Abstract
Abscisic acid (ABA) is a phytohormone regulating important functions in higher plants, notably responses to abiotic stress. Recently, chemical or physical stimulation of human granulocytes was shown to induce production and release of endogenous ABA, which activates specific cell functions. Here we provide evidence that ABA stimulates several functional activities of the murine microglial cell line N9 (NO and tumor necrosis factor-α production, cell migration) through the second messenger cyclic ADP-ribose and an increase of intracellular calcium. ABA production and release occur in N9 cells stimulated with bacterial lipopolysaccharide, phorbol myristate acetate, the chemoattractant peptide f-MLP, or β-amyloid, the primary plaque component in Alzheimer disease. Finally, ABA priming stimulates N9 cell migration toward β-amyloid. These results indicate that ABA is a pro-inflammatory hormone inducing autocrine microglial activation, potentially representing a new target for anti-inflammatory therapies aimed at limiting microglia-induced tissue damage in the central nervous system.
Microglial cells are the monocyte/macrophage equivalent of the central nervous system and represent the first line of defense in the brain, by removing infectious agents and damaged cells (1). Microglia can also release a variety of trophic factors and cytokines able to regulate the communication between neuronal and other glial cells and can contribute to tissue repair and neuroprotection (2–4). Pathologic microglial activation, however, confers neurotoxic properties to these cells, thereby causing neuronal degeneration (5). Excessive activation of microglia, under conditions of chronic inflammation, can contribute to the pathogenesis of neurodegenerative diseases, such as multiple sclerosis and Alzheimer and Parkinson diseases, by producing and releasing a number of potentially cytotoxic substances, including pro-inflammatory cytokines and NO (4, 6–8). Therefore, identification of the molecular mechanisms underlying microglial activation might lead to the development of new anti-inflammatory drugs for the treatment of these diseases.
Abscisic acid (ABA)2 is a plant hormone regulating important biological functions in higher plants, including response to abiotic stress, control of stomatal closure, regulation of seed dormancy, and germination (9). Recently, ABA was shown to behave as an endogenous pro-inflammatory hormone in human granulocytes (10), stimulating several functional activities of these cells (migration, phagocytosis, reactive oxygen species, and NO production) through a signaling cascade that involves a protein kinase A-mediated ADP-ribosyl cyclase phosphorylation and consequent overproduction of the universal Ca2+ mobilizer cyclic ADP-ribose (cADPR) (11). This mechanism leads to an increase of the intracellular Ca2+ concentration, which is ultimately responsible for granulocyte activation (10).
The facts that microglial cells play a defensive role in the central nervous system similar to that of granulocytes in other tissues and that cADPR has been described as the second messenger involved in the activation of microglia induced by lipopolysaccharide (LPS) (12) prompted us to investigate the effect of ABA in these cells.
Indeed, exogenous ABA, at concentrations ranging from 250 nm to 20 μm, elicits functional activation of murine N9 cells, stimulating TNF-α release and cell migration through activation of the ADP-ribosyl cyclase CD38 and overproduction of cADPR. Moreover, N9 cells produce and release ABA when stimulated with LPS, amyloid β-peptide (βA), phorbol myristate acetate (PMA), or the chemoattractant peptide f-MLP. These results indicate that ABA behaves as an endogenous, pro-inflammatory hormone in murine microglia and provide a new target for future investigations into the role of this hormone in inflammatory and degenerative diseases of the central nervous system accompanied by microglial activation.
EXPERIMENTAL PROCEDURES
Materials—Fura-2AM and the protein kinase A inhibitor (peptide sequence 14–22, myristoylated, I-PKA) were obtained from Calbiochem (Milano, Italy). The IB4 anti-CD38 monoclonal antibody (mAb) was kindly provided by Prof. F. Malavasi (Torino, Italy). The cDNA for mouse CD38 was a generous gift from Dr. E. Ferrero (Torino, Italy). The anti-phosphoserine mAb (clone PSR-45, catalog number P3430) was obtained from Sigma. TNF-α was obtained from ICN Biomedical Inc, (Milano, Italy). Iscove's modified Dulbecco's medium was purchased from Cambrex BioScience (Milano, Italy). (±)-cis,trans-ABA and all other chemicals were obtained from Sigma.
Cell Cultures—Murine N9 microglial cells, originally developed by Prof. P. Ricciardi-Castagnoli (13) and kindly provided by Prof. M. Matteoli (Milan, Italy) were grown as described (12). The TNF-α-sensitive murine fibroblast cell line L929 was obtained from the American Type Culture Collection (Manassas, VA) and cultured as described (12).
Fluorometric Determination of the [Ca2+]i—Adherent N9 cells were seeded on 20-mm-diameter coverslips. After 24 h, the cells were loaded with 10 μm Fura-2AM for 45 min at 37 °C in complete medium, washed once with Hanks' balanced salt solution (HBSS; catalog number H8264; Sigma) and stimulated with 20 μm ABA in the same solution or in Ca2+-free HBSS (catalog number H6648). The [Ca2+]i was evaluated as described previously (14). Alternatively, the cells were cultured in complete medium in the presence or absence of 20 μm ABA for 1.5, 4, or 24 h. In the last 45 min, the cells were loaded with 10 μm Fura-2AM at 37 °C in complete medium, and the [Ca2+]i was measured as reported previously (14).
N9 cells transfected with wild type or with mutagenized murine CD38 were seeded onto 8-well Lab Tek chambered coverglasses (Nalge Nunc Int., Naperville, IL) in complete medium, loaded with 10 μm calcium green (Invitrogen) for 2 h at 37 °C, and then washed twice with HBSS. Confocal microscopy acquisitions were performed on a Leica TCS SL confocal microscope (Leica Microsystems, Milan, Italy) equipped with argon/helium-neon laser sources and a HC PL APO 20.0 × 0.7NA air objective. Fluorescence emission was monitored on regions of interest of 10–15 cells for 10 min after the addition of 20 μm ABA. Stack profiles of acquisitions were then obtained and compared using Leica confocal software.
Determination of Intracellular cADPR Levels—Adherent N9 cells (3 × 107 cells/assay, i.e. ∼5 mg of protein), cultured in complete medium, were incubated at 37 °C for 0.5 or 24 h, without (control) or with 20 μm ABA. The cells were recovered by trypsin treatment and washed once with HBSS, and the cell pellets were processed as described (12). The cADPR content was measured on the neutralized cell extracts by a sensitive enzymatic cyclic assay (15).
Assays of CD38 Enzymatic Activity—The ectocellular cyclase activity of CD38 was measured on NGD+, an analog of NAD+, because the product cyclic GDP-ribose (cGDPR) is not a substrate of the hydrolase (16). Intact N9 cells (10 mg/ml) were resuspended in 0.3 ml of HBSS containing 0.2 mm NGD+ at 37 °C in the absence (control) or in the presence of 0.2, 2, or 20 μm ABA. At various times (0, 10, 30, and 60 min), 60-μl aliquots of the incubations were withdrawn and centrifuged at 5000 × g for 30 s; supernatants were recovered and deproteinized with 5% (v/v) trichloroacetic acid.
N9 cells (1 mg/ml), transfected with murine wild type (WT)-CD38, S10A-CD38, S19A-CD38, or S10A/S19A-CD38 were resuspended in 0.35 ml of HBSS containing 0.2 mm NGD+ at 37 °C in the absence (control) or in the presence of 20 μm ABA. At various times (5, 10, and 20 min), 100-μl aliquots of the incubations were withdrawn and treated as described above.
HPLC analyses of nucleotides to measure the production of cGDPR were performed as described (17). Protein content on aliquots of each incubation, saved prior to trichloroacetic acid addition, was measured according to Bradford (18).
Determination of Intracellular cAMP Levels—N9 cells were seeded in 35 × 10-mm dishes (0.7 × 106 cells/dish). After 48 h, the cells were recovered by trypsin treatment and preincubated (or not) in complete medium in the presence of 2 μg/ml pertussis toxin (PTX) for 1 h. The medium was then replaced with HBSS, and the cells were preincubated for 5 min at 25 °C in the presence of 10 μm of the cAMP phosphodiesterase inhibitor 4-(3-butoxy-4-methoxy-benzyl)imidazolidin-2-one (Ro 20-1724; Sigma, catalog number B8279). After 1 min from the addition of 20 μm ABA, the incubation was stopped by removal of HBSS, and the addition of 200 μl of ice-cold perchloric acid (0.6 m), and the cells were removed by scraping. The cell extracts were centrifuged to remove proteins, and the cAMP content was measured on the neutralized cell extracts (19) by a radioimmunoassay (GE Healthcare). The results were expressed as pmol cAMP/106 cells.
Immunopurification of CD38 and Western Blot Analyses—N9 cells (5 × 107 for each assay), cultured in complete medium, were incubated at 37 °C for 10 min in the absence (control) or in the presence of 20 μm ABA. The cells were recovered by scraping and immunopurification of CD38, and subsequent Western blot analyses were performed as described previously (12). Immunodetection of CD38 was obtained with an anti-phosphoserine mAb and with the IB4 anti-CD38 mAb, followed by incubation with a secondary anti-mouse IgG antibody (GE Healthcare), according to the instructions of the Immobilon Western kit (Millipore, Billerica, MA).
Site-directed Mutagenesis—Amino acid substitutions (S10A, A19A, and S10A/S19A) were generated using the QuikChange multi-site-directed mutagenesis kit (Stratagene, San Diego, CA) according to the manufacturer's protocol. The CD38 plasmid was purified using PureLink™ HiPure plasmid filter kit (Invitrogen) and sequenced by TibMolbiol (Genova, Italy).
Transfection of N9 cells was performed using the Nucleofector System (Amaxa GmbH, Cologne, Germany), with the cell line nucleofector kit T according to the manufacturer's instructions (program A-023). Harvested N9 cells (5 × 106 cells/sample) were transfected with 2 μg of pcDNA3.1 plasmid containing murine WT-CD38, S10A-CD38, S19A-CD38, or S10A/S19A-CD38.
Immediately after transfection, the cells were resuspended in 15 ml of Iscove's modified Dulbecco's medium supplemented with fetal calf serum (5%), penicillin (50 units/ml), streptomycin (50 μg/ml), and 50 nm β-mercaptoethanol and incubated in a humidified 5% CO2 atmosphere at 37 °C for 24 h. The cells were used for all experiments 24 h after transfection.
Determination of Nitric Oxide Production—N9 cells were seeded in 24-well plates (0.25 × 106/well) and incubated for 4 h in 2 ml complete medium at 37 °C in the absence (control) or in the presence of 0.25 or 20 μm ABA. As a positive control, N9 were incubated for 4 or 24 h in the presence of 100 ng/ml LPS. At the end of the incubations, 500-μl aliquots of each supernatant were recovered, and nitrite production was measured as described (12).
Assay of TNF-α Release—N9 cells were seeded in 24-well plates (3 × 105/well) and incubated for various times (0, 0.5, 2, 6, and 24 h) in complete medium at 37 °C in the absence (control) or in the presence of 0.25 or 20 μm ABA. TNF-α released into the medium was assayed by measuring its apoptotic effect on the murine fibroblast cell line L929 (20), as described (12).
Chemokinesis and Chemotaxis—N9 cells were resuspended at 107/ml in chemotaxis buffer (HBSS, phosphate-buffered saline, and 5% albumin, 39:16:1). Chemokinesis and chemotaxis assays were performed using 96-well ChemoTx system microplates (Neuroprobe, Inc., Gaithersburg, MD) with a 8-μm pore polycarbonate filter. For chemotaxis assays, ABA was diluted at different concentrations in chemotaxis buffer (0.2, 2, 20, or 200 μm) and added to the bottom wells. Cell suspensions (25 μl) were placed directly on top of the filter, and the plates were incubated for 90 min at 37 °C. For chemokinesis assays, the cells were preincubated for different times (10, 30, or 60 min or 24 h) in the absence (control) or in the presence of 0.25 or 20 μm ABA and then placed in the upper well of chemotaxis chambers; migration toward the bottom wells, containing chemotaxis buffer or 15 μm βA in chemotaxis buffer, was measured. The transmigrated cells were collected following ChemoTx system instructions, transferred into a 96-well plate and quantified by adding 60 μl of a solution containing 0.2% Nonidet P-40 and 1 μm SYTOX Green. After 20 min of incubation at 37 °C, fluorescence was recorded with a fluorescence plate reader (Fluostar Optima, BMG Labtechnologies GmbH, Offenburg Germany; excitation, 485 nm; emission, 520 nm). A standard curve was obtained by placing serial dilutions of the cell suspension in the bottom wells.
The results are expressed as chemotaxis index (CI = number of cells migrated toward chemoattractant/number of cells migrated toward medium) and chemokinesis index (KI = number of ABA-treated cells migrated toward medium/number of untreated cells migrated toward medium).
Scatchard Plot Analysis of ABA-binding Sites—N9 cells (2 × 105/determination) were incubated in triplicate at 20 °C for 60 min in 100 μl of HBSS, with or without excess unlabeled ABA (to determine nonspecific binding), in the presence of increasing concentrations of [3H]ABA (GE Healthcare). At the end of the incubation time, the cells were centrifuged (30 s at 5,000 × g), the supernatant was discarded, the cell pellets were washed once in excess HBSS at 0 °C by centrifugation, and the radioactivity of the cells was determined on a Packard β-counter. Specific binding was calculated as the difference between the total binding of [3H]ABA to the cells and the [3H]ABA binding in the presence of excess unlabeled ABA (nonspecific binding).
Detection of ABA—N9 cells (9 × 107 adherent cells in 36 ml of medium/assay) were incubated in the absence (control) or in the presence of LPS (100 ng/ml) for 72 h and 10 μm βA, added (or not) during the last 24 h of incubation, and f-MLP (1 μm) or PMA (0.1 μg/ml) for 1 h. At the end of the incubations, the cells were recovered and centrifuged at 200 × g for 5 min. Aliquots (4 ml) of the supernatants were added to 16 ml of distilled methanol. The cell pellets were resuspended in 500 μl of water, 50-μl aliquots were saved for protein determination, and 2 ml of distilled methanol were added to the rest of the cell lysates. Trace amounts of [3H]ABA (3 × 103 cpm) were added as internal standard to each methanol extract for calculation of the extraction yield, and free ABA was extracted as described previously (21). Briefly, the methanol was evaporated under vacuum, and the aqueous phase was acidified (at pH 3.0) with trifluoroacetic acid and extracted three times with 2 volumes of diethylether. The diethylether was evaporated under vacuum, and the extract was redissolved in 250 μl of Tris-buffered saline, pH 7.4. For measurement by enzyme-linked immunosorbent assay, the extracts were assayed with a sensitive and specific kit (Agdia, Elkhart, IN) according to the manufacturer's instructions. Measurements by HPLC-mass spectrometry were performed as described (10).
Statistical Analyses—All of the results were tested by paired t test. p values <0.05 were considered significant.
RESULTS
ABA Induces a cADPR-mediated, Sustained Ca2+ Response in N9 Microglial Cell Line—In human granulocytes, ABA induces a sustained increase of the [Ca2+]i through a signaling pathway sequentially involving a PTX-sensitive G protein-receptor complex, PKA activation, ADP-ribosyl cyclase (ADPRC) phosphorylation, and activation, consequent cADPR overproduction, and, eventually, a cADPR-dependent Ca2+ influx (10). Similarly to what observed in human granulocytes, incubation of Fura-2-loaded N9 cells with 20 μm (±)-cis,trans-ABA (ABA) in HBSS buffer resulted in a sustained [Ca2+]i rise. After 10 min from the addition of ABA, the [Ca2+]i increased from a basal value of 82 ± 5 to 160 ± 18 nm (n = 5, p < 0.01) (Fig. 1, trace 1). Single-cell recordings showed a similar kinetics of [Ca2+]i increase (not shown), indicating that the steady increase of the fluorescence did not result from the sum of individual spiking signals occurring in single cells. Prolonging the incubation time to 90 min or up to 4 h resulted in a limited further increase of the [Ca2+]i to 194 ± 18 and to 227 ± 18 nm, respectively. After 24 h from the addition of ABA, the [Ca2+]i remained elevated over basal values (170 ± 4 nm). Thus, most of the [Ca2+]i increase induced by ABA on N9 cells occurs during the first 10 min of incubation, reaching a plateau over the subsequent 2–4 h. Finally, 250 nm ABA was sufficient to induce a detectable [Ca2+]i increase to 142 ± 10 nm in 10 min of incubation (not shown).
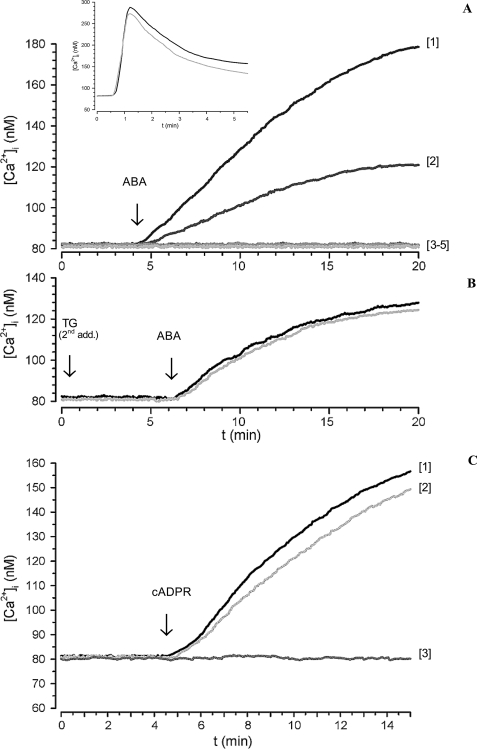
FIGURE 1.
ABA-induced increase of the [Ca2+]i in FURA-2AM-loaded N9 cells. A, ABA (20 μm) was added to cells in HBSS (trace 1) or to cells preincubated either with 8-Br-cADPR (100 μm for 1 h; trace 2), with I-PKA (1 μm for 90 min; trace 3), or with PTX (2 μg/ml for 1 h; trace 4) or to cells in Ca2+-free HBSS containing 0.1 mm EGTA (trace 5). Inset, thapsigargin (10 μm) was added to cells in HBSS (gray trace) or in Ca2+-free HBSS, containing 0.1 mm EGTA (black trace). B, thapsigargin (10 μm) was added to cells in HBSS and the rapid, ensuing [Ca2+]i increase was monitored by fluorescence microscopy (not shown). The cells were then incubated without (black trace) or with 100 μm 8-Br-cADPR (gray trace) for 45 min at 37 °C. A second addition of thapsigargin (TG second addition) was performed under continuous fluorescence recording followed after 6 min by the addition of 20 μm ABA. C, cADPR (100 μm) was added to cells in HBSS (trace 1) or to cells preincubated with 8-Br-cADPR (100 μm for 1 h; trace 2) or to cells in Ca2+-free HBSS, containing 0.1 mm EGTA (trace 3). The traces shown (≥20 cells/field) are from individual experiments, representative of results obtained from at least three experiments. The fluorescence values were acquired every 0.5 s.
When 20 μm ABA was added to N9 cells in the presence of 0.1 mm EGTA in Ca2+-free HBSS, the sustained [Ca2+]i elevation was abrogated (Fig. 1, trace 5). The addition of thapsigargin, an inhibitor of SERCA pumps (22), to N9 cells in the presence or absence of 0.1 mm EGTA, triggered a similar [Ca2+]i increase (inset to Fig. 1A), demonstrating that EGTA did not empty the ER Ca2+ stores. Thus, influx of extracellular calcium is responsible for the generation of the ABA-induced [Ca2+]i increase.
To determine whether cADPR was involved in the ABA-triggered Ca2+ response, intact N9 cells were preincubated with the membrane-permeant cADPR antagonist 8-Br-cADPR (23) and then stimulated with 20 μm ABA. 8-Br-cADPR strongly reduced (by 70%) the ABA-induced [Ca2+]i increase (Fig. 1, trace 2), indicating a causal role of intracellular cADPR in the ABA-promoted Ca2+ response. The presence of a PKA-specific inhibitor (a cell-permeant, myristoylated peptide, I-PKA) abrogated the Ca2+ increase triggered by ABA, indicating a causal role of PKA in the ABA-induced Ca2+ elevation (Fig. 1, trace 3). Finally, the ABA-induced [Ca2+]i increase was abrogated in the presence of PTX (Fig. 1, trace 4). A slow and sustained [Ca2+]i increase, abrogated by EGTA and strongly reduced by 8-Br-cADPR, was also observed in ABA-treated granulocytes, where it was shown to be dependent on the increase of the intracellular level of cADPR ([cADPR]i) (10). A slow and sustained [Ca2+]i increase has been already reported in several human and murine cell types in response to both exogenously added and endogenously produced cADPR (24–27).
To investigate the role of intracellular Ca2+ stores in the ABA-induced [Ca2+]i increase in N9 cells, these stores were depleted by incubation of the cells with thapsigargin, an inhibitor of SERCA pumps (22). After verifying that a second addition of thapsigargin was without effect (Fig. 1B), indicating that the stores were indeed empty, the addition of ABA induced a [Ca2+]i increase (Fig. 1B, black trace) quantitatively lower than that observed in TG-untreated cells (Fig. 1A, trace 1) that was not inhibited by preincubation of the cells with 8-Br-cADPR (Fig. 1B, gray trace). These results suggest that ABA induces an extracellular Ca2+ influx that, in turn, triggers a cADPR-dependent intracellular Ca2+ release. The ABA-induced [Ca2+]i increase (Fig. 1A, trace 1), thus appears to be the sum of a Ca2+ influx (Fig. 1B, black trace) and of an 8-Br-cADPR-inhibitable intracellular Ca2+ release.
To try to reproduce the Ca2+ signaling induced by ABA through the use of its second messenger, we added exogenous cADPR to N9. cADPR can enter several cell types across multiple nucleoside transporters, both equilibrative and concentrative (28–30). cADPR (100 μm) was added to Fura2-loaded N9 cells; as shown in Fig. 1C, exogenous cADPR induced a sustained [Ca2+]i increase, which was prevented by extracellular EGTA and was slightly inhibited (by ∼15%, n = 4) by 8-Br-cADPR. Thus, exogenous cADPR induces a similar Ca2+ response in N9 as ABA, indicating that both the 8-Br-cADPR-sensitive Ca2+ release and the extracellular Ca2+ influx are cADPR-dependent effects, although the contribution of intracellular Ca2+ release to the overall Ca2+ increase is higher for ABA compared with exogenous cADPR.
Involvement of cADPR in the ABA-triggered [Ca2+]i elevation was confirmed by the rise of the [cADPR]i levels following incubation of N9 cells with 20 μm ABA for 30 min; the [cADPR]i increased from a basal value of 2.06 ± 0.40 pmol/mg to 6.88 ± 0.95 pmol/mg (n = 3, p < 0.01). After 24 h of incubation with 20 μm ABA, the [cADPR]i remained elevated (5.70 ± 0.78 pmol/mg) (n = 3, p < 0.01). When N9 cells were treated for 1 h with 20 μm ABA and then further incubated for 23 h in the absence of ABA, the [cADPR]i remained elevated (5.02 ± 0.81 pmol/mg, n = 3), indicating that the continuous presence of ABA was not needed to maintain the increased [cADPR]i.
Role of PKA in the ABA-induced Stimulation of GDP-ribosyl Cyclase Activity in N9 Cells—The ABA-induced increase of the [cADPR]i in N9 microglial cells prompted the investigation of the effect of ABA on the ADPRC activity expressed in these cells by CD38 (12). Exposure of intact N9 cells to ABA resulted in a concentration-dependent increase of the ectocellular GDP-ribosyl cyclase (GDPRC) activity on NGD+ (a NAD+ analog yielding the poorly hydrolyzable cyclic product cGDPR). The GDPRC activity of N9 cells increased by 30, 41, and 50% 10 min after the addition of 0.25, 2.0, and 20 μm ABA, respectively (n = 5 for 0.25 and 2.0 μm, p < 0.01; n = 15 for 20 μm, p < 0.001) (Fig. 2A). After 24 h of incubation with 20 μm ABA, the GDPRC activity remained elevated compared with untreated cells (3.6 ± 0.5 and 2.4 ± 0.6 pmol cGDPR/min/mg protein, respectively; n = 3, p < 0.01). In parallel, N9 cells were incubated for 1 h with 20 μm ABA and then further incubated for 23 h in the absence of ABA; the GDPRC activity was as high as that in cells continuously exposed to ABA for 24 h (3.7 ± 0.5 pmol cGDPR/min/mg protein, n = 3). This result, together with the increased [cADPR]i 24 h after “priming” of the cells with ABA for 1 h (see above), indicates that continuous presence of ABA is not required for long term activation of the cyclase. In contrast to what was observed on cyclase activity, the cADPR hydrolase activity was not increased by ABA treatment at any of the time points investigated. Moreover, expression of CD38 was not significantly modified 24 h after ABA priming of N9 cells, as evaluated by reverse transcription-PCR (not shown).
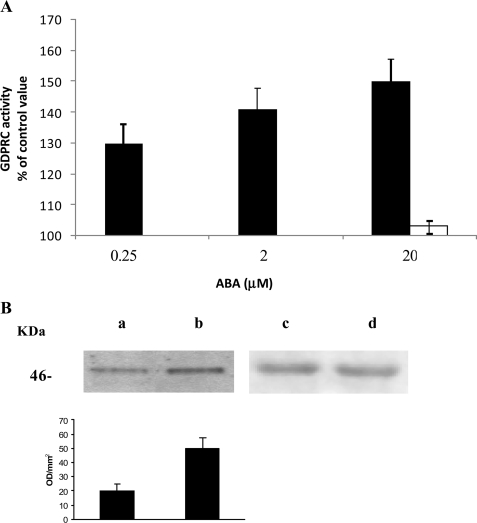
FIGURE 2.
GDP-ribosyl cyclase activity and CD38 phosphorylation in ABA-stimulated N9 cells. A, after addition of the indicated concentrations of ABA to N9 cells (3 × 107), the GDP-ribosyl cyclase activity was determined on intact cells (black bars), as described under “Experimental Procedures.” The cells were also preincubated with I-PKA (1 μm for 90 min) before the addition of 20 μm ABA (white bar). The results are expressed as cGDPR production relative to untreated controls (2.5 ± 0.7 pmol cGDPR/min/mg protein) and are the means ± S.D. of at least five experiments. B, CD38 was immunopurified (see “Experimental Procedures”) from cells preincubated for 10 min without (lanes a and c) or with 20 μm ABA (lanes b and d). Western blots were stained with an anti-phosphoserine mAb (lanes a and b) or with an anti-CD38 mAb (lanes c and d). The graph displayed below the Western blots shows the bioluminescence intensity of the anti-phosphoserine mAb-stained bands, as quantified using the Chemidoc System; the values are expressed as optical density/mm2 and are the means ± S.D. of three experiments.
In human granulocytes, ABA-induced stimulation of A(G)-DPRC activities was shown to be caused by an increase of the [cAMP]i, leading to a PKA-catalyzed phosphorylation of CD38 (10). Thus, the [cAMP]i was evaluated following incubation of N9 cells with 20 μm ABA for 1 min; the [cAMP]i increased from a basal value of 3.66 ± 0.43 pmol/106 cells to 5.64 ± 0.87 pmol/106 cells (n = 3, p < 0.01). In line with its complete inhibition of the ABA-induced [Ca2+]i increase (Fig. 1), PTX abrogated the ABA-induced [cAMP]i increase. N9 cells were also preincubated in the presence or absence of 1 μm I-PKA prior to the addition of 20 μm ABA. I-PKA prevented the ABA-induced cyclase activation (Fig. 2A), in line with its complete inhibition of the ABA-induced [Ca2+]i rise (Fig. 1). To verify CD38 phosphorylation by PKA following exposure of N9 cells to ABA, CD38 was immunopurified from N9 cells incubated for 10 min in the presence or absence (controls) of 20 μm ABA. Western blot analysis of CD38, as developed with an anti-phosphoserine mAb, showed a higher extent of CD38 phosphorylation in N9 cells incubated with ABA as compared with controls (Fig. 2B). These results, together with the inhibition afforded by I-PKA on the ABA-induced [Ca2+]i increase (Fig. 1), demonstrate that stimulation of ADPRC activity by ABA is mediated by a PKA-induced phosphorylation of CD38, similarly to what was observed in human granulocytes (10).
The NetPhos 2.0 server (Center for Biological Sequence Analysis, Technical University of Denmark) predicts two serine residues as possible phosphorylation sites in the intracellular domain of murine CD38: Ser19, scoring the highest phosphorylation potential (0.976), and Ser10, with a phosphorylation potential of 0.713. Three mutants of murine CD38 were generated by site-directed mutagenesis: S10A, S19A, and the double mutant S10A/S19A. N9 cells transfected with WT murine CD38 or with each mutant were then incubated with or without ABA, and the GDPRC activity was measured. GDPRC activity was similar in CD38-transfected cells, WT or mutagenized (0.13 ± 0.02, 0.15 ± 0.02, 0.11 ± 0.01 and 0.14 ± 0.02 nmol cGDPR/min/mg in WT, S10A, S19A, and S10A/S19A, respectively), and this activity was ∼50-fold higher compared with GDPRC activity in untransfected cells (2.5 ± 0.7 pmol cGDPR/min/mg). Thus, the GDPRC activity of native, untransfected CD38 was irrelevant compared with the activity of the transfected protein, enabling the investigation of the effect of CD38 mutation on GDPRC activation, without interference by native CD38. The [cADPR]i in the CD38-transfected cells was increased ∼2-fold compared with untransfected controls (4.3 ± 0.4, 4.2 ± 0.5, and 3.9 ± 0.6 pmol/mg in WT, S10A, and S19A, respectively, compared with a basal level of 2.06 ± 0.40 pmol/mg in untransfected cells; see above). The limited increase of the [cADPR]i as compared with the 50-fold increase of the GDPRC activity in CD38-transfected versus untransfected cells, is likely due to the similar GDPRC-to-cADPR hydrolase ratio (∼1.4) in the transfected and in the untransfected cells. As shown in Fig. 3A, ABA-induced cyclase activation (∼40% increase over unstimulated values) was observed in cells transfected with WT-CD38 but not in cells transfected with each one of its mutants (S10A or S19A) or with the double mutant (S10A/S19A). This result indicates that both Ser10 and Ser19 need to be phosphorylated by PKA to ensure cyclase activation by ABA. Conversely, no activation of the hydrolase activity was observed following ABA treatment of CD38-transfected cells. The higher [cADPR]i in CD38-transfected cells determined an increased basal [Ca2+]i in these cells compared with untransfected cells (∼100 versus 80 nm; Figs. 1A and 3B).
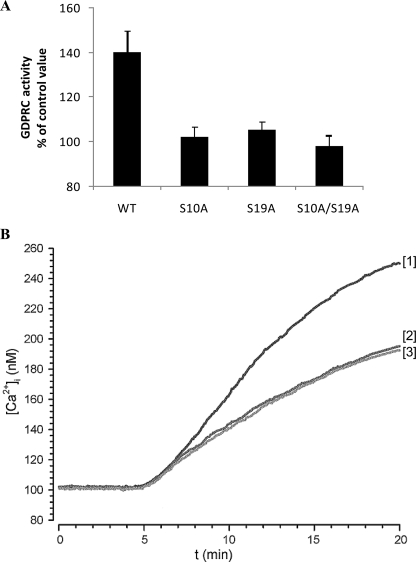
FIGURE 3.
Effect of CD38 site-directed mutagenesis on ABA-induced GDPRC activation and [Ca2+]i increase. N9 cells were transfected with WT CD38 or with the CD38 mutants S10A, S19A or with the double mutant S10A/S19A. A, after addition (or not, control) of 20 μm ABA, the GDP-ribosyl cyclase activity was determined on intact cells. The results are expressed as cGDPR production relative to untreated controls and are the means ± S.D. of four experiments. B, adherent N9 cells, transfected with WT CD38 (trace 1) or with the CD38 mutants S10A (trace 2) or S19A (trace 3) were loaded with FURA-2AM, and the [Ca2+]i changes were monitored as described in Ref. 12. Neither cell type in the absence of ABA showed any [Ca2+]i increase (not shown). The individual traces are representative of results obtained from four experiments.
Analysis of the [Ca2+]i increase induced by ABA in N9 cells transfected with WT and with mutagenized CD38 showed a significantly higher Ca2+ response in WT-CD38 transfected cells (Fig. 3B) compared with untransfected cells (Fig. 1A). Both Ser mutations (S10A or S19A) resulted in a decrease of the Ca2+ response (Fig. 3B) to values similar to those observed in untransfected cells (Fig. 1A). These results support the conclusion that mutation of either Ser10 or Ser19 abrogates the ABA-induced [Ca2+]i increase attributable to transfected CD38, leaving the [Ca2+]i response caused by endogenous CD38 unaltered.
The basal value of the [cADPR]i in CD38-transfected cells was higher compared with the value in untransfected cells (see above). cADPR has been demonstrated to be the second messenger responsible for NO and TNF-α overproduction in LPS-stimulated N9 microglial cells (12). In CD38-transfected N9 (WT, S10A, or S19A, all expressing a similar cyclase activity), basal release of NO (estimated from the accumulation of the end products of NO reduction, nitrites, and nitrates in the culture medium (31) and TNF-α was indeed significantly higher compared with that in untransfected controls and similar in all CD38-transfected cells (1.5 ± 0.2- and 4.1 ± 0.3-fold increase over controls, for NO and TNF-α, respectively; n = 4). Untargeted cell migration across chemotactic chamber filters was also increased 1.6 ± 0.2-fold in CD38-transfected N9 cells, compared with untransfected cells (n = 4).
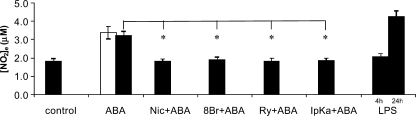
ABA Induces Production and Release of NO and TNF-α by N9 Cells—The ABA-induced increase of the [cADPR]i and of the [Ca2+]i in N9 cells prompted us to investigate whether ABA could stimulate NO and TNF-α release by N9 cells. As shown in Fig. 4, both 0.25 and 20 μm ABA induced an increase of ∼80% of the nitrites and nitrates concentration in the cell supernatant, as compared with untreated cultures (from a basal value of 1.81 ± 0.42 to 3.40 ± 0.5 and 3.22 ± 0.44 μm, respectively; n = 9, p < 0.001; Fig. 4). These values are in the range of nitrite and nitrate production observed upon stimulation of N9 cells for 4 or 24 h with LPS (Fig. 4), a known activator of NO production in microglia. The causal role of the CD38/cADPR system in activating NO production and release in ABA-treated N9 cells was demonstrated by the strong inhibition of nitrites and nitrates release obtained in the presence of 8-Br-cADPR, ryanodine (both potent cADPR antagonists) or nicotinamide (an inhibitor of ADPRC). Moreover, the ABA-induced nitrite and nitrate release was prevented by I-PKA (Fig. 4).
FIGURE 4.
ABA induces NO release from N9 cells. N9 cells (2.5 × 105/sample) were incubated with 0.25 μm (white bar) or with 20 μm (black bar) ABA for 4 h. The cells were preincubated with 8-Br-cADPR (100 μm for 90 min), with nicotinamide (10 mm for 5 min), with ryanodine (50 μm for 1 h), or with I-PKA (1 μm for 90 min) and then stimulated with 20 μm ABA for 4 h. As a positive control, N9 cells were also incubated with 100 ng/ml LPS for 4 or 24 h. The culture supernatants were recovered and centrifuged, and their nitrite concentrations were assayed as described under “Experimental Procedures.” The results are the means ± S.D. of nine different experiments. *, p < 0.001.
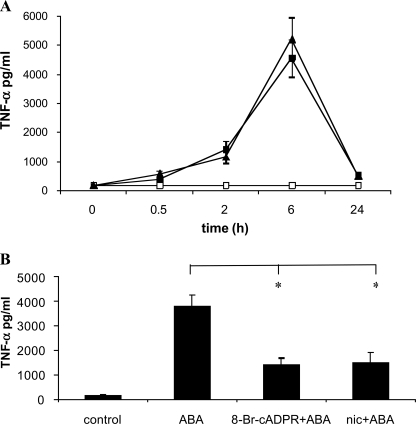
In addition to enhancing NO generation and release, ABA also stimulated TNF-α release from N9 cells. TNF-α release was highest 6 h after cell stimulation with ABA and 250 nm ABA proved to be as effective as 20 μm ABA in stimulating TNF-α release (Fig. 5A). Pretreatment of the cells with 8-Br-cADPR or nicotinamide significantly reduced the ABA-triggered TNF-α release (Fig. 5B), indicating involvement of the CD38/cADPR system also in the stimulation by ABA of TNF-α release from N9 cells.
FIGURE 5.
ABA induces TNF-α release from N9 cells. A, N9 cells were incubated without (white squares) or with 0.25 μm (black triangles) or 20 μm (black squares) ABA. At the times indicated, the culture supernatants were recovered and centrifuged, and the TNF-α concentration was determined as described under “Experimental Procedures.” The results are the means ± S.D. of three different experiments. B, N9 cells were preincubated in the absence (control) or in the presence of 8-Br-cADPR (100 μm for 90 min) or of nicotinamide (nic) (10 mm for 5 min) and then stimulated with 20 μm ABA for 6 h. The TNF-α concentration in the culture supernatants was determined, and results shown are the means ± S.D. of seven different experiments. *, p < 0.001.
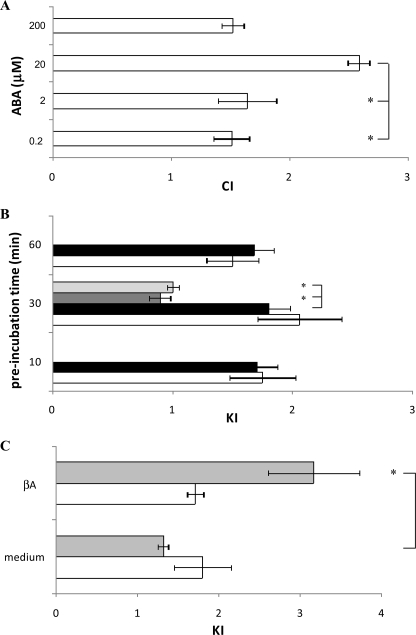
ABA Stimulates Chemotaxis and Chemokinesis in N9 Cells—Different concentrations of ABA, placed in the bottom well of a chemotaxis migration chamber, induced a bell-shaped chemotactic response in N9 cells, with maximal migration being recorded toward 20 μm ABA (Fig. 6A). A bell-shaped CI curve is not unprecedented, because it has been also observed in the chemotactic response of granulocytes to ABA (10) and to NAD+ (27), of dendritic cells to ATP (32), and in the chemotactic response of microglia to nerve growth factor (33) and to f-MLP, W peptide, and Aβ42 (34).
FIGURE 6.
ABA-induced chemotaxis and chemokinesis in N9 cells. Migration of N9 cells through 8-μm pore membranes was measured in ChemoTx chambers. A, cell migration toward a solution containing ABA at the concentrations indicated is expressed as CI. The results shown are the means ± S.D. from three different determinations (p < 0.001, compared with CI values toward medium). *, p < 0.01. B, N9 cells were incubated for the time indicated on the y axis with either 0.25 μm (white bars) or with 20 μm (black bars) ABA and then placed in the ChemoTx chamber to migrate toward medium for 90 min. The cells were preincubated with 8-Br-cADPR (100 μm for 90 min; gray bar) or with nicotinamide (10 mm for 5 min; light gray bar) before exposure to 20 μm ABA for 30 min. The results are expressed as KI and are the means ± S.D. from seven different experiments (p < 0.001, compared with KI values of untreated N9 cells). *, p < 0.001. C, N9 cells were preincubated for 30 min (white bars) or for 24 h (gray bars) with 20 μm ABA and then challenged to migrate toward medium or toward 15 μm βA. Untreated N9 migrated similarly toward medium and toward βA (KI = 1.1; not shown). The results are expressed as KI and are the means ± S.D. from four different experiments (*, p < 0.01).
To assess the effect of ABA on chemokinesis (untargeted cell movement), N9 cells were preincubated with 0.25 or 20 μm ABA for 10, 30, or 60 min and then placed on top of the filter of a chemotaxis chamber containing buffer in the bottom well. Cell migration through the filter was increased in ABA-treated cells compared with untreated controls, with no significant difference being observed at the various incubation times explored (Fig. 6B), in line with the rapid activation (within minutes) of the hormone signaling pathway via CD38 phosphorylation (Fig. 2) and activation and the consequent [Ca2+]i increase (Fig. 1). Preincubation of the cells with 8-Br-cADPR or with nicotinamide, prior to exposure to 20 μm ABA for 30 min, abrogated chemokinesis induced by ABA (Fig. 6B). Next, we investigated whether exogenous cADPR, which induced a [Ca2+]i increase (Fig. 1C), could stimulate chemokinesis of microglial cells. Indeed, preincubation of N9 cells with 100 μm cADPR stimulated untargeted cell movement (KI values of 1.23 ± 0.02 and 2.84 ± 0.70, after 30 min and 24 h of preincubation, respectively; not shown), confirming involvement of the cADPR/[Ca2+]i signaling in the migration response of N9 to ABA.
To test whether ABA priming of N9 resulted in an increased sensitivity of the cells to the known chemoattractant amyloid βA, similarly to what reported for LPS priming (34), N9 cells were preincubated with either 0.25 or 20 μm ABA for 30 min or for 24 h, and cell migration toward βA was evaluated. As shown in Fig. 6C, the cells pretreated with 20 μm ABA for 30 min migrated similarly toward medium or βA. Conversely, a 24-h pretreatment with 20 μm ABA greatly enhanced chemotaxis toward βA (CI values were 3.2 versus 1.3, toward βA and medium, respectively). Similar results were obtained upon preincubation of cells with 0.25 μm ABA (not shown). Thus, ABA priming of N9 cells stimulates cell migration toward βA, the primary plaque component in Alzheimer disease (35).
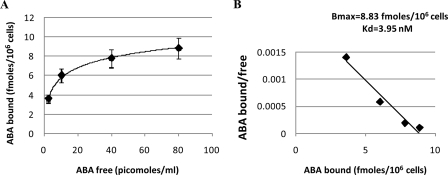
Binding of ABA to N9 Cells—Presence of high affinity ABA-binding sites in N9 cells was investigated with radioactive ABA. Intact N9 cells were incubated with [3H]ABA at increasing concentrations, in the presence or absence of excess unlabeled ABA. Scatchard plot analysis of the results indicated the presence of ∼5.4 × 103 high affinity binding sites/cell, with a Kd of 4 nm (Fig. 7), both values being similar to those reported in human granulocytes (10).
FIGURE 7.
ABA-binding sites on N9 cell. N9 cells were incubated in the presence of increasing concentrations of [3H]ABA, with or without excess unlabeled ABA. Radioactivity of washed cell pellets was determined on a Packard β-counter. A, ABA binding (n = 4). B, Scatchard plot analysis of ABA-binding sites. The results are the means ± S.D. from three different experiments.
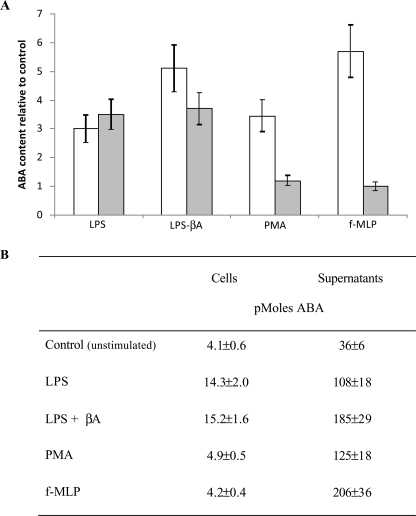
ABA Content in N9 Cells—ABA was detected by HPLC-mass spectrometry in acid extracts of unstimulated N9 cells at 0.31 ± 0.07 pmol/mg protein (n = 5). The intracellular ABA concentration ([ABA]i) increased in N9 cells challenged with LPS; in cells incubated for 72 h in the presence of 100 ng/ml LPS, the [ABA]i increased to 1.32 ± 0.29 pmol/mg protein (n = 5, p < 0.0001). Comparable results were obtained with a sensitive enzyme-linked immunosorbent assay specific for (+)-cis,trans-ABA, which was utilized for subsequent experiments (Fig. 8). The ABA concentration in the medium ([ABA]m) of N9 cells incubated for 72 h in the presence or absence (controls) of LPS was evaluated; the [ABA]m of LPS-treated cells increased 3-fold over controls. Interestingly, when cells were primed with LPS for 48 h, to induce overexpression of the βA receptor (34) and then exposed to βA for further 24 h, the [ABA]m further increased to values 5-fold higher than controls. Similarly, the [ABA]i was also increased by stimulation with βA (3.7-fold over control cells). Finally, when N9 cells were incubated for 1 h in the presence of f-MLP or PMA, the [ABA]m was also increased 5.7- and 3.4-fold, respectively, over untreated cells (Fig. 8).
FIGURE 8.
Effect of LPS, βA, PMA, and f-MLP on ABA content in N9 cells. N9 cells were incubated in complete medium with LPS (100 ng/ml) for 72 h, where 10 μm βA was added (or not) during the last 24 h of incubation, or with PMA (0.1 μg/ml for 1 h) or with f-MLP (1 μm for 1 h). The ABA content in cells (3 × 107/determination; gray bars) and in supernatants (36 ml of total volume; white bars) was determined by enzyme-linked immunosorbent assay. A, the results are expressed as ABA content relative to control, unstimulated cultures. B, total amount of ABA in the cell extracts and in the culture supernatants. The results shown are the means ± S.D. (n = 3).
DISCUSSION
N9 is a murine microglial cell line extensively utilized by researchers in the field of neurobiology, because it is generally considered a reliable in vitro model for microglia; it enables the study of microglial function in the absence of contaminating cell types, which are present in primary cultures and may complicate interpretation of the relative contribution of each cell type to the observed functional response (36). The results presented here indicate that the plant hormone ABA behaves as a pro-inflammatory hormone in N9 cells, similarly to what has been recently observed in human granulocytes. Exogenous ABA stimulates release of NO and TNF-α, chemotaxis and chemokinesis in N9 cells, and cell stimulation with LPS, PMA, or f-MLP induces ABA production and release, indicating that ABA is an endogenous microglial hormone.
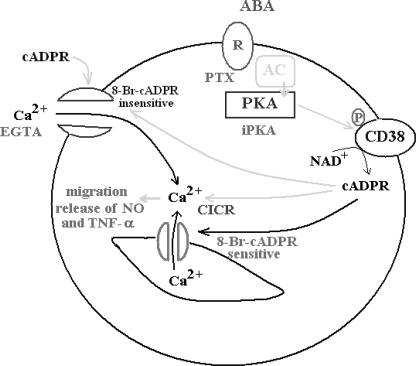
The ABA signaling pathway in N9 microglial cells is summarized in Fig. 9. Extracellular ABA binds to a PTX-sensitive receptor-G protein complex, as demonstrated by presence of high affinity binding sites for ABA on intact N9 cells (Fig. 7) and by the inhibition of the ABA-induced increase of the [cAMP]i (see “Results”) and of the [Ca2+]i exerted by PTX (Fig. 1). The enhanced [cAMP]i levels activate PKA, leading to CD38 phosphorylation (Fig. 2B) and to the rapid up-regulation of its A(G)DPRC activity (Fig. 2A), which is prevented by the PKA-specific inhibitor (Fig. 2A). Both Ser10 and Ser19 of the short intracellular domain of CD38 need to be phosphorylated to obtain cyclase activation, as monitored extracellularly on intact cells (Fig. 3). Because CD38 has been demonstrated to occur as a dimer on the cell plasma membrane (37–40), the presence of four phosphorylated Ser residues on the cytoplasmic tails of a CD38 dimer apparently results in a conformational change capable of affecting the ectocellular catalytic domain.
FIGURE 9.
Schematic representation of the mechanism of the ABA-induced [Ca2+]i increase in N9 cells. Interaction of ABA with a plasma membrane PTX-sensitive receptor/G protein complex triggers the activation of AC, overproduction of cAMP, and stimulation of PKA, which can be prevented by the specific inhibitor (iPKA). PKA phosphorylates CD38 stimulating cADPR overproduction, which induces extracellular Ca2+ influx through an 8-Br-cADPR-insensitive plasma membrane Ca2+ channel and activates intracellular Ca2+ release through an 8-Br-sensitive Ca2+-induced Ca2+-release (CICR). Exogenously added cADPR also induces an 8-Br-cADPR-insensitive Ca2+ influx. The increased [Ca2+]i levels stimulate functional responses (cell migration, release of NO, and TNF-α).
The ABA-induced [Ca2+]i increase is due to extracellular Ca2+ influx, because it is abrogated by EGTA (Fig. 1A, trace 5). However, a contribution of a cADPR-dependent Ca2+-induced Ca2+ release to the overall ABA-induced Ca2+ response is demonstrated by its partial inhibition by 8-Br-cADPR (Fig. 1A, trace 2) and by the fact that depletion of intracellular Ca2+ stores by thapsigargin results in a significantly lower Ca2+ increase (Fig. 1B, black trace). Abrogation by the PKA inhibitor of the ABA-induced Ca2+ response (Fig. 1A, trace 3) is in agreement with the fact that cADPR, produced downstream of PKA activation, opens a plasma membrane Ca2+ channel (Fig. 9).
These results demonstrate that, in N9 cells, a cADPR-dependent influx of extracellular Ca2+ precedes and is necessary for the cADPR-mediated intracellular Ca2+ release (Fig. 9). The same holds true for human granulocytes: (i) thapsigargin-treated human granulocytes also respond to ABA with a Ca2+ influx, resulting in a Ca2+ increase lower than that observed when the intracellular Ca2+ stores are not pre-emptied, and (ii) exogenous cADPR induces a Ca2+ influx.3 This new mechanistic information does not contradict previously published results (10); rather, it provides further evidence supporting a role for cADPR in both a Ca2+ influx and intracellular Ca2+ release in ABA-treated cells.
A cADPR-dependent Ca2+ entry was reported in human T-lymphocytes (41), in neutrophils (42), and, more recently, in murine insulinoma cells, human T lymphocytes, and neutrophils, where cADPR has been shown to activate 8-Br-cADPR-sensitive TRPM2 channels (43–45). A role for TRPM2 in the cADPR-mediated, ABA-induced Ca2+ influx in N9 cells seems unlikely because 8-Br-cADPR does not inhibit the extracellular Ca2+ influx in thapsigargin-treated, ABA-stimulated N9 cells (Fig. 1B, gray trace).
