Abstract
Plasma concentrations of biologically active vitamin D (1,25-(OH)2D) are tightly controlled via feedback regulation of renal 1α-hydroxylase (CYP27B1; positive) and 24-hydroxylase (CYP24A1; catabolic) enzymes. In pregnancy, this regulation is uncoupled, and 1,25-(OH)2D levels are significantly elevated, suggesting a role in pregnancy progression. Epigenetic regulation of CYP27B1 and CYP24A1 has previously been described in cell and animal models, and despite emerging evidence for a critical role of epigenetics in placentation generally, little is known about the regulation of enzymes modulating vitamin D homeostasis at the fetomaternal interface. In this study, we investigated the methylation status of genes regulating vitamin D bioavailability and activity in the placenta. No methylation of the VDR (vitamin D receptor) and CYP27B1 genes was found in any placental tissues. In contrast, the CYP24A1 gene is methylated in human placenta, purified cytotrophoblasts, and primary and cultured chorionic villus sampling tissue. No methylation was detected in any somatic human tissue tested. Methylation was also evident in marmoset and mouse placental tissue. All three genes were hypermethylated in choriocarcinoma cell lines, highlighting the role of vitamin D deregulation in this cancer. Gene expression analysis confirmed a reduced capacity for CYP24A1 induction with promoter methylation in primary cells and in vitro reporter analysis demonstrated that promoter methylation directly down-regulates basal promoter activity and abolishes vitamin D-mediated feedback activation. This study strongly suggests that epigenetic decoupling of vitamin D feedback catabolism plays an important role in maximizing active vitamin D bioavailability at the fetomaternal interface.
In eutherian mammals, proper fetal growth and development are dependent on the formation and function of the placenta. In combination with the maternal decidua, cells of the placenta (primarily trophoblasts) regulate the exchange of nutrients, growth factors, oxygen, and waste between maternal and fetal circulations. The placenta also protects the developing pregnancy from the maternal immune system (1).
Although mammalian placentas share a basic common function in modulating exchange at the fetomaternal interface, it is clear that the human placenta is unique in structure and function (2). Animal models may therefore be of limited value in fully dissecting processes underlying certain aspects of human pathologies, such as pre-eclampsia and gestational trophoblastic disease (3). An examination of human placentation is therefore critical in understanding its unique function and potential role in disease.
The biologically active form of vitamin D (1,25-(OH)2D)4 is a potent secosteroid hormone. Its wide ranging actions include regulating calcium and phosphorus homeostasis, immune system modulation, cellular differentiation, and apoptosis (4–6). It also has potent antiproliferative effects (7–9). Vitamin D deficiency has been linked to several adverse pregnancy outcomes, including those associated with placental insufficiency (including pre-eclampsia) (10, 11).
Although concentrations of 1,25-(OH)2D are low in the early embryo, circulating maternal levels are elevated (∼2-fold) during pregnancy (reviewed in Ref. 12), and it is clear that a large proportion of maternal circulating 1,25-(OH)2D is derived from decidua and placental cells (13). There is inconsistent evidence regarding transplacental transfer of 1,25-(OH)2D from mother to fetus (14, 15), but it is clear that the immediate precursor, 25-(OH)2D, crosses from maternal to fetal circulations (16). At birth, fetal and maternal levels of 25-(OH)2D are comparable, and indirect measures suggest that 1,25-(OH)2D may also be correlated (17).
Several components of the vitamin D pathway are strongly expressed in human uterine decidua and placental tissues from early pregnancy (18, 19). VDR (vitamin D receptor)-bound 1,25-(OH)2D regulates key target genes associated with implantation and trophoblast functioning (including specific cytokines and homeobox genes (18–20). 1,25-(OH)2D has been shown to directly modulate the production of human chorioric gonadotrophin in human trophoblasts, and reduced expression and activity of 1-hydroxylase has been reported in differentiating trophoblast cells undergoing syncitialization in vitro (18). A supportive role of vitamin D in placental function is also evident by its influence on human placental lactogen, estradiol, and progesterone gene transcription (21, 22). In addition, vitamin D, in the form of 1,25-(OH)2D can function as an intracrine regulator of antibacterial responses in human trophoblasts that may lead to a novel mode of activation of innate immune responses in the placenta (23). Given its potent immunosuppressive effects, 1,25-(OH)2D may also play a role in implantation tolerance (19). The vitamin D metabolic pathway involves multiple enzymatic reactions, each of which is a potential target for epigenetic regulation. The CYP27B1 (1α-hydroxylase) gene product, 1α-hydroxylase, is directly responsible for the production of 1,25-(OH)2D from 25-(OH)2D. Largely expressed in the kidney, it is also expressed in decidual cells and the placenta (24). In contrast, 1,25-(OH)2D-hydroxylase (24-hydroxylase), encoded by the CYP24A1 (vitamin D 24-hydroxylase) gene, plays a pivotal role in the attenuation of vitamin D responsiveness by catalyzing synthesis of less active metabolites from both 1,25-(OH)2D and 25-(OH)2D (25–28). The CYP24A1 gene is up-regulated in several cancer cell types, where it attenuates vitamin D-mediated growth inhibition (29). However, other cancers appear to show decreased activity of this gene (4). Interestingly, a lack of expression of CYP24A1 has been directly associated with DNA methylation of the upstream promoter and first exon regions in tumor-derived endothelial cells in mice and rat osteoblastic ROS17/2.8 cells (30, 31).
The importance of epigenetic regulation in placental development and function is becoming increasingly apparent. General disruption of DNA methylation using drug treatment has been shown to disrupt placental development and also inhibit invasiveness in trophoblast cell models (32, 33). In addition to the well documented parent-of-origin specific imprinting of genes in the placenta (reviewed in Ref. 34), specific epigenetic silencing of nonimprinted genes has also recently been reported (35–38). This is likely to play a role in many aspects of placental functioning.
Very little is known about the epigenetic regulation of enzymes modulating vitamin D homeostasis at the fetomaternal interface. Given the likely importance of 1,25-(OH)2Din proper placental development and functioning, fetal development, and the modulation of maternal immune response to the developing fetus, we investigated epigenetic regulation of vitamin D metabolism in human placental tissue in general and in specific cell subtypes.
EXPERIMENTAL PROCEDURES
Tissue Isolation—Tissues were collected for research purposes with approval from the Human Research Ethics Committees at the Royal Women's Hospital (03/51), Mercy Hospital for Women (R07/15), and Monash Medical Centre (07084C). For human placental tissue sampling, a core of full-thickness tissue was isolated from two randomly chosen sites. Tissue sections from other species were washed briefly in saline before processing by maceration with a scalpel blade prior to immediate DNA isolation. CVS samples were collected and processed as previously described (37).
First trimester human cytotrophoblast cells were isolated as per Ref. 39, and purity of preparations was determined using antibodies to the trophoblast-specific cytokeratin-7 (1:100 dilution, clone OV-TL 12/30; DakoCytomation, Glostrup, Denmark) using immunocytochemistry. Cells were fixed in ethanol, rehydrated, and treated with 0.3% hydrogen peroxide in methanol for 10 min to block endogenous peroxidase activity. Cells were then washed with Tris-buffered saline and incubated with nonimmune block for 30 min. Primary antibody was applied and incubated at 48 °C for 16 h followed by biotinylated horse anti-mouse IgG (1:200) for 30 min and then streptavidinbiotin-peroxidase complex ABC (DakoCytomation) according to the manufacturer's instructions. Peroxidase activity was visualized by application of diaminobenzidine substrate (DakoCytomation) for 3–5 min. Cells were counterstained with Harris hematoxylin (Sigma), air-dried, and mounted. Only cell preparations in which 95% of the cells were positive for cytokeratin-7 were used for subsequent experiments.
For CVS samples (8–10 weeks of gestation), contaminating maternal blood and decidua were removed, and ∼5–10 mg of cleaned villi was separated from the diagnostic sample, washed in isotonic saline, pelleted by centrifugation, and stored at –20 °C until DNA extraction and bisulfite modification. A cell culture from each villi sample was established by finely macerating the villi and resuspending in 1.0 ml of 0.25% trypsin with incubation of the suspension at 37 °C for 45 min. Cells were then resuspended in 7 ml of RPMI 1640 medium (SAFC Biosciences), pelleted at 560 × g, and resuspended in a small volume of AmnioMAX medium (Invitrogen) containing 0.5 μg/ml amphotericin B. The cell suspension was then inoculated onto glass coverslips in 35-mm Petri dishes with subsequent incubation at 37 °C in 5% CO2 in humidified air. Wells were then flooded with 2 ml of AmnioMAX medium the following day and were grown to around 95% confluence, trypsinized, pelleted, and stored at –20 °C until DNA extraction and bisulfite modification. Cultured cells were isolated following 14–18 days in culture with a maximum of three passages.
Cell Culturing and Transfection—JEG-3 cells (HTB-36 (40)), BeWo (CCL-98 (41)), and JAR cells (HTB-144 (42)), were maintained in RPMI plus 10% fetal calf serum at 37 °C in 5% CO2. The SV40-transformed trophoblast HIPEC-65 cell line was maintained as previously described (43). Cells were grown for a minimal number of passages (<8) and were split (1:5–1:8) prior to confluence and harvesting of genomic DNA or transfection analysis.
For transfection experiments, a total of 8 × 103 JAR or HIPEC65 cells were plated 24 h prior to transfection in each well of a 96-well plate (96F Nunclon Delta white microwell). Transfection was carried out using Lipofectamine LTX reagent (Invitrogen). Luciferase and Renilla activity was measured 24 h post-transfection using the DualGLO assay system (Promega, Madison, WI) on a FluorSTAR Optima microplate reader (BMG Labtech, Offenburg, Germany). For vitamin D induction experiments, cells were treated with 10 or 100 nm 1,25-(OH)2D (Sigma) 12 h post-transfection, with luciferase activity monitored after overnight incubation. Endometrial stromal cells and decidua were isolated as previously described (44).
Genomic DNA and Methylation Analysis—Genomic DNA isolation and bisulfite DNA sequencing was performed as previously described (37, 38). DNA samples were processed using the Methyl Easy™ bisulfite modification kit (Human Genetic Signatures, Sydney, Australia) kit according to the manufacturer's instructions. Amplification was performed on converted genomic DNA using primers directed to bisulfite modified DNA: VDRbisF1, 5′-AATATTTTTTGTTCTTTAAGTGTTAAGTA; VDRbisF2, 5′-TAAATTTTAGTGTTTTTTAGTGTTTTAGTT; VDRBisR1, 5′-CTAACCTAATCAACCCAAATAAAAATA; CYP24BisF1, 5′-GGAAAGGGGGTTTTAAGAAATