3GIN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table and two supplemental figures.
Footnotes
The abbreviations used are: SR, sarcoplasmic reticulum; CBD, calcium binding domains; NCX, Na,Ca-exchanger; PDB, Protein Data Bank; WT, wild type; r.m.s.d., root mean square deviation; MES, 4-morpholineethanesulfonic acid; HEDTA, N-(2hydroxyethyl)ethylenediamine-N,N′,N′-triacetic acid.
References
- 1.Bers, D. M. (2002) Nature 415 198–205 [DOI] [PubMed] [Google Scholar]
- 2.Del Monte, F., and Hajjar, R. J. (2008) Heart Fail. Rev. 13 151–162 [DOI] [PubMed] [Google Scholar]
- 3.Nicoll, D. A., Ren, X., Ottolia, M., Phillips, M., Paredes, A. R., Abramson, J., and Philipson, K. D. (2007) Ann. N. Y. Acad. Sci. 1099 1–6 [DOI] [PubMed] [Google Scholar]
- 4.Mercado-Besserer, G., Ottolia, M., Nicoll, D. A., Chaptal, V., Cascio, D., Philipson, K. D., and Abramson, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18467–18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicoll, D. A., Sawaya, M. R., Kwon, S., Cascio, D., Philipson, K. D., and Abramson, J. (2006) J. Biol. Chem. 281 21577–21581 [DOI] [PubMed] [Google Scholar]
- 6.Hilge, M., Aelen, J., and Vuister, G. W. (2006) Mol. Cell. 22 15–25 [DOI] [PubMed] [Google Scholar]
- 7.Johnson, E., Bruschweiler-Li, L., Showalter, S. A., Vuister, G. W., Zhang, F., and Brüschweiler, R. (2008) J. Mol. Biol. 377 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 9.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458–464 [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 11.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 12.Nicoll, D. A., Ottolia, M., Lu, L., Lu, Y., and Philipson, K. D. (1999) J. Biol. Chem. 274 910–917 [DOI] [PubMed] [Google Scholar]
- 13.Dyck, C., Omelchenko, A., Elias, C. L., Quednau, B. D., Philipson, K. D., Hnatovich, M., and Hryshko, L. V. (1999) J. Gen. Physiol. 114 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton, C., Thompson, S., and Epel, D. (2004) Cell Calcium 35 427–431 [DOI] [PubMed] [Google Scholar]
- 15.Hilgemann, D. W., and Lu, C. C. (1998) Methods Enzymol. 293 267–280 [DOI] [PubMed] [Google Scholar]
- 16.Matsuoka, S., Nicoll, D. A., Hryshko, L. V., Levitsky, D. O., Weiss, J. N., and Philipson, K. D. (1995) J. Gen. Physiol. 105 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyman, L., Mikhasenko, H., Hiller, R., and Khananshvili, D. (2009) J. Biol. Chem. 284 6185–6193 [DOI] [PubMed] [Google Scholar]
- 18.Schwartz, E. M., and Benzer, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.