The ABA-triggered signaling pathway shows striking similarities with the plant and sponge ABA signaling pathways. In plants, ABA triggers PLC activation via a G protein-linked receptor (46) and stimulates ADPRC, leading to cADPR overproduction, [Ca2+]i increase, gene transcription, and stomatal closure (47). In sponges, ABA triggers PKA-dependent ADPRC phosphorylation and cADPR-mediated [Ca2+]i increase, resulting in stimulation of water filtration and oxygen consumption (21, 48).
The functional effects induced by ABA on microglial cells (overproduction and release of NO and TNF-α, stimulation of cell migration) lie downstream of the CD38/cADPR/[Ca2+]i signaling pathway, because all are inhibited in the presence of 8-Br-cADPR (Figs. 4, 5B, and 6B), ryanodine (Fig. 4), nicotinamide (Figs. 4, 5B, and 6B), or I-PKA (Fig. 4). A role of cADPR as the second messenger in microglial activation is not unprecedented, because it has been already described in the LPS-induced functional activation of N9 cells, resulting in release of NO and of TNF-α (12). In light of the presence of high affinity (Kd, 4 nm) ABA-binding sites on intact N9 cells (Fig. 7), ABA release by LPS-stimulated N9 cells (Fig. 8) might indeed contribute to the functional activation of these cells through an autocrine-positive feedback mechanism.
Together with LPS, PMA and f-MLP also induce ABA release from N9 cells (Fig. 8). The fact that the [ABA]i is higher in LPS-treated cells than in untreated cells is likely the result of stimulation of ABA synthesis over the 72-h incubation with LPS, resulting in both ABA release into the medium and accumulation of intracellular ABA. Conversely, the [ABA]i is not increased in cells stimulated for 1 h with PMA or with f-MLP, despite the increased release of ABA into the medium, possibly because of the short incubation time, which does not allow accumulation of intracellular ABA to become apparent. In any case, the fact that several stimuli known to induce glial (as well as inflammatory cell) activation also stimulate ABA release from these cells points to a general role of ABA in the inflammatory response of microglial cells.
Microglial cells are believed to play an important pathogenetic role in Alzheimer disease, because amyloid plaques contain a variety of inflammation-related proteins (complement factors, acute-phase proteins, pro-inflammatory cytokines) and clusters of activated microglia (49). Deposits of fibrillar βA, the main component of the Alzheimer disease plaque, attract microglial cells and activate them to produce and release inflammatory mediators, some of which are involved in a feedback mechanism inducing further microglial recruitment and activation, eventually leading to localized neuronal damage (35). Our results demonstrate that ABA is a new member of this family of inflammatory mediators involved in the cross-talk between microglia and Aβ deposits. Specifically, ABA-activated N9 cells acquire the capacity to migrate toward βA (Fig. 6C) and N9 cells, pretreated with LPS to induce overexpression of the βA receptor (34), release ABA upon exposure to βA (Fig. 8). Thus, the following positive feedback mechanism between ABA and βA, maintaining microglial cells in an activated state, can be outlined; activated microglial cells release ABA, which stimulates chemotaxis toward βA, which in turn up-regulates ABA release. The fact that ABA also stimulates microglial release of NO and TNF-α (Figs. 4 and 5A), which are known to play a pivotal role in neuronal damage (50–53), suggests a central role for this hormone in the pathogenesis of microglia-dependent nervous tissue damage.
These results may suggest a new target for anti-inflammatory therapy, e.g. through the use of ABA antagonists, aimed at preventing or reducing microglial activation in those conditions (ischemia, neurodegenerative diseases) where neuroinflammation is believed to significantly contribute to brain tissue damage (54–57). Indeed, ABA has been detected in the mammalian brain (58, 59), suggesting a possible role for ABA in the central nervous system, as already proposed for other inflammatory mediators (60, 61).
This work was supported in part by grants from the Associazione Italiana per la Ricerca sul Cancro, Italian Ministry of Education; University and Scientific Research Grants MIUR-PRIN 2005, MIUR FIRB RBAUO19A3C, MIUR FIRB RBNE01ERXR, MIUR FIRB RBLA039LSF, and MIUR FIRB RBIP06LSS2; grants from Regione Liguria, and grants from the University of Genova and Fondazione Cassa di Risparmio di Genova e Imperia.
Footnotes
The abbreviations used are: ABA, abscisic acid; cADPR, cyclic ADP-ribose; LPS, lipopolysaccharide; HBSS, Hanks' balanced salt solution; cGDPR, cyclic GDP-ribose; mAb, monoclonal antibody; βA, amyloid β peptide; PKA, cyclic AMP-activated protein kinase; I-PKA, protein kinase A inhibitor; ADPRC, ADP-ribosyl cyclase; GDPRC, GDP-ribosyl cyclase; PTX, pertussis toxin; CI, chemotaxis index; KI, chemokinesis index; TNF, tumor necrosis factor; PMA, phorbol myristate acetate; WT, wild type; HPLC, high pressure liquid chromatography; f-MLP, formyl-methionyl-leucyl-phenylalanine.
S. Bruzzone, unpublished data.
References
- 1.Farber, K., and Kettenmann, H. (2005) Brain Res. Brain Res. Rev. 48 133–143 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, A., Kann, O., Ohlemeyer, C., Hanisch, U. K., and Kettenmann, H. (2003) J. Neurosci. 23 4410–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura, Y., Ohmaki, M., Murakami, K., and Yoneda, Y. (2003) Brain Res. 962 122–128 [DOI] [PubMed] [Google Scholar]
- 4.Town, T., Nikolic, V., and Tan, J. (2005) J. Neuroinflammation 2 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paris, D., Town, T., and Mullan, M. (2000) Neurosci. Lett. 278 5–8 [DOI] [PubMed] [Google Scholar]
- 6.Gebicke-Haerter, P. J. (2001) Microsc. Res. Tech. 54 47–58 [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Scarano, F., and Baltuch, G. (1999) Annu. Rev. Neurosci. 22 219–240 [DOI] [PubMed] [Google Scholar]
- 8.Aschner, M., Allen, J. W., Kimelberg, H. K., LoPachin, R. M., and Streit, W. J. (1999) Annu. Rev. Pharmacol. Toxicol. 39 151–173 [DOI] [PubMed] [Google Scholar]
- 9.Grill, E., and Himmelbach, A. (1998) Curr. Opin. Plant Biol. 1 412–418 [DOI] [PubMed] [Google Scholar]
- 10.Bruzzone, S., Moreschi, I., Usai, C., Guida, L., Damonte, G., Salis, A., Scarfì, S., Millo, E., De Flora, A., and Zocchi, E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, H. C. (2002) Cyclic ADP-ribose and NAADP: Structures, Metabolism and Functions, Kluwer Academic Publishers, Norwell, MS
- 12.Franco, L., Bodrato, N., Moreschi, I., Usai, C., Bruzzone, S., Scarfì, S., Zocchi, E., and De Flora, A. (2006) J. Neurochem. 99 165–176 [DOI] [PubMed] [Google Scholar]
- 13.Righi, M., Mori, L., De Libero, G., Sironi, M., Biondi, A., Mantovani, A., Donini, S. D., and Ricciardi-Castagnoli, P. (1989) Eur. J. Immunol. 19 1443–1448 [DOI] [PubMed] [Google Scholar]
- 14.Zocchi, E., Daga, A., Usai, C., Franco, L., Guida, L., Bruzzone, S., Costa, A., Marchetti, C., and De Flora, A. (1998) J. Biol. Chem. 273 8017–8024 [DOI] [PubMed] [Google Scholar]
- 15.Graeff, R., and Lee, H. C. (2002) Biochem. J. 367 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graeff, R. M., Walseth, T. F., Fryxell, K., Branton, W. D., and Lee, H. C. (1994) J. Biol. Chem. 269 30260–30267 [PubMed] [Google Scholar]
- 17.Franco, L., Zocchi, E., Usai, C., Guida, L., Bruzzone, S., Costa, A., and De Flora, A. (2001) J. Biol. Chem. 24 21642–42168 [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. M. (1976) Anal. Biochem. 72 248–252 [DOI] [PubMed] [Google Scholar]
- 19.Bruzzone, S., De Flora, A., Usai, C., Graeff, R., and Lee, H. C. (2003) Biochem. J. 375 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vercammen, D., Vandenabeele, P., Beyaert, R., Declercq, W., and Fiers, W. (1997) Cytokine 9 801–808 [DOI] [PubMed] [Google Scholar]
- 21.Zocchi, E., Carpaneto, A., Cerrano, C., Bavestrello, G., Giovine, M., Bruzzone, S., Guida, L., Franco, L., and Usai, C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14859–14864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inesi, G., and Sagara, Y. (1992) Arch. Biochem. Biophys. 298 313–317 [DOI] [PubMed] [Google Scholar]
- 23.Walseth, T. F., and Lee, H. C. (1993) Biochim. Biophys. Acta 1178 235–242 [DOI] [PubMed] [Google Scholar]
- 24.Podestà, M., Zocchi, E., Pitto, A., Usai, C., Franco, L., Bruzzone, S., Guida, L., Bacigalupo, A., Scadden, D. T., Walseth, T. F., De Flora, A., and Daga, A. (2000) FASEB J. 14 680–690 [DOI] [PubMed] [Google Scholar]
- 25.Verderio, C., Bruzzone, S., Zocchi, E., Fedele, E., Schenk, U., De Flora, A., and Matteoli, M. (2001) J. Neurochem. 78 1–13 [DOI] [PubMed] [Google Scholar]
- 26.Moreschi, I., Bruzzone, S., Nicholas, R. A., Fruscione, F., Sturla, L., Benvenuto, F., Usai, C., Meis, S., Kassack, M. U., Zocchi, E., and De Flora, A. (2006) J. Biol. Chem. 281 31419–31429 [DOI] [PubMed] [Google Scholar]
- 27.Bruzzone, S., Moreschi, I., Guida, L., Usai, C., Zocchi, E., and De Flora, A. (2006) Biochem. J. 393 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guida, L., Bruzzone, S., Sturla, L., Franco, L., Zocchi, E., and De Flora, A. (2002) J. Biol. Chem. 277 47097–47105 [DOI] [PubMed] [Google Scholar]
- 29.Guida, L., Franco, L., Bruzzone, S., Sturla, L., Zocchi, E., Basile, G., Usai, C., and De Flora, A. (2004) J. Biol. Chem. 279 22066–22075 [DOI] [PubMed] [Google Scholar]
- 30.Podestà, M., Benvenuto, F., Pitto, A., Figari, O., Bacigalupo, A., Bruzzone, S., Guida, L., Franco, L., Paleari, L., Bodrato, N., Usai, C., De Flora, A., and Zocchi, E. (2005) J. Biol. Chem. 280 5343–5349 [DOI] [PubMed] [Google Scholar]
- 31.Lee, H., Jeong, J., Son, E., Mosa, A., Cho, G. J., Choi, W. S., Ha, J. H., Kim, I. K., Lee, M. G., Kim, C. Y., and Suk, K. (2004) J. Neuroimmunol. 156 88–95 [DOI] [PubMed] [Google Scholar]
- 32.Idzko, M., Dichmann, S., Ferrari, D., Di Virgilio, F., la Sala, A., Girolomoni, G., Panther, E., and Norgauer, J. (2002) Blood 100 925–932 [DOI] [PubMed] [Google Scholar]
- 33.De Simone, R., Ambrosini, E., Carnevale, D., Ajmone-Cat, M. A., and Minghetti, L. (2007) J. Neuroimmunol. 190 53–60 [DOI] [PubMed] [Google Scholar]
- 34.Cui, Y. H., Le, Y., Gong, W., Proost, P., Van Damme, J., Murphy, W. J., and Wang, J. M. (2002) J. Immunol. 168 434–442 [DOI] [PubMed] [Google Scholar]
- 35.Rogers, J., and Lue, L. F. (2001) Neurochem. Int. 39 333–340 [DOI] [PubMed] [Google Scholar]
- 36.Saura, J. (2007) J. Neuroinflammation 4 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruzzone, S., Guida, L., Franco, L., Zocchi, E., Corte, G., and De Flora, A. (1998) FEBS Lett. 433 275–278 [DOI] [PubMed] [Google Scholar]
- 38.Khoo, K. M., and Chang, C. F. (1998) Biochem. Mol. Biol. Int. 44 841–850 [DOI] [PubMed] [Google Scholar]
- 39.Berruet, L., Muller-Steffner, H., and Schuber, F. (1998) Biochem. Mol. Biol. Int. 46 847–855 [PubMed] [Google Scholar]
- 40.Moreno-García, M. E., Partida-Sánchez, S., Primack, J., Sumoza-Toledo, A., Muller-Steffner, H., Schuber, F., Oppenheimer, N., Lund, F. E., and Santos-Argumedo, L. (2004) Eur. J. Biochem. 271 1025–1034 [DOI] [PubMed] [Google Scholar]
- 41.Guse, A. H., Berg, I., da Silva, C. P., Potter, B. V., and Mayr, G. W. (1997) J. Biol. Chem. 272 8546–8550 [DOI] [PubMed] [Google Scholar]
- 42.Partida-Sánchez, S., Cockayne, D. A., Monard, S., Jacobson, E. L., Oppenheimer, N., Garvy, B., Kusser, K., Goodrich, S., Howard, M., Harmsen, A., Randall, T. D., and Lund, F. E. (2001) Nat. Med. 7 1209–1216 [DOI] [PubMed] [Google Scholar]
- 43.Togashi, K., Hara, Y., Tominaga, T., Higashi, T., Konishi, Y., Mori, Y., and Tominaga, M. (2006) EMBO J. 25 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beck, A., Kolisek, M., Bagley, L. A., Fleig, A., and Penner, R. (2006) FASEB J. 20 962–964 [DOI] [PubMed] [Google Scholar]
- 45.Lange, I., Penner, R., Fleig, A., and Beck, A. (2008) Cell Calcium 44 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, X., Yue, Y., Li, B., Nie, Y., Li, W., Wu, W. H., and Ma, L. (2007) Science 315 1712–1716 [DOI] [PubMed] [Google Scholar]
- 47.Wu, Y., Kuzma, J., Marechal, E., Graeff, R., Lee, H. C., Foster, R., and Chua, N. H. (1997) Science 278 2126–2130 [DOI] [PubMed] [Google Scholar]
- 48.Zocchi, E., Basile, G., Cerrano, C., Bavestrello, G., Giovine, M., Bruzzone, S., Guida, L., Carpaneto, A., Magrassi, R., and Usai, C. (2003) J. Cell Sci. 116 629–636 [DOI] [PubMed] [Google Scholar]
- 49.Eikelenboom, P., Veerhuis, R., Scheper, W., Rozemuller, A. J., Van Gool, W. A., and Hoozemans, J. J. (2006) J. Neural. Transm. 113 1685–1695 [DOI] [PubMed] [Google Scholar]
- 50.Dickson, D. W., Lee, S. C., Mattiace, L. A., Yen, S. H., and Brosnan, C. (1993) Glia 7 75–83 [DOI] [PubMed] [Google Scholar]
- 51.Chao, C. C., Hu, S., and Peterson, P. K. (1995) Crit. Rev. Neurobiol. 9 189–205 [PubMed] [Google Scholar]
- 52.Kreutzeberg, G. W. (1996) Trends Neurosci. 19 312–318 [DOI] [PubMed] [Google Scholar]
- 53.Ghoshal, A., Das, S., Ghosh, S., Mishra, M. K., Sharma, V., Koli, P., Sen, E., and Basu, A. (2007) Glia 55 483–496 [DOI] [PubMed] [Google Scholar]
- 54.Gebicke-Haerter, P. J., Van Calker, D., Norenberg, W., and Illes, P. (1996) Neurochem. Int. 29 1–12 [PubMed] [Google Scholar]
- 55.Tan, J., Town, T., Paris, D., Placzek, A., Parker, T., Crawford, F., Yu, H., Humphrey, J., and Mullan, M. (1999) J. Neuroimmunol. 97 77–85 [DOI] [PubMed] [Google Scholar]
- 56.Hirsch, E. C., Breidert, T., Rousselet, E., Hunot, S., Hartmann, A., and Michel, P. P. (2003) Ann. N. Y. Acad. Sci. 991 214–228 [DOI] [PubMed] [Google Scholar]
- 57.Wilms, H., Zecca, L., Rosenstiel, P., Sievers, J., Deuschl, G., and Lucius, R. (2007) Curr. Pharm. Des. 13 1925–1928 [DOI] [PubMed] [Google Scholar]
- 58.Le Page-Degivry, M. T., Bidard, J. N., Rouvier, E., Bulard, C., and Lazdunski, M. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 1155–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen, F. S. C., MacTaggart, J. M., Wang, L. C. H., and Westly, J. C. (1988) Agric. Biol. Chem. 52 1273–1274 [Google Scholar]
- 60.Adler, M. W., and Rogers, T. J. (2005) J. Leukocyte Biol. 78 1204–1209 [DOI] [PubMed] [Google Scholar]
- 61.Vitkovic, L., Bockaert, J., and Jacque, C. (2000) J. Neurochem. 74 457–471 [DOI] [PubMed] [Google Scholar]