Abstract
Continuous bone remodeling in response to mechanical loading is critical for skeletal integrity, and interstitial fluid flow is an important stimulus for osteoblast/osteocyte growth and differentiation. However, the biochemical signals mediating osteoblast anabolic responses to mechanical stimulation are incompletely understood. In primary human osteoblasts and murine MC3T3-E1 cells, we found that fluid shear stress induced rapid expression of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNAs; these genes encode transcriptional regulators that maintain skeletal integrity. Fluid shear stress increased osteoblast nitric oxide (NO) synthesis, leading to activation of cGMP-dependent protein kinase (PKG). Pharmacological inhibition of the NO/cGMP/PKG signaling pathway blocked shear-induced expression of all four fos family genes. Induction of these genes required signaling through MEK/Erk, and Erk activation was NO/cGMP/PKG-dependent. Treating cells with a membrane-permeable cGMP analog partly mimicked the effects of fluid shear stress on Erk activity and fos family gene expression. In cells transfected with small interfering RNAs (siRNA) specific for membrane-bound PKG II, shear- and cGMP-induced Erk activation and fos family gene expression was nearly abolished and could be restored by transducing cells with a virus encoding an siRNA-resistant form of PKG II; in contrast, siRNA-mediated repression of the more abundant cytosolic PKG I isoform was without effect. Thus, we report a novel function for PKG II in osteoblast mechanotransduction, and we propose a model whereby NO/cGMP/PKG II-mediated Erk activation and induction of c-fos, fra-1, fra-2, and fosB/ΔfosB play a key role in the osteoblast anabolic response to mechanical stimulation.
Mechanical stress is a primary determinant of bone growth and remodeling; the strength of bone increases with weight bearing and muscular activity and decreases with unloading and disuse (1, 2). Weight bearing and locomotion stimulate interstitial fluid flow through the bone canalicular system, and the resultant shear stress is thought to be a major mechanism whereby mechanical forces stimulate bone growth (1–4). Fluid shear stress activates various signal transduction pathways and initiates an anabolic response in osteocytes and osteoblasts, leading to changes in gene expression and increased cell proliferation and differentiation (1, 5). As part of this response, a rapid and transient increase in intracellular calcium, nitric oxide (NO),2 and prostaglandin E2 occurs, and transcription of genes such as c-fos, cox-2, and igf-1/2 is induced (1, 2, 5).
The transcription factor complex AP1, composed of Fos and Jun proteins, plays an essential role in bone development and post-natal skeletal homeostasis (6). De-regulated c-fos expression in mice interferes with normal bone development and induces osteosarcomas, whereas c-Fos-deficient mice develop osteopetrosis because of an early arrest in osteoclast differentiation (7, 8). Mice overexpressing Fra-1, Fra-2, or ΔFosB (a splice variant of FosB) exhibit a progressive increase in bone mass because of enhanced osteoblast differentiation and bone formation in the presence of normal osteoclast activity (6, 9, 10). In contrast, conditional loss of Fra-1, Fra-2, or JunB leads to defective osteoblast differentiation and severe osteopenia (6, 11, 12). Mechanical loading of long bones or vertebral bodies in rodents rapidly induces c-fos mRNA in osteoblasts/osteocytes (13–16). The same response occurs in cultured osteoblasts exposed to diverse mechanical stimuli such as fluid shear stress, stretch, pulsed ultrasound, or gravitational force (17–24). The mechanism(s) regulating mechanically induced c-fos expression are poorly understood, and inhibition of calcium influx and NO or prostaglandin synthesis appears to affect c-fos induction differentially, depending on the type of mechanical stimulation (14, 15, 17, 19, 21).
NO synthesis inhibitors prevent new bone formation induced by mechanical stimulation (14, 25), and endothelial NO synthase (eNOS, type III NOS) appears to be the predominant NOS isoform expressed in bone (26). eNOS-deficient mice demonstrate reduced post-natal bone mass because of defects in osteoblast number and maturation and impaired bone formation in response to mechanical stimulation and estrogens (27–29). Osteoblasts exhibit a biphasic response to NO donors, with low doses promoting proliferation and differentiation, and high doses inducing apoptosis (30–32). Low doses of NO donors alleviate ovariectomy-induced bone loss in rats, and a randomized, placebo-controlled trial in healthy post-menopausal women demonstrated increased bone formation in subjects receiving the NO donor isosorbide mononitrate (33, 34). These data indicate an important role of NO in osteoblast biology, but little is known about signaling downstream of NO in osteoblasts (35).
One of the major intracellular targets of NO is soluble guanylate cyclase (sGC), which is activated on NO binding to the heme prosthetic group of the enzyme. The resultant increase in cGMP affects multiple target proteins, including cGMP-dependent protein kinases (PKGs), phosphodiesterases, and cyclic nucleotide-gated ion channels (36, 37). Type I PKG is cytosolic and widely expressed, with high levels in smooth muscle cells and platelets, whereas type II PKG is membrane-anchored, with high levels in brain and intestinal mucosa (37). Studies in PKG I- and PKG II-deficient mice have demonstrated a multitude of functions for PKG I, but there are few established functions for PKG II (37). PKG II-deficient mice are dwarfs because of a severe defect in endochondral ossification at the growth plates, secondary to impaired chondrocyte proliferation and differentiation (38, 39). Both PKG I and II are expressed in osteoblasts, but their physiological significance in these cells is largely unknown (38, 40, 41).
We previously showed that NO and cGMP regulate c-fos transcription in different cell types (40, 42–47). In this study, we found that fluid shear -stress induces c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression in osteoblasts via NO/cGMP, and we define a new function for PKG II as a regulator of gene expression and activator of the extracellular signal-regulated kinases (Erk)-1/2 in mechanically stimulated osteoblasts.
EXPERIMENTAL PROCEDURES
Materials—Antibodies against eNOS, Erk1/2, and α-tubulin and phospho-specific antibodies for eNOS (Ser(P)1177) and Erk1 (Tyr(P)204, clone E-4) were from Santa Cruz Biotechnology; an antibody specific for dually phosphorylated Erk1 (Thr(P)202/Tyr(P)204) was from Promega. Antibodies specific for vasodilator-stimulated phosphoprotein (VASP) phosphorylated on Ser239 (clone 16C2) and against the C terminus of PKG I were from Calbiochem/EMD; a PKG II-specific antibody was from Abgent. The calcium ionophore A23187, the intracellular calcium chelator 1,2-bis(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid, the MAPK/Erk kinase (MEK) inhibitor U0126, and the p38 inhibitor SB203580 were from Calbiochem/EMD. The cGMP agonist 8-(4-chlorophenylthio)-GMP (8-pCPT-cGMP) and cGMP antagonist 8-(4-chlorophenylthio)-β-phenyl-1,N2-ethenoguanosine-3′,5′-cyclic monophosphorothioate, Rp isomer (Rp)-8-pCPT-PET-cGMPS) were from Biolog. The NOS inhibitor N-nitro-l-arginine methyl ester (l-NAME), the sGC inhibitor 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one (ODQ), the NO donor 3-(2-hydroxy-2-nitroso-1-propylhydrazino)-1-propanamine (PAPA-NONOate), and an antibody against the β1 subunit of sGC were from Cayman.
Cell Culture and Characterization of Human Primary Osteoblasts—Murine MC3T3-E1 osteoblastic cells with high differentiation potential (clone 4, hereafter referred to as MC3T3 cells) and UMR106 rat osteosarcoma cells were from the American Tissue Culture Collection. MC3T3 cells were grown in minimal essential medium without ascorbic acid and UMR106 cells in Dulbecco's modified Eagle's medium; both cell lines were maintained in the presence of 10% fetal bovine serum (FBS) and used at <12 passages. Human umbilical vein endothelial cells were isolated and cultured as described previously (48). Human primary osteoblasts (hPOBs) were established from trabecular bone obtained from surgical specimens of patients undergoing joint replacement for degenerative joint disease, according to an institutionally approved human subjects protocol as described previously (41). Primary cultures were maintained for a maximum of five passages in Dulbecco's modified Eagle's medium with 10% FBS and exhibited histochemical staining for alkaline phosphatase activity in >85% of cells; in addition, cells demonstrated a >16-fold induction of osteocalcin mRNA in response to 1,25-dihydroxyvitamin D3 (10 nm) (41, 49).
Exposure of Cells to Fluid Shear Stress—Cells were plated on 38 × 75-mm NaOH-etched glass slides at 0.5–1 × 106 cells/slide in minimal essential medium or Dulbecco's modified Eagle's medium containing 10% FBS. After 18 h, they were serum-deprived for 24 h, and the slides were transferred to a parallel plate flow chamber (Cytodyne Inc., San Diego). Minimal essential medium with 1 mg/ml fatty acid-free bovine serum albumin (Sigma) was injected into the chamber using a syringe pump at a flow rate that generated 12 dynes/cm2 of shear for up to 20 min (50). The flow chamber, media, and accompanying apparatus were maintained at 37 °C throughout the experiment. Sham-treated cells served as “static controls”; they were grown under identical conditions and mounted into the flow chamber but were not subjected to shear stress. After shear stress, cells were left in the chamber without flow for the indicated time before harvesting.
Reverse Transcription (RT)-PCR—RNA was extracted using TriReagent™ (Molecular Research Center, Inc.); 1 μg of total RNA was subjected to reverse transcription using Superscript™ reverse transcriptase (Invitrogen) and PCR was performed on 5 or 10% of the RT product as described (51). The PCR primers for amplification of murine and human glyceraldehyde-3-phosphate dehydrogenase (gapd), c-fos, fra-1, fra-2, fosB/ΔfosB, pkg I, and pkg II mRNA are described in supplemental Table 1. Semi-quantitative PCR conditions for c-fos and gapd were 30 s of denaturation at 95 °C, 30 s annealing at 65 °C, and 30 s extension at 72 °C for 23 cycles; for fra-1/2 and fosB annealing and extension were 45 s at 60 °C for 25 cycles. Control experiments with variable amounts of input cDNA demonstrated a linear increase in a single PCR product over a >20-fold range. Quantitative RT-PCR was performed using an MX3000 real time PCR detection system (Stratagene) and IQ™ SYBR Green Supermix (Bio-Rad). Melting curves after 40 cycles confirmed a single PCR product for each primer pair. Standard curves were generated by plotting Ct values versus the amount of input RNA, and demonstrated similar amplification efficiencies for all primers. Relative changes in mRNA expression were analyzed using the 2–ΔΔCt method, with gapd serving as an internal reference to correct for differences in RNA extraction or reverse transcription efficiencies (52).
Quantitation of NOx and cGMP—Nitric oxide production was monitored based on nitrite and nitrate accumulation in the medium using a two-step colorimetric assay kit according to the manufacturer's protocol (Active Motif), but the assay was scaled down 10-fold, and samples were read using a NanoDrop™ spectrophotometer (NanoDrop Technologies, Inc.). Experiments were performed in phenol red-free medium, with 5 × 105 cells/slide in the parallel plate flow chamber described above. cGMP production was measured in cell lysates using a competitive enzyme immunoassay from Cayman according to the manufacturer's protocol.
DNA and siRNA Transfections—MC3T3 cells (1.5 × 106) were transfected with 2 μg of expression vector encoding human VASP in 100 μl of solution V using the Amaxa Nucleoporator program D-24. After overnight recovery in full growth medium, cell from several transfections were pooled, and equal numbers of cells were plated on glass slides for shear experiments.
The sequence targeted by siRNA in the C terminus of PKG Iα/β was 5′-CCGGACAUUUAAAGACAGCAA-3′ (siRNA PKG-1). The sequences targeted in PKG II were 5′-CTGCTTGGAAGTGGAATACTA-3′ (siRNA PKG-2a) and 5′-CCGGGTTTCTTGGGTAGTCAA-3′ (siRNA PKG-2b). siRNA oligoribonucleotides, including a control siRNA targeting green fluorescent protein (GFP), were produced by Qiagen. MC3T3 cells were plated at 1.3 × 105 cells per well of a 6-well dish or at 5 × 105 cells per glass slide and were transfected 18 h later (at ∼40% confluency) with 100 pmol of siRNA and 3 μl of Lipofectamine 2000™ (Invitrogen) in 1 ml of 10% FBS-containing media. The medium was replaced 5 h later, and 18 h later, cells were transferred to medium containing 0.1% FBS.
Adenovirus Infection—Adenoviral vectors encoding either β-galactosidase (LacZ) or rat PKG II (53) were produced using the pAd/CMV/V5-DEST™ Gateway® system (Invitrogen). Cells were infected in full growth medium at an multiplicity of infection of ∼30.
Western Blot Analyses, Cell Fractionation, and PKG Activity Assay—Western blots were generated using horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence as described (45). To determine PKG II activity, MC3T3 cells were extracted by Dounce homogenization, and nuclei and cell debris were removed by centrifugation at 300 × g for 10 min, and the supernatant was subjected to centrifugation at 50,000 × g for 30 min to generate “cytosolic” (supernatant) and “membrane” (pellet) fractions. PKG II activity in the membrane fraction was determined in the presence of 1% Triton-X-100 and 500 mm NaCl, as described previously (40). For Western blotting, membranes were solubilized in 1% Triton X-100, 60 mm β-octyl glucoside. Films were scanned using ImageJ software (nih.gov).
Statistical Analyses—Pairwise comparison of data groups was done by two-tailed Student's t test and comparison of multiple groups by analysis of variance, with a Dunnett's post test analysis to the control group; a p value of < 0.05 was considered statistically significant. Results shown in bar graphs represent the mean ± S.D. of at least three independent experiments, unless stated otherwise. All other results are representative experiments that were reproduced at least three times.
RESULTS
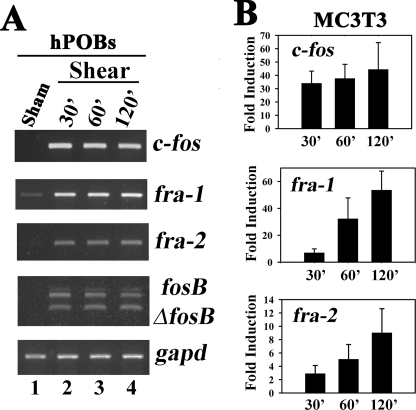
Fluid Shear Stress Induces c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA Expression in Osteoblasts—Osteoblast c-fos and fosB/ ΔfosB mRNA and protein are induced rapidly by mechanical loading in rat vertebrae and mouse limb bones in vivo and by fluid shear stress in osteoblast cell lines or primary rodent osteoblasts (13–18, 54). Genetic experiments have implicated fra-1 and fra-2 in regulating osteoblast differentiation and bone mass (6). We found that c-fos, fra-1, fra-2, and fosB/ΔfosB mRNAs were strongly induced 30 min after the onset of flow in both MC3T3 cells and hPOBs and remained elevated for at least 2 h (Fig. 1A shows hPOBs, results for MC3T3 cells are shown in supplemental Fig. 1A). The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (gapd) was not affected by fluid shear in either cell type. Real time RT-PCR analysis in MC3T3 cells showed that c-fos mRNA increased 30–40-fold at all time points, and that fra-1 and fra-2 mRNA levels increased progressively up to 50- and 10-fold, respectively, at 2 h compared with sham-treated control cells (Fig. 1B). Basal mRNA levels of all four fos genes were somewhat variable in sham-treated cells, explaining the relatively large standard deviations in Fig. 1B; this may be due to the sensitivity of the cells to even small changes in fluid flow that can occur during mounting of the slides. Similar results were obtained in UMR106 rat osteosarcoma cells (data not shown).
FIGURE 1.
Effect of fluid shear stress on osteoblast c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression. A, serum-deprived hPOBs were incubated in a parallel plate flow chamber for the indicated times; cells were exposed to laminar flow for the first 20 min (lanes 2–4) or kept under static conditions (lane 1, Sham). At the indicated times, which include the initial 20 min of laminar flow, total cytoplasmic mRNA was isolated, and c-fos, fra-1, fra-2, fosB/ ΔfosB, or gapd mRNA levels were determined by semi-quantitative RT-PCR as described under “Experimental Procedures.” PCR products were separated by nondenaturing agarose gel electrophoresis and visualized by ethidium bromide staining. Sham-treated cells were harvested at 30 min, but similar results were obtained with sham-treated cells harvested at 120 min. B, MC3T3 cells were treated as described for hPOBs in A, but c-fos, fra-1, and fra-2 mRNA levels were quantified by real time RT-PCR and normalized relative to gapd mRNA levels as described under “Experimental Procedures”; the relative mRNA levels found in static controls were assigned a value of 1. p < 0.05 for the comparison between shear-stressed and sham-treated cells for all time points.
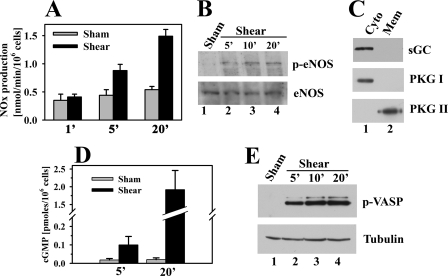
Fluid Shear Stress Induces NO and cGMP Production, eNOS, and VASP Phosphorylation—Mechanical stimulation of osteoblasts or osteocytes induces NO synthesis and increased shear rates correlate with increased NO production (55–58). To determine the amount of NO produced in our system, we measured the concentration of stable NO oxidation products (nitrites and nitrates, referred to as NOx) in media collected 3 min after the end of flow, allowing calculation of the rate of NOx production during this 3-min period (Fig. 2A). In MC3T3 cells exposed to 5 min of fluid shear, NOx production nearly doubled compared with sham-treated cells, and it reached 1.5 ± 0.1 nmol/min/106 cells after 20 min. In contrast, 20 min of fluid shear stress increased NOx production to 3.4 ± 0.4 nmol/min/106 cells in human umbilical vein endothelial cells (supplemental Fig. 1B). The rate of NOx production in MC3T3 cells is comparable with that of primary mouse osteoblasts exposed to pulsating fluid flow (58). The NOx concentration in the medium of shear-stressed MC3T3 cells continued to rise up to 40 min after the cessation of flow, suggesting a sustained activation of NOS (supplemental Fig. 1C).
FIGURE 2.
Effect of fluid shear stress on osteoblast NO and cGMP production, eNOS and VASP phosphorylation. A, MC3T3 cells were placed in the flow chamber as described in Fig. 1; they were kept either under static conditions (gray bars) or were exposed to laminar flow (black bars) for the indicted times. At the end of flow (or static incubation), cells were kept in the chamber for 3 additional min, and nitrate plus nitrite (NOx) concentrations were measured in the media collected from the chamber. Thus, NOx production was measured over a 3-min interval after the cessation of flow. p < 0.05 was the comparison between shear-stressed and sham-treated cells for the 5- and 20-min time points. B, cells were kept under static conditions for 20 min or were exposed to fluid shear stress for the indicated times, and cell lysates were analyzed by SDS-PAGE/Western blotting using antibodies specific for eNOS phosphorylated on Ser1177 (upper panel) or recognizing eNOS irrespective of its phosphorylation state (lower panel). C, cells were extracted by Dounce homogenization and fractionated by differential centrifugation; cytosolic (Cyto, lane 1) and membrane (Mem, lane 2) fractions were analyzed by SDS-PAGE/Western blotting using antibodies specific for soluble guanylate cyclase β1 subunit (sGC, upper panel), PKG I (middle panel), or PKG II (lower panel). D, cells were placed into the flow chamber and incubated for 15 min in the presence of 0.5 mm IBMX; cells were then either kept under static conditions (gray bars) or were exposed to fluid shear stress (black bars), both in the presence of IBMX. Cells were harvested at the indicated times, and the intracellular cGMP concentration was determined as described under “Experimental Procedures.” E, MC3T3 cells expressing human VASP were treated as described in B, but Western blots were probed with antibodies specific for VASP phosphorylated on Ser239 (upper panel) or α-tubulin (lower panel).
To determine whether fluid shear stress increased eNOS activity in MC3T3 cells, we used an antibody specific for eNOS phosphorylated on Ser1177; phosphorylation of this site by multiple kinases increases enzyme activity (59). Little Ser1177 phosphorylation was detectable in sham-treated cells, but as early as 5 min after the onset of flow, phosphorylation increased and remained elevated at 20 min (Fig. 2B, upper panel; the lower panel shows total eNOS expression that was not changed by fluid shear).
To document presence of the full NO/cGMP/PKG signal transduction pathway in osteoblasts, we examined MC3T3 cells for sGC and PKG. We found that sGC protein was easily detectable in cytosolic extracts of MC3T3 cells, and that PKG I and II were present in cytosolic and membrane fractions of MC3T3 cells, respectively (Fig. 2C). Fig. 2D shows that the increased NO in shear-stressed MC3T3 cells dramatically increased the intracellular cGMP concentration; cGMP was measured in cells treated with the phosphodiesterase inhibitor isobutylmethylxanthine (IBMX) to block cGMP catalysis. To determine whether the shear-induced increase in cGMP activated PKG in intact osteoblasts in the absence of phosphodiesterase inhibitors, we used a phospho-specific antibody to examine VASP phosphorylation on Ser259, a preferred PKG phosphorylation site (61). VASP Ser259 phosphorylation was undetectable in sham-treated cells but increased within 5 min of initiating fluid shear stress and reached a maximum level at 10–20 min (Fig. 2E shows MC3T3 cells transfected with a vector expressing human VASP, but similar results were obtained in hPOBs, which express endogenous VASP, as shown in supplemental Fig. 1D).
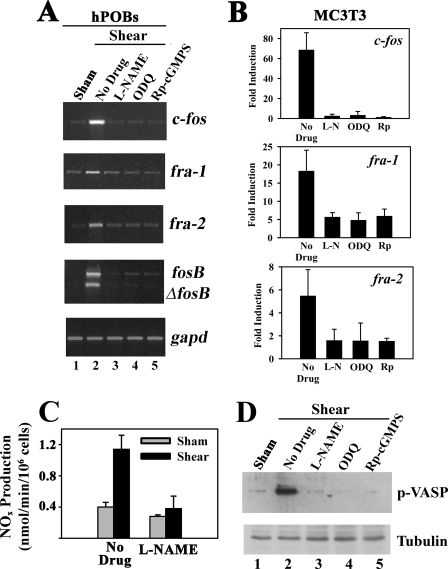
Inhibition of NO/cGMP Signaling Blocks Shear-induced c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA Expression—The experiments in Figs. 1 and 2 show that fluid shear stress leads to rapid NOx and cGMP accumulation, PKG activation, and induction of fos family members but do not establish a causal relationship between NO/cGMP/PKG signaling and increased gene expression. To address this question, we treated MC3T3 cells and hPOBs for 1 h with l-NAME (to inhibit NOS), ODQ (to inhibit sGC), or (Rp)-8-pCPT-PET-cGMPS (to inhibit PKG) prior to fluid shear stress. We found that all three agents severely reduced shear induction of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA, without affecting gapd mRNA (Fig. 3A shows semi-quantitative RT-PCR results for hPOBs, and Fig. 3B summarizes real time PCR results for c-fos, fra-1 and fra-2 in MC3T3 cells). The three inhibitors almost completely prevented shear induction of c-fos and fra-2 mRNAs; however, there was some residual shear induction of fra-1 mRNA, suggesting that some NO/cGMP/PKG-independent mechanism may contribute to the mechanical stimulation of fra-1. Control experiments demonstrated that the drugs inhibited NO/cGMP/PKG signaling effectively; l-NAME reduced the rate of NOx production in fluid shear-stressed cells to the level found in sham-treated control cells (Fig. 3C); all three agents blocked shear-induced VASP Ser259 phosphorylation (Fig. 3D); and ODQ and (Rp)-8-pCPT-PET-cGMPS blocked VASP phosphorylation in cells treated with the NO donor PAPA-NONOate (data not shown). These results suggest that fluid shear stress induction of fos family genes requires PKG activation by NO/cGMP.
FIGURE 3.
Inhibition of NO/cGMP signaling prevents shear-induced c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression. A, serum-deprived hPOBs were placed in a flow chamber and were incubated for 1 h with either culture medium alone (lanes 1 and 2) or with medium containing 4 mm l-NAME (lane 3), 10 μm ODQ (lane 4), or 100 μm (Rp)-8-pCPT-PET-cGMPS ((Rp)-cGMPS, lane 5). Cells were then either kept under static conditions (lane 1) or were exposed to laminar flow for 20 min (lanes 2–5). Ten minutes after the cessation of flow, total RNA was extracted, and c-fos, fra-1, fra-2, fosB/ΔfosB, or gapd mRNA levels were determined by semi-quantitative RT-PCR as described in Fig. 1A. B, MC3T3 cells were treated as described for hPOBs in A, but c-fos, fra-1, and fra-2 mRNA levels were quantified after 60 min by real time RT-PCR as described in Fig. 1B. p < 0.05 for the comparison between control cells receiving no drug and cells treated with l-NAME (L-N), ODQ, or (Rp)-cGMPS (Rp). C, MC3T3 cells were kept under static conditions (gray bars) or were exposed to laminar flow for 20 min (black bars); 5 min after the cessation of flow, media were collected from the chamber, and nitrate plus nitrite (NOx) concentrations were measured. D, MC3T3 cells expressing human VASP were incubated for 1 h with medium alone (lanes 1 and 2) or with medium containing 4 mm l-NAME (lane 3), 10 μm ODQ (lane 4), or 100 μm (Rp)-8-pCPT-PET-cGMPS (Rp-cGMPS, lane 5). Cells were kept under static conditions (lane 1) or were exposed to 20 min of fluid shear stress (lanes 2–5), and 10 min after the cessation of flow, cell lysates were analyzed by SDS-PAGE/Western blotting using antibodies specific for VASP phosphorylated on Ser239 (upper panel) or α-tubulin (lower panel).
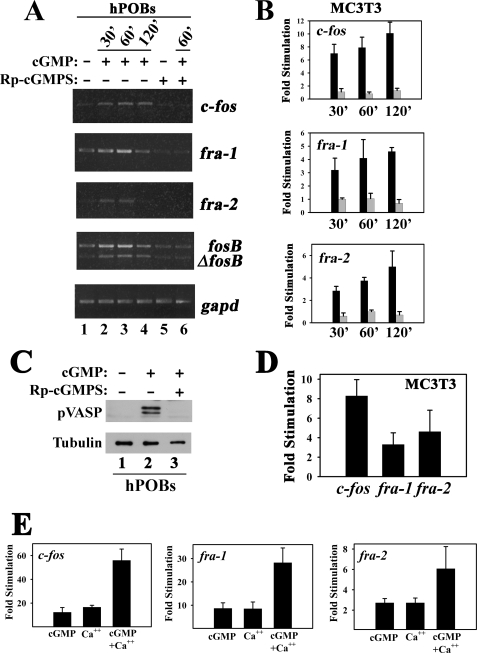
cGMP Partly Mimics the Effects of Fluid Shear Stress on c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA Expression, and cGMP Cooperates with Calcium—To determine whether direct PKG activation can reproduce the effects of fluid shear stress on fos family gene expression, we treated cells with a membrane-permeable cGMP agonist. Exposure of serum-starved MC3T3 cells or hPOBs to 8-pCPT-cGMP for 30 min, 1 h, or 2 h increased c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression, but when cells were pretreated with the PKG inhibitor (Rp)-8-pCPT-PET-cGMPS, the effects of 8-pCPT-cGMP on all four fos family genes were abolished (Fig. 4A shows hPOBs, and Fig. 4B summarizes real time RT-PCR results for c-fos, fra-1, and fra-2 in MC3T3 cells, with black and gray bars representing cells treated with 8-pCPT-cGMP in the absence and presence of the PKG inhibitor, respectively). (Rp)-8-pCPT-PET-cGMPS prevented VASP Ser259 phosphorylation in cells treated with 8-pCPT-cGMP, demonstrating effective inhibition of PKG activity by the cGMP antagonist (Fig. 4C).
FIGURE 4.
cGMP partly mimics the effects of fluid shear stress on osteoblast c-fos, fra-1, fra-2, and fosB/ ΔfosB mRNA expression. A, serum-deprived hPOBs were incubated for 1 h in the absence (lanes 1–4) or presence of 100 μm (Rp)-8-pCPT-PET-cGMPS (Rp-cGMPS, lanes 5 and 6) before adding 50 μm 8-pCPT-cGMP (cGMP) to all cells for the indicated times. c-fos, fra-1, fra-2, fosB/ΔfosB or gapd mRNA levels were determined by semi-quantitative RT-PCR as described in Fig. 1A. B, MC3T3 cells were incubated in the absence (black bars) or presence (gray bars) of (Rp)-8-pCPT-PET-cGMPS and then stimulated with 8-pCPT-cGMP as described in A for hPOBs, but c-fos, fra-1, and fra-2 mRNA levels were quantified by real time RT-PCR. Relative mRNA levels (normalized to gapd) measured in mock-treated cells were assigned a value of 1. p < 0.05 for the comparison between cells treated with (Rp)-cGMPS plus cGMP versus cGMP alone for all time points. C, hPOBs were preincubated in the absence (lanes 1 and 2) or presence (lane 3) of 100 μm (Rp)-8-pCPT-PET-cGMPS for 1 h and treated with 50 μm 8-pCPT-cGMP (lanes 2 and 3) for 30 min. VASP phosphorylation on Ser259 was assessed using a phosphorylation site-specific antibody; a duplicate Western blot was probed with an anti-α-tubulin antibody. D, MC3T3 cells were treated for 1 h with 10 μm sodium nitroprusside, and c-fos, fra-1, and fra-2 mRNA levels were quantified by real time RT-PCR. E, MC3T3 cells were treated for 1 h with either cGMP (100 μm 8-pCPT-cGMP), calcium ionophore (0.3 μm A23187), or both agents, and fos family gene expression was measured as in B.
When MC3T3 cells were treated with the NO donor sodium nitroprusside to activate PKG through sGC stimulation, c-fos, fra-1 and fra-2 mRNAs were induced to a similar extent as seen with 8-pCPT-cGMP (compare Fig. 4D to Fig. 4B). However, we noticed that sodium nitroprusside and 8-pCPT-cGMP induced the fos genes to a lesser extent than observed in fluid shear-stressed MC3T3 cells (compare the y axis in Fig. 4, B and D, with that in Fig. 1B), suggesting that the NO/cGMP/PKG signaling pathway cooperates with other pathways to induce fos family genes in mechanically stimulated osteoblasts.
Fluid shear stress rapidly increases the intracellular calcium concentration in MC3T3 cells and primary osteoblasts/osteocytes through calcium release from intracellular stores and calcium influx via mechano-sensitive and/or L-type voltage-gated calcium channels; calcium chelation prevents fluid shear stress-induced c-fos and fosB/ΔfosB expression (17, 19, 54, 62–65). We confirmed these latter results, and we found that treating MC3T3 cells with 1,2-bis(2-aminophenoxy) ethane-N, N,N′,N′-tetraacetic acid or EGTA to chelate intra- or extracellular calcium, respectively, also reduced fluid shear stress induction of fra-1 and -2 mRNA (supplemental Fig. 2A). We previously showed that calcium and cGMP cooperate to increase c-fos mRNA expression (40, 47). Therefore, we examined the effect of the calcium ionophore A23187 alone and in combination with 8-pCPT-cGMP on the expression of all four fos family genes in MC3T3 cells and hPOBs. Increasing the intracellular calcium concentration with A23187 stimulated expression of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA to a similar extent as treatment with 8-pCPT-cGMP; together the effect was at least additive (Fig. 4E and supplemental Fig. 2B show results for MC3T3 cells, but similar results were obtained in hPOBs). Combined treatment with calcium ionophore plus cGMP induced c-fos and fra-1/2 mRNAs to levels comparable with those found in shear-stressed osteoblasts (compare Fig. 4E and Fig. 1B). These results suggest that induction of fos family genes in shear-stressed osteoblasts is due to a combined effect of increased intracellular calcium and cGMP, although calcium and cGMP signaling may be partly interdependent, as discussed below.
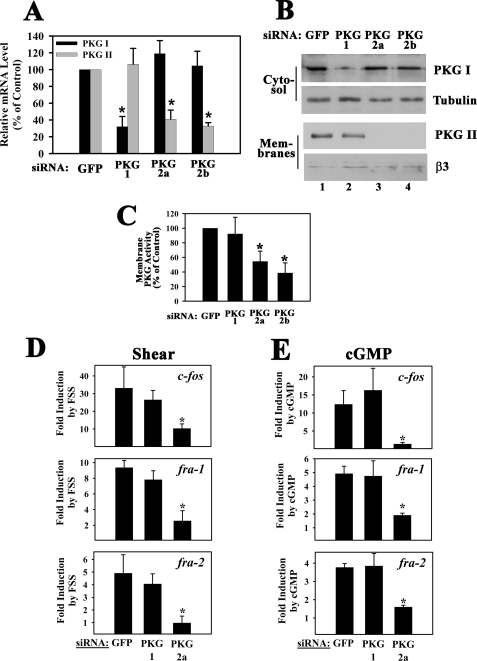
Shear- and cGMP-induced c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA Expression Is Mediated by PKG II and Not PKG I—Having established that fluid shear stress induces fos family gene expression in osteoblasts via NO/cGMP/PKG signaling, we examined whether gene induction occurred via PKG I, PKG II, or both. Because there are no specific inhibitors for PKG I versus II, we used an siRNA approach. In MC3T3 cells transfected with an siRNA targeting a C-terminal PKG I sequence (siRNA PKG-1, targeting both PKG Iα and PKG Iβ splice variants), PKG I mRNA levels were reduced by 68%, compared with the levels found in control siRNA-transfected cells, whereas PKG II mRNA levels were unaffected (Fig. 5A; PKG I and II mRNA levels are shown in black and gray bars, respectively). Using an antibody specific for the common C terminus of PKG Iα and Iβ, we found that PKG I protein in cytosolic extracts was reduced in proportion to the mRNA knockdown (Fig. 5B, top panel; the 2nd panel shows a duplicate Western blot probed with a tubulin antibody to demonstrate equal loading). In MC3T3 cells transfected with siRNA sequences targeting PKG II (siRNAs PKG-2a and -2b), PKG II mRNA levels were reduced by 60 and 68%, respectively, compared with control siRNA-transfected cells, but PKG I mRNA levels were not altered (Fig. 5A). PKG II protein levels in the membranes of cells transfected with control or PKG I-specific siRNAs were similar, but levels were below detection in cells transfected with PKG II-specific siRNAs (Fig. 5B, 3rd panel; loading was shown by re-probing the blot with an antibody specific for β3 integrin, as shown in the bottom panel). We measured similar PKG activities in membrane preparations of control siRNA- and PKG I siRNA-transfected MC3T3 cells and significantly reduced activity in PKG II siRNA-transfected cells (Fig. 5C). The residual membrane-bound PKG activity in PKG II siRNA-transfected cells is likely explained by the higher sensitivity of the activity assay compared with Western blotting. Because siRNA knockdown of PKG I did not affect membrane-associated PKG activity, and Western blots failed to detect PKG I in membrane preparations, we conclude that membrane-associated PKG activity in MC3T3 cells represents predominantly PKG II, which is consistent with results in other cell types (40, 53, 66, 67).
FIGURE 5.
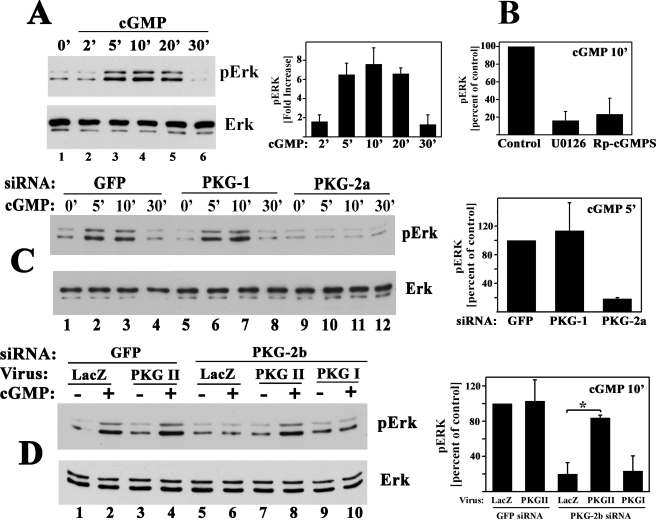
Effect of siRNA-mediated PKG I or PKG II knockdown on shear- and cGMP-induced c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression. MC3T3 cells were transfected with siRNAs specific for GFP (control), or received siRNAs targeting sequences in PKG I (siRNA PKG-1) or PKG II (siRNAs PKG-2a and -2b) as described under “Experimental Procedures.” A, at 48 h after transfection, mRNA levels of PKG I (black bars) and PKG II (gray bars) were quantified by real time PCR; levels were normalized to gapd mRNA, which was not affected by any of the siRNAs. Relative PKG mRNA levels measured in GFP (control) siRNA-transfected cells were assigned a value of 100%. *, p < 0.05 for the comparison between control siRNA and PKG siRNA-treated cells. B, in parallel experiments, cells were extracted by Dounce homogenization and fractionated by differential centrifugation; cytosolic fractions (upper two panels) and membrane fractions (lower two panels) were analyzed by SDS-PAGE/Western blotting using an antibody specific for the C terminus of PKG I common to PKG Iα and Iβ (upper panel), an anti-α-tubulin antibody (2nd panel), an antibody specific for PKG II (3rd panel), and an anti-β3 integrin antibody (lowest panel). C, siRNA-transfected cells were fractionated as described in B, and PKG activity was determined in the membrane fractions as described under “Experimental Procedures.” *, p < 0.05 for the comparison between control siRNA and PKG siRNA-treated cells. D, cells transfected with the siRNAs targeting GFP, PKG I, or PKG II (PKG-2a siRNA) were kept either under static conditions or were subjected to fluid shear stress for 20 min. Cells were harvested 60 min after the onset of flow, and c-fos, fra-1, and fra-2 mRNA levels were determined by quantitative RT-PCR; relative mRNA levels (normalized to gapd) measured in GFP siRNA-transfected static control cells were assigned a value of 1. E, cells transfected with the indicated siRNAs were either mock-treated or were treated with 50μm 8-pCPT-cGMP for 1 h, and c-fos, fra-1, or fra-2 mRNA levels were determined by quantitative RT-PCR. Relative mRNA levels (normalized to gapd) measured in GFP siRNA-transfected mock-treated cells were assigned a value of 1. *, p < 0.05 for the comparison between GFP and PKG II siRNA-treated cells (D and E).
When we examined the effects of siRNA-mediated down-regulation of PKG I and PKG II on shear-induced fos family gene expression in MC3T3 cells, we were surprised to find that knockdown of PKG I had no significant effect on gene expression, whereas knockdown of PKG II significantly diminished fluid shear stress-induced c-fos, fra-1, and fra-2 mRNA expression (Fig. 5D; results for PKG-2a siRNA are shown, but similar results were obtained with PKG-2b siRNA). We also examined 8-pCPT-cGMP-induced fos family gene expression in siRNA-transfected MC3T3 cells. Neither basal nor cGMP-induced c-fos, fra-1 or fra-2 mRNA levels were altered in PKG I siRNA-transfected cells, whereas cGMP-induced mRNA levels were severely reduced in cells transfected with the PKG II-specific siRNA (Fig. 5E). Similar results were obtained for fosB/ΔfosB (supplemental Fig. 3). Thus, fluid shear stress- and cGMP-induced c-fos, fra-1, fra-2 and fosB/ΔfosB mRNA expression in osteoblasts requires PKG II but not PKG I.
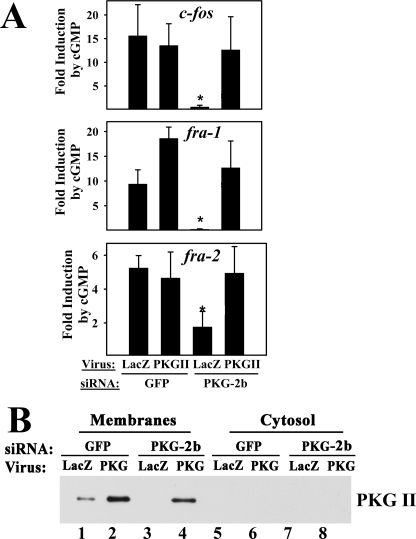
To confirm these results, we used a second PKG II-specific siRNA (PKG-2b) and an adenoviral vector encoding rat PKG II, which is resistant to the effects of this siRNA. In MC3T3 cells transfected with the mouse PKG II-specific siRNA, infection with rat PKG II virus, but not with a control LacZ virus, restored cGMP-induced fos family gene expression to the levels found in control GFP siRNA-transfected cells (Fig. 6A). Viral transduction of PKG II in control siRNA-transfected cells did not further enhance cGMP-induced c-fos or fra-2 mRNA expression, suggesting that endogenous PKG II levels, although low, were not rate-limiting. However, cGMP stimulation of fra-1 mRNA was enhanced in cells with elevated PKG II levels (Fig. 6A, p < 0.05 for the comparison of GFP siRNA-treated cells infected with LacZ versus PKG II virus). Viral PKG II expression was demonstrated by Western blotting of membranes prepared from PKG II virus-infected MC3T3 cells (Fig. 6B; again, endogenous PKG II levels in PKG II siRNA-transfected cells were below detection).
FIGURE 6.
Rescue of PKG II siRNA-transfected cells with a virus encoding siRNA-resistant PKG II. A, MC3T3 cells were transfected with either GFP siRNA or the mouse PKG II-specific siRNA (PKG-2b), and 24 h later, cells were infected with either control virus encoding β-galactosidase (LacZ) or virus encoding siRNA-resistant rat PKG II as indicated. Forty eight hours later, cells were either mock-treated or were treated with 8-pCPT-cGMP for 1 h, and fos family gene expression was determined as described in Fig. 5E. *, p < 0.05 for the comparison between GFP versus PKG II siRNA-treated cells infected with LacZ virus and the comparison between PKG II siRNA-treated cells infected with LacZ versus PKG II virus. B, cells were transfected with GFP siRNA (lanes 1, 2, 5, and 6) or PKG-2b siRNA (lanes 3, 4, 7, and 8) and were infected with LacZ virus (odd lanes) or PKG II virus (even lanes) as described in A. Levels of endogenous murine PKG II and virally expressed rat PKG II were examined by Western blotting of membrane and cytosolic proteins.
Induction of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA Expression by Fluid Shear Stress Is MEK/Erk-dependent—Mechanical stimulation enhances osteoblast proliferation via activation of the MEK/Erk pathway (20, 54, 62, 68). Whether MEK inhibition prevents c-fos mRNA expression appears to depend on the type of mechanical stimulus and/or the source of osteoblasts, with variable results also reported for the effect of p38 MAPK inhibitors on mechanically induced c-fos expression (20–22, 24, 54). We therefore investigated the effects of MEK and p38 inhibitors in fluid shear-stressed osteoblasts.
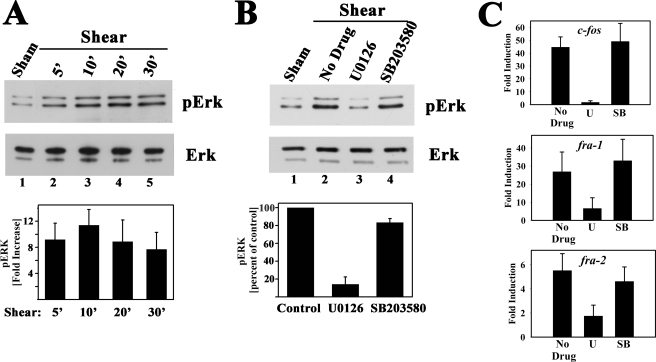
We found that fluid shear stress rapidly increased Erk1/2 phosphorylation on a site that correlates with Erk1/2 activation (Fig. 7A shows results in MC3T3 cells, but similar results were obtained in hPOBs and UMR106 cells). Maximal induction of Erk phosphorylation after 10 min of shear stress was 11.4-fold compared with sham-treated cells. Pretreating cells with the MEK inhibitor U0126 blocked Erk phosphorylation and almost completely prevented shear induction of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNAs (Fig. 7, B and C; U0126 inhibition of fosB/ ΔfosB mRNA induction was measured by semi-quantitative RT-PCR; data not shown). The p38 inhibitor SB203580, however, did not affect Erk or fos family genes (Fig. 7, B and C; SB203580 inhibited UV stress-induced p38 activation in control experiments; data not shown). A similar effect of U0126 on fluid shear stress-induced fosB mRNA expression was reported in primary rat osteoblasts (54).
FIGURE 7.
Induction of c-fos, fra-1, and fra-2 mRNA expression by fluid shear stress is MEK/Erk-dependent. A, serum-deprived MC3T3 cells were kept under static conditions for 30 min (sham-treated, lane 1) or were subjected to laminar flow for 5 min (lane 2), 10 min (lane 3), or 20 min (lanes 4 and 5); cells in lane 5 were kept in the flow chamber for an additional 10 min after the 20- min exposure to flow. Cell lysates were analyzed by Western blotting using a phospho-Erk1/2-specific antibody (clone E-4, upper panel); duplicate blots were probed with an antibody recognizing Erk1/2 irrespective of their phosphorylation state (lower panel). The bar graph below summarizes results of three independent experiments; phospho-Erk1 bands were scanned, and the intensity of the band found in sham-treated cells was assigned a value of 1. B, serum-deprived MC3T3 cells were preincubated for 1 h in the flow chamber under static conditions with either culture medium alone or with medium containing 10 μm U0126 or 10 μm SB203580, and cells were exposed to laminar flow for 10 min; Erk phosphorylation was assessed as described in A, with phospho-Erk levels in shear-stressed cells incubated with media alone assigned a value of 100%. C, cells were treated as described in B; they were exposed to laminar flow for 20 min and were harvested 10 min later for determination of c-fos, fra-1, fra-2, and gapd mRNA levels by quantitative RT-PCR, as described in Fig. 1B. *, p < 0.05 for the comparison between cells treated with U0126 (U) versus control cells receiving no drug (in B and C). S, SB203580.
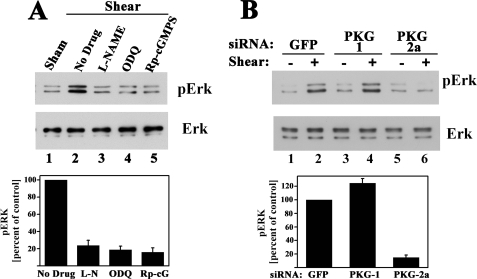
Fluid Shear Stress-induced Erk Activation Requires NO/cGMP/PKG II Signaling—Because fluid shear stress induction of the four fos family genes required signaling through NO/cGMP/PKG II and the MEK/Erk pathway, we asked whether Erk activation occurred through NO/cGMP/PKG II. Pretreating osteoblasts with l-NAME, ODQ, or (Rp)-8-pCPT-PET-cGMPS prevented shear-induced Erk1/2 phosphorylation in MC3T3 cells (Fig. 8A); similar results were obtained in hPOBs (supplemental Fig. 4A; l-NAME not shown in hPOBs). Fluid shear stress-induced Erk phosphorylation was abolished in MC3T3 cells transfected with a PKG II-specific siRNA, but transfection of a PKG I-specific siRNA was without effect (Fig. 8B; results similar to those shown for PKG-2a siRNA were obtained with PKG-2b siRNA). Thus, Erk activation by shear stress, like shear-induced fos family gene expression, required NO/cGMP activation of PKG II.
FIGURE 8.
Fluid shear stress-induced Erk activation requires NO/cGMP/PKG II signaling. MC3T3 cells were serum-deprived for 24 h prior to the indicated treatments, and Erk phosphorylation was assessed as described in Fig. 7. A, cells were treated for 1 h with 4 mm l-NAME, 10 μm ODQ, or 100 μm (Rp)-8-pCPT-PET-cGMPS (Rp-cGMPS) as indicated, prior to a 10-min exposure to laminar flow (lanes 2–5); cells in lane 1 were kept under static conditions (sham-treated). In the bar graph below, three independent experiments are summarized. p < 0.05 for the comparison between control cells receiving no drug and cells treated with l-NAME, ODQ, or (Rp)-cGMPS. B, cells were transfected with control siRNAs specific for GFP or received siRNAs targeting PKG I (siRNA PKG-1) or PKG II (siRNA PKG-2a) as described in Fig. 5. Cells were either kept under static conditions (–) or were exposed to laminar flow (+) for 10 min. The bar graph summarizes two independent experiments. p < 0.05 for the comparison between cells transfected with PKG-2a siRNA versus GFP siRNA.
NO/cGMP Activation of PKG II Is Sufficient to Activate Erk1/2—We next examined whether direct PKG activation by 8-pCPT-cGMP affected Erk phosphorylation in osteoblasts. In serum-starved MC3T3 cells, 50 μm 8-pCPT-cGMP increased Erk1/2 phosphorylation 7.6-fold at 10 min, with phospho-Erk levels returning to base line at 30 min (Fig. 9A; comparable results were obtained in hPOBs, shown in supplemental Fig. 4B). In cells treated with A23187 to increase intracellular calcium, Erk phosphorylation increased to a similar extent as observed with 8-pCPT-cGMP, and the effect of both agents appeared additive (supplemental Fig. 2C). These results are consistent with the combined effects of calcium and cGMP on c-fos, fra-1, fra-2, and fosB/ ΔfosB mRNA expression (Fig. 4E and supplemental Fig. 2, A and B).
FIGURE 9.
NO/cGMP activation of PKG II is sufficient to activate Erk1/2. Erk phosphorylation was assessed in serum-deprived MC3T3 cells as described in Fig. 7A. A, cells were treated with 50 μm 8-pCPT-cGMP for the indicated times. The bar graph summarizes three independent experiments. B, cells were preincubated for 1 h in medium alone or in medium containing 10 μm U0126 or 10 μm SB203580 as indicated, prior to receiving 50 μm 8-pCPT-cGMP (cGMP) for 10 min; phospho-Erk levels in cells treated with cGMP alone were assigned a value of 100%. C, cells were transfected with a control siRNA specific for GFP (lanes 1–4), or an siRNA targeting PKG I (siRNA PKG-1, lanes 5–8), or PKG II (siRNA PKG-2a, lanes 9–12) as described in Fig. 5 and were treated with 50 μm 8-pCPT-cGMP for the indicated times. The bar graph on the right summarizes phospho-Erk1 levels measured in cells treated with cGMP for 5 min; p < 0.05 was used for the comparison between cells transfected with PKG-2a siRNA versus GFP siRNA. D, cells were transfected with either GFP siRNA (lanes 1–4) or the mouse PKG II-specific siRNA PKG-2b (lanes 5–10); 8 h later, cells were infected with control virus (LacZ, lanes 1, 2, 5, and 6), virus encoding siRNA-resistant rat PKG II (lanes 3, 4, 7, and 8), or virus encoding PKG I (lanes 9 and 10). Forty eight hours later, cells were treated with 8-pCPT-cGMP for 10 min. The bar graph on the right summarizes cGMP-induced phospho-Erk1 levels; *, p < 0.05 for the comparison between LacZ and PKG II virus-infected cells transfected with PKG-2a siRNA.
Inhibition of MEK by U0126 or inhibition of PKG by (Rp)-8-pCPT-PET-cGMPS blocked cGMP-induced Erk phosphorylation in MC3T3 cells (Fig. 9B). ODQ inhibited Erk phosphorylation induced by the NO donor PAPA-NONOate, and (Rp)-8-pCPT-PET-cGMPS inhibited cGMP-induced Erk phosphorylation in hPOBs (supplemental Fig. 4B).
Consistent with PKG II mediating cGMP effects on fos family gene expression (Figs. 5 and 6), siRNA-mediated suppression of PKG II prevented cGMP-induced Erk activation, whereas suppression of PKG I was without effect (Fig. 9C). To confirm the role of PKG II in regulating Erk1/2, we transfected cells with the mouse-specific siRNA PKG-2b and restored PKG II expression by infecting cells with adenovirus encoding rat PKG II. Viral expression of PKG II restored cGMP-induced Erk phosphorylation in PKG-2b siRNA-transfected cells (Fig. 9D; viral expression of rat PKG II is shown in Fig. 6B). In contrast, infecting cells with a LacZ- or PKG I-expressing adenovirus was without effect (viral expression of PKG I was confirmed by Western blotting and activity assay; data not shown). We conclude that cGMP activation of PKG II is necessary and sufficient to mediate Erk activation in osteoblasts and that PKG I cannot replace PKG II in this function.
DISCUSSION
We found that fluid shear stress induced a rapid and sustained increase in c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA in human primary osteoblasts, MC3T3, and UMR106 cells. These fos family genes encode members of the AP-1 transcription factor family that regulate osteoblast proliferation and differentiation (6, 8, 10, 11). We determined that mechanical stimulation of all four fos family genes was mediated by the NO/cGMP/PKG II signaling pathway and required PKG II-dependent Erk activation (Fig. 10).
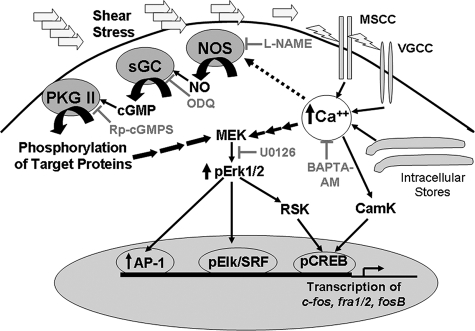
FIGURE 10.
NO/cGMP and calcium signaling in osteoblast mechanotransduction. Osteoblast exposure to fluid shear stress leads to a rapid increase in intracellular calcium through activation of mechanosensitive and voltage-gated calcium channels (MSCC and VGCC, respectively), and possibly through calcium release from intracellular stores (17, 54, 62, 63, 69). An initial burst of NO synthesis may require calcium activation of NO synthase (NOS), but sustained NO synthesis is calcium-independent and may involve activation of NO synthase by phosphorylation (57, 59). NO activates sGC and the resulting cGMP activates membrane-bound PKG II, which phosphorylates substrates leading to activation of the MEK/Erk pathway. The MEK/Erk pathway increases transcription of c-fos, fra-1, fra-2, and fosB/ΔfosB through the following: (i) RSK-mediated phosphorylation and activation of the CREB; (ii) direct phosphorylation of Elk that forms a complex with and activates serum-response factor (SRF); and (iii) recruitment of AP-1 (Fos/Jun) complexes to the promoters (79). Calcium activates the MEK/Erk pathway through Ras/Raf activation of MEK, and can directly activate CREB through calmodulin-dependent protein kinase (CamK) (89). Inhibitors used in this study are indicated in gray. BAPTA-AM, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid.
One of the earliest effects of fluid shear stress on osteoblasts is an increase in the intracellular calcium concentration. In both MC3T3 cells and human osteoblasts, the increase is detectable within seconds of flow onset, and is sustained for at least several minutes (17, 63, 69). Increased intracellular calcium leads to activation of constitutively expressed NO synthases, and increased NO production in osteoblasts occurs within 5 min of flow onset (55, 56, 70) (Fig. 2A). However, only the initial burst of NO synthesis is calcium-dependent, whereas the later sustained phase of NO production is calcium-independent (57). Our results suggest that the sustained stimulation of NO synthase activity may occur through eNOS phosphorylation by multiple kinases, as observed in fluid shear-stressed endothelial cells (59, 71).
We found that fluid shear stress stimulates osteoblasts to produce sufficient NO to increase the intracellular cGMP concentration and activate PKG, based on increased VASP phosphorylation on Ser259, a site targeted by both PKG I and II. Using an siRNA approach, we showed that PKG II, but not PKG I, is required for shear-induced c-fos, fra-1, fra-2 and fosB/ΔfosB mRNA expression. This was surprising, because osteoblasts contain ∼5-fold more cytosolic PKG activity (PKG I) than membrane-bound PKG activity (PKG II),3 and PKG I and II share many substrates in vitro (72). However, PKG II may be uniquely situated for activation by shear-induced NO. In neuronal cells, PKG II, NO synthase, and sGC localize to specialized synaptic membrane domains that contain glutamate receptors (66, 73). A similar co-localization of eNOS, sGC, and PKG II may occur in osteoblast membranes, near mechanoreceptors that transduce biophysical stimuli into biochemical signals. Possible mechanoreceptors include mechanosensitive calcium channels, G-proteins activated by membrane deformation, and integrin adhesions and cytoskeletal structures (3, 5, 74). We previously showed in osteoblasts that PKG II phosphorylates and inactivates glycogen synthase kinase-3, leading to increased DNA binding activity of CAAT enhancer-binding protein β (C/EBP-β); the latter cooperates with CREB in mediating cGMP regulation of c-fos (40, 75). In contrast, cGMP stimulation of interleukin-6 transcription in osteoblasts requires PKG I (41). Thus, PKG I and II have different roles in osteoblasts, consistent with our finding that PKG II, but not PKG I, mediates mechanotransduction.
Mice with a targeted deletion of PKG II and rats with a spontaneous deletion in the PKG II gene develop dwarfism because of impaired chondroblast differentiation (38, 76). In chondroblasts, PKG II regulates the switch from proliferation to hypertrophic differentiation by inhibiting the transcription factor Sox9 and by phosphorylating and inactivating glycogen synthase kinase-3; the latter suppresses β-catenin degradation, leading to increased differentiation (39, 77). Our results suggest that defective PKG II regulation of c-fos, fra-1, fra-2, and fosB/ ΔfosB in osteoblasts and/or chondroblasts may contribute to the developmental bone defects observed in PKG II-deficient rodents. No obvious skeletal abnormalities have been observed in PKG I-deficient mice, but their life span is too short to determine whether PKG I contributes to post-natal skeletal homeostasis (78).
We found that fluid shear stress induction of c-fos, fra-1, fra-2, and fosB/ΔfosB mRNAs required signaling through the MEK/Erk but not the p38 MAPK pathway. These findings are consistent with previous work showing that the MEK inhibitor U0126 largely prevents c-fos induction in MC3T3 cells exposed to gravitational force and fosB/ΔfosB induction in mechanically stimulated primary rat osteoblasts, whereas the p38 inhibitor SB203580 has no effect (20, 24, 54). The MEK/Erk signal transduction pathway regulates transcription of all four fos family genes by targeting common cis-acting promoter elements, i.e. the serum-response element, cAMP-response element, and phorbol ester-response element(s) (AP-1-binding sites) (Fig. 10) (79). Erk phosphorylates the ternary complex factor Elk-1, causing it to associate with and activate serum-response factor at the serum-response element, and Erk activation recruits c-Jun/Fra-2-containing AP-1 complexes to the fra-1 and fra-2 promoters (80–85). The Erk-activated kinases of the RSK family phosphorylate and activate CREB, which plays a major role in fluid shear stress-induced transcription of fosB/ΔfosB (54, 84, 86). Erk regulation of c-fos, fra-1, fra-2, and fosB/ΔfosB expression may explain why Erk is essential for osteoblast growth and differentiation (20, 68, 87).
The mechanism of Erk1/2 activation in mechanically stimulated osteoblasts is only partly understood. In fluid shear-stressed MC3T3 cells, Erk activation is calcium-dependent and inhibited by chelation of intra- or extracellular calcium; whether it requires calcium entry through mechano-sensitive cation channels and/or L-type voltage-gated calcium channels is unclear (Fig. 10) (54, 62, 63). We found that fluid shear stress-induced Erk1/2 phosphorylation was blocked by pharmacological inhibition of either NO/cGMP synthesis or PKG activity, and siRNA experiments demonstrated that both shear- and cGMP-induced Erk activation was mediated by PKG II. Possible targets of PKG II leading to Erk activation include MEK, focal adhesion kinase, and Src family kinases. Consistent with our results, Kapur et al. (88) found that l-NAME blocked fluid shear stress-induced Erk phosphorylation in hPOBs, leading to decreased shear-induced DNA synthesis, but signaling downstream of NO was not examined. Chelation of extra- or intracellular calcium prevents fluid shear stress-induced NO synthesis at early time points (57) and blocks Erk activation (54, 62, 63) and c-fos, fra-1, fra-2, and fosB/ΔfosB mRNA expression (supplemental Fig. 2A) (17, 54). These data suggest a linear pathway from calcium through NO/cGMP/PKG II to Erk activation and expression of fos family genes (Fig. 10). However, we found that calcium and cGMP cooperate to increase expression of the four fos genes. Calcium is known to regulate the c-fos promoter through multiple mechanisms, including Ras/Raf/MEK/Erk/RSK- and calcium/calmodulin-dependent protein kinase phosphorylation of CREB (89) (Fig. 10).
Fra-1, Fra-2, and ΔFosB are important for osteoblast proliferation and differentiation, as demonstrated in vitro, and by the dramatic osteosclerotic or osteoporotic phenotypes of mice that either overexpress or lack these proteins, respectively (6, 8, 10, 11). c-Fos is required for osteoblast cell cycle control, and all four fos genes are coordinately regulated during osteoblast differentiation (90–92). Fos proteins cooperate with the osteoblast-specific transcription factor Runx/Cbfa1 to regulate genes involved in osteoblast differentiation and extracellular matrix synthesis (93, 94). In addition, c-Fos, together with CREB and C/EBP-β, mediate fluid shear stress induction of cyclooxygenase-2, a gene implicated in the adaptive response of bone to mechanical stimulation (95). Induction of c-fos and fosB/ΔfosB mRNA by mechanical loading of bones in intact animals correlates with increased bone formation, and c-fos mRNA induction and bone formation in vivo are at least partly prevented by NO synthase inhibitors, consistent with the positive effects of NO donors on osteoblast growth and differentiation in vitro and bone formation in vivo (14, 16, 25, 30, 31, 33, 34, 54). We conclude that NO/cGMP/PKG II-mediated induction of c-fos, fra-1, fra-2, and fosB/ΔfosB likely plays an essential role in the anabolic response of bone to mechanical stimulation.
Supplementary Material
Acknowledgments
We thank W. Bugbee for providing us with operative bone specimens; G. Firestein and D. Boyle for assistance with primary osteoblast cultures; and S. Shattil for providing the β3 integrin antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-AR051300 (to R. B. P., H. R., S. Z., and J. -C. Y.) and Training Grant T32-HL007261 (to N. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table I and Figs. 1–4.
Footnotes
The abbreviations used are: NO, nitric oxide; 8-pCPT-cGMP, 8-(4-chlorophenylthio)-cGMP; CREB, cAMP-response element-binding protein; eNOS, endothelial nitric-oxide synthase; Erk, extracellular signal-regulated kinase; gapd, glyceraldehyde-3-phosphate dehydrogenase; hPOB, human primary osteoblasts; IBMX, isobutylmethylxanthine; l-NAME, N-nitro-l-arginine methyl ester; ODQ, 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; PAPA-NONOate, 3-(2-hydroxy-2-nitroso-1-propylhydrazino)-1-propanamine; PKG, cGMP-dependent protein kinase; (Rp)-8-pCPT-PET-cGMPS, 8-(4-chlorophenylthio)-β-phenyl-1,N2-ethenoguanosine-3′,5′-cyclic monophosphorothioate (Rp isomer); sGC, soluble guanylate cyclase; siRNA, small interfering RNA; RT, reverse transcription; GFP, green fluorescent protein.
S. Zhuang and R. Pilz, unpublished results.
References
- 1.Hughes-Fulford, M. (2004) Sci. STKE 2004, RE12. [DOI] [PubMed]
- 2.Ehrlich, P. J., and Lanyon, L. E. (2002) Osteoporos. Int. 13 688–700 [DOI] [PubMed] [Google Scholar]
- 3.Orr, A. W., Helmke, B. P., Blackman, B. R., and Schwartz, M. A. (2006) Dev. Cell 10 11–20 [DOI] [PubMed] [Google Scholar]
- 4.Hillsley, M. V., and Frangos, J. A. (1994) Biotechnol. Bioeng. 43 573–581 [DOI] [PubMed] [Google Scholar]
- 5.Rubin, J., Rubin, C., and Jacobs, C. R. (2006) Gene (Amst.) 367 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner, E. F., and Eferl, R. (2005) Immunol. Rev. 208 126–140 [DOI] [PubMed] [Google Scholar]
- 7.Ruther, U., Garber, C., Komitowski, D., Muller, R., and Wagner, E. F. (1987) Nature 325 412–416 [DOI] [PubMed] [Google Scholar]
- 8.Grigoriadis, A. E., Schellander, K., Wang, Z. Q., and Wagner, E. F. (1993) J. Cell Biol. 122 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jochum, W., David, J. P., Elliott, C., Wutz, A., Plenk, H., Jr., Matsuo, K., and Wagner, E. F. (2000) Nat. Med. 6 980–984 [DOI] [PubMed] [Google Scholar]
- 10.Sabatakos, G., Sims, N. A., Chen, J., Aoki, K., Kelz, M. B., Amling, M., Bouali, Y., Mukhopadhyay, K., Ford, K., Nestler, E. J., and Baron, R. (2000) Nat. Med. 6 985–990 [DOI] [PubMed] [Google Scholar]
- 11.Eferl, R., Hoebertz, A., Schilling, A. F., Rath, M., Karreth, F., Kenner, L., Amling, M., and Wagner, E. F. (2004) EMBO J. 23 2789–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenner, L., Hoebertz, A., Beil, T., Keon, N., Karreth, F., Eferl, R., Scheuch, H., Szremska, A., Amling, M., Schorpp-Kistner, M., Angel, P., and Wagner, E. F. (2004) J. Cell Biol. 164 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moalli, M. R., Caldwell, N. J., Patil, P. V., and Goldstein, S. A. (2000) J. Bone Miner. Res. 15 1346–1353 [DOI] [PubMed] [Google Scholar]
- 14.Chow, J. W., Fox, S. W., Lean, J. M., and Chambers, T. J. (1998) J. Bone Miner. Res. 13 1039–1044 [DOI] [PubMed] [Google Scholar]
- 15.Lean, J. M., Mackay, A. G., Chow, J. W., and Chambers, T. J. (1996) Am. J. Physiol. 270 E937–E945 [DOI] [PubMed] [Google Scholar]
- 16.Raab-Cullen, D. M., Thiede, M. A., Petersen, D. N., Kimmel, D. B., and Recker, R. R. (1994) Calcif. Tissue Int. 55 473–478 [DOI] [PubMed] [Google Scholar]
- 17.Chen, N. X., Ryder, K. D., Pavalko, F. M., Turner, C. H., Burr, D. B., Qiu, J., and Duncan, R. L. (2000) Am. J. Physiol. 278 C989–C997 [DOI] [PubMed] [Google Scholar]
- 18.Pavalko, F. M., Chen, N. X., Turner, C. H., Burr, D. B., Atkinson, S., Hsieh, Y. F., Qiu, J., and Duncan, R. L. (1998) Am. J. Physiol. 275 C1591–C1601 [PubMed] [Google Scholar]
- 19.Peake, M. A., Cooling, L. M., Magnay, J. L., Thomas, P. B., and El Haj, A. J. (2000) J. Appl. Physiol. 89 2498–2507 [DOI] [PubMed] [Google Scholar]
- 20.Hatton, J. P., Pooran, M., Li, C. F., Luzzio, C., and Hughes-Fulford, M. (2003) J. Bone Miner. Res. 18 58–66 [DOI] [PubMed] [Google Scholar]
- 21.Granet, C., Vico, A. G., Alexandre, C., and Lafage-Proust, M. H. (2002) Cell. Signal. 14 679–688 [DOI] [PubMed] [Google Scholar]
- 22.Kletsas, D., Basdra, E. K., and Papavassiliou, A. G. (2002) J. Cell Physiol. 190 313–321 [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald, J., and Hughes-Fulford, M. (1999) FASEB J. 13 553–557 [DOI] [PubMed] [Google Scholar]
- 24.Naruse, K., Miyauchi, A., Itoman, M., and Mikuni-Takagaki, Y. (2003) J. Bone Miner. Res. 18 360–369 [DOI] [PubMed] [Google Scholar]
- 25.Fox, S. W., Chambers, T. J., and Chow, J. W. (1996) Am. J. Physiol. 270 E955–E960 [DOI] [PubMed] [Google Scholar]
- 26.MacPherson, H., Noble, B. S., and Ralston, S. H. (1999) Bone (NY) 24 179–185 [DOI] [PubMed] [Google Scholar]
- 27.Aguirre, J., Buttery, L., O'Shaughnessy, M., Afzal, F., de Marticorena, I. F., Hukkanen, M., Huang, P., Maclntyre, I., and Polak, J. (2001) Am. J. Pathol. 158 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergula, A. P., Haidekker, M. A., Huang, W., Stevens, H. Y., and Frangos, J. A. (2004) Bone (NY) 34 562–569 [DOI] [PubMed] [Google Scholar]
- 29.Armour, K. E., Armour, K. J., Gallagher, M. E., Godecke, A., Helfrich, M. H., Reid, D. M., and Ralston, S. H. (2001) Endocrinology 142 760–766 [DOI] [PubMed] [Google Scholar]
- 30.Mancini, L., Moradi-Bidhendi, N., Becherini, L., Martineti, V., and MacIntyre, I. (2000) Biochem. Biophys. Res. Commun. 274 477–481 [DOI] [PubMed] [Google Scholar]
- 31.Hikiji, H., Shin, W. S., Oida, S., Takato, T., Koizumi, T., and Toyo-oka, T. (1997) FEBS Lett. 410 238–242 [DOI] [PubMed] [Google Scholar]
- 32.Chae, H. J., Park, R. K., Chung, H. T., Kang, J. S., Kim, M. S., Choi, D. Y., Bang, B. G., and Kim, H. R. (1997) J. Pharm. Pharmacol. 49 897–902 [DOI] [PubMed] [Google Scholar]
- 33.Wimalawansa, S. J., De, M. G., Gangula, P., and Yallampalli, C. (1996) Bone (NY) 18 301–304 [DOI] [PubMed] [Google Scholar]
- 34.Jamal, S. A., Cummings, S. R., and Hawker, G. A. (2004) J. Bone Miner. Res. 19 1512–1517 [DOI] [PubMed] [Google Scholar]
- 35.van't Hof, R. J., and Ralston, S. H. (2001) Immunology 103 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munzel, T., Feil, R., Mulsch, A., Lohmann, S. M., Hofmann, F., and Walter, U. (2003) Circulation 108 2172–2183 [DOI] [PubMed] [Google Scholar]
- 37.Schlossmann, J., Feil, R., and Hofmann, F. (2005) Front. Biosci. 10 1279–1289 [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer, A., Aszòdi, A., Seidler, U., Ruth, P., Hofmann, F., and Fässler, R. (1996) Science 274 2082–2086 [DOI] [PubMed] [Google Scholar]
- 39.Chikuda, H., Kugimiya, F., Hoshi, K., Ikeda, T., Ogasawara, T., Shimoaka, T., Kawano, H., Kamekura, S., Tsuchida, A., Yokoi, N., Nakamura, K., Komeda, K., Chung, U. I., and Kawaguchi, H. (2004) Genes Dev. 18 2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Y., Zhuang, S., Cassenaer, S., Casteel, D. E., Gudi, T., Boss, G. R., and Pilz, R. B. (2003) Mol. Cell. Biol. 23 4066–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broderick, K. E., Zhang, T., Rangaswami, H., Zeng, Y., Zhao, X., Boss, G. R., and Pilz, R. B. (2007) Mol. Endocrinol. 21 1148–1162 [DOI] [PubMed] [Google Scholar]
- 42.Pilz, R. B., Suhasini, M., Idriss, S., Meinkoth, J. L., and Boss, G. R. (1995) FASEB J. 9 552–558 [DOI] [PubMed] [Google Scholar]
- 43.Gudi, T., Huvar, I., Meinecke, M., Lohmann, S. M., Boss, G. R., and Pilz, R. B. (1996) J. Biol. Chem. 271 4597–4600 [DOI] [PubMed] [Google Scholar]
- 44.Idriss, S. D., Gudi, T., Casteel, D. E., Kharitonov, V. G., Pilz, R. B., and Boss, G. R. (1999) J. Biol. Chem. 274 9489–9493 [DOI] [PubMed] [Google Scholar]
- 45.Gudi, T., Lohmann, S. M., and Pilz, R. B. (1997) Mol. Cell. Biol. 17 5244–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gudi, T., Casteel, D. E., Vinson, C., Boss, G. R., and Pilz, R. B. (2000) Oncogene 19 6324–6333 [DOI] [PubMed] [Google Scholar]
- 47.Gudi, T., Hong, G. K. P., Vaandrager, A. B., Lohmann, S. M., and Pilz, R. B. (1999) FASEB J. 13 2143–2152 [PubMed] [Google Scholar]
- 48.Kuchan, M. J., Jo, H., and Frangos, J. A. (1994) Am. J. Physiol. 267 C753–C758 [DOI] [PubMed] [Google Scholar]
- 49.Gallagher, J. A. (2003) Methods Mol. Med. 80 3–18 [DOI] [PubMed] [Google Scholar]
- 50.Stevens, H. Y., and Frangos, J. A. (2003) Methods Mol. Med. 80 381–398 [DOI] [PubMed] [Google Scholar]
- 51.Casteel, D. E., Zhuang, S., Gudi, T., Tang, J., Vuica, M., Desiderio, S., and Pilz, R. B. (2002) J. Biol. Chem. 277 32003–32014 [DOI] [PubMed] [Google Scholar]
- 52.Zeng, Y., Zhuang, S., Gloddek, J., Tseng, C. C., Boss, G. R., and Pilz, R. B. (2006) J. Biol. Chem. 281 16951–16961 [DOI] [PubMed] [Google Scholar]
- 53.Jarchau, T., Haeusler, C., Markert, T., Poehler, D., Vandekerckhove, J., De Jonge, H. R., Lohmann, S. M., and Walter, U. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9426–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue, D., Kido, S., and Matsumoto, T. (2004) J. Biol. Chem. 279 49795–49803 [DOI] [PubMed] [Google Scholar]
- 55.Bacabac, R. G., Smit, T. H., Mullender, M. G., Dijcks, S. J., Van Loon, J. J., and Klein-Nulend, J. (2004) Biochem. Biophys. Res. Commun. 315 823–829 [DOI] [PubMed] [Google Scholar]
- 56.Johnson, D. L., McAllister, T. N., and Frangos, J. A. (1996) Am. J. Physiol. 271 E205–E208 [DOI] [PubMed] [Google Scholar]
- 57.McAllister, T. N., and Frangos, J. A. (1999) J. Bone Miner. Res. 14 930–936 [DOI] [PubMed] [Google Scholar]
- 58.Bakker, A. D., Soejima, K., Klein-Nulend, J., and Burger, E. H. (2001) J. Biomech. 34 671–677 [DOI] [PubMed] [Google Scholar]
- 59.Fleming, I., and Busse, R. (2003) Am. J. Physiol. 284 R1–R12 [DOI] [PubMed] [Google Scholar]
- 60.Deleted in proof
- 61.Smolenski, A., Bachmann, C., Reinhard, K., Hoenig-Liedl, P., Jarchau, T., Hoschuetzky, H., and Walter, U. (1998) J. Biol. Chem. 273 20029–20035 [DOI] [PubMed] [Google Scholar]
- 62.Liu, D., Genetos, D. C., Shao, Y., Geist, D. J., Li, J., Ke, H. Z., Turner, C. H., and Duncan, R. L. (2008) Bone (NY) 42 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You, J., Reilly, G. C., Zhen, X., Yellowley, C. E., Chen, Q., Donahue, H. J., and Jacobs, C. R. (2001) J. Biol. Chem. 276 13365–13371 [DOI] [PubMed] [Google Scholar]
- 64.Donahue, S. W., Donahue, H. J., and Jacobs, C. R. (2003) J. Biomech. 36 35–43 [DOI] [PubMed] [Google Scholar]
- 65.Ajubi, N. E., Klein-Nulend, J., Alblas, M. J., Burger, E. H., and Nijweide, P. J. (1999) Am. J. Physiol. 276 E171–E178 [DOI] [PubMed] [Google Scholar]
- 66.Serulle, Y., Zhang, S., Ninan, I., Puzzo, D., McCarthy, M., Khatri, L., Arancio, O., and Ziff, E. B. (2007) Neuron 56 670–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaandrager, A. B., Edixhoven, M., Bot, A. G. M., Kroos, M. A., Jarchau, T., Lohmann, S., Genieser, H.-G., and De Jonge, H. R. (1997) J. Biol. Chem. 272 11816–11823 [DOI] [PubMed] [Google Scholar]
- 68.Jiang, G. L., White, C. R., Stevens, H. Y., and Frangos, J. A. (2002) Am. J. Physiol. 283 E383–E389 [DOI] [PubMed] [Google Scholar]
- 69.Jacobs, C. R., Yellowley, C. E., Davis, B. R., Zhou, Z., Cimbala, J. M., and Donahue, H. J. (1998) J. Biomech. 31 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein-Nulend, J., Helfrich, M. H., Sterck, J. G., MacPherson, H., Joldersma, M., Ralston, S. H., Semeins, C. M., and Burger, E. H. (1998) Biochem. Biophys. Res. Commun. 250 108–114 [DOI] [PubMed] [Google Scholar]
- 71.Boo, Y. C., and Jo, H. (2003) Am. J. Physiol. 285 C499–C508 [DOI] [PubMed] [Google Scholar]
- 72.Gamm, D. M., Francis, S. H., Angelotti, T. P., Corbin, J. D., and Uhler, M. D. (1995) J. Biol. Chem. 270 27380–27388 [DOI] [PubMed] [Google Scholar]
- 73.Wang, H. G., Lu, F. M., Jin, I., Udo, H., Kandel, E. R., de Vente, J., Walter, U., Lohmann, S. M., Hawkins, R. D., and Antonova, I. (2005) Neuron 45 389–403 [DOI] [PubMed] [Google Scholar]
- 74.Gudi, S., Nolan, J. P., and Frangos, J. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2515–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao, X., Zhuang, S., Chen, Y., Boss, G. R., and Pilz, R. B. (2005) J. Biol. Chem. 280 32683–32692 [DOI] [PubMed] [Google Scholar]
- 76.Chikuda, H., Kugimiya, F., Hoshi, K., Ikeda, T., Ogasawara, T., Kamekura, S., Ogata, N., Nakamura, K., Chung, U. I., and Kawaguchi, H. (2005) J. Bone Miner. Metab. 23 200–204 [DOI] [PubMed] [Google Scholar]
- 77.Kawasaki, Y., Kugimiya, F., Chikuda, H., Kamekura, S., Ikeda, T., Kawamura, N., Saito, T., Shinoda, Y., Higashikawa, A., Yano, F., Ogasawara, T., Ogata, N., Hoshi, K., Hofmann, F., Woodgett, J. R., Nakamura, K., Chung, U. I., and Kawaguchi, H. (2008) J. Clin. Investig. 118 2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pfeifer, A., Klatt, P., Massberg, S., Ny, L., Sausbier, M., Hirneib, C., Wang, G.-X., Korth, M., Aszòdi, A., Andersson, K.-E., Krombach, F., Mayerhofer, A., Ruth, P., Fassler, R., and Hofmann, F. (1998) EMBO J. 17 3045–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hazzalin, C. A., and Mahadevan, L. C. (2002) Nat. Rev. Mol. Cell Biol. 3 30–40 [DOI] [PubMed] [Google Scholar]
- 80.Gille, H., Kortenjann, M., Strahl, T., and Shaw, P. E. (1996) Mol. Cell. Biol. 16 1094–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, Y., and Prywes, R. (2000) Oncogene 19 1379–1385 [DOI] [PubMed] [Google Scholar]
- 82.Adiseshaiah, P., Li, J., Vaz, M., Kalvakolanu, D. V., and Reddy, S. P. (2008) Biochem. Biophys. Res. Commun. 371 304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adiseshaiah, P., Papaiahgari, S. R., Vuong, H., Kalvakolanu, D. V., and Reddy, S. P. (2003) J. Biol. Chem. 278 47423–47433 [DOI] [PubMed] [Google Scholar]
- 84.Adiseshaiah, P., Peddakama, S., Zhang, Q., Kalvakolanu, D. V., and Reddy, S. P. (2005) Oncogene 24 4193–4205 [DOI] [PubMed] [Google Scholar]
- 85.Murakami, M., Ui, M., and Iba, H. (1999) Cell Growth & Differ. 10 333–342 [PubMed] [Google Scholar]
- 86.Mayr, B., and Montminy, M. (2001) Nat. Rev. Mol. Cell Biol. 2 599–609 [DOI] [PubMed] [Google Scholar]
- 87.Lai, C. F., Chaudhary, L., Fausto, A., Halstead, L. R., Ory, D. S., Avioli, L. V., and Cheng, S. L. (2001) J. Biol. Chem. 276 14443–14450 [DOI] [PubMed] [Google Scholar]
- 88.Kapur, S., Baylink, D. J., and Lau, K. H. (2003) Bone (NY) 32 241–251 [DOI] [PubMed] [Google Scholar]
- 89.Shaywitz, A. J., and Greenberg, M. E. (1999) Annu. Rev. Biochem. 68 821–861 [DOI] [PubMed] [Google Scholar]
- 90.Sunters, A., Thomas, D. P., Yeudall, W. A., and Grigoriadis, A. E. (2004) J. Biol. Chem. 279 9882–9891 [DOI] [PubMed] [Google Scholar]
- 91.Demiralp, B., Chen, H. L., Koh, A. J., Keller, E. T., and McCauley, L. K. (2002) Endocrinology 143 4038–4047 [DOI] [PubMed] [Google Scholar]
- 92.McCabe, L. R., Banerjee, C., Kundu, R., Harrison, R. J., Dobner, P. R., Stein, J. L., Lian, J. B., and Stein, G. S. (1996) Endocrinology 137 4398–4408 [DOI] [PubMed] [Google Scholar]
- 93.Lian, J. B., Stein, G. S., Stein, J. L., and van Wijnen, A. J. (1998) J. Cell Biochem. 30 S62–S72 [DOI] [PubMed] [Google Scholar]
- 94.D'Alonzo, R. C., Selvamurugan, N., Karsenty, G., and Partridge, N. C. (2002) J. Biol. Chem. 277 816–822 [DOI] [PubMed] [Google Scholar]
- 95.Ogasawara, A., Arakawa, T., Kaneda, T., Takuma, T., Sato, T., Kaneko, H., Kumegawa, M., and Hakeda, Y. (2001) J. Biol. Chem. 276 7048–7054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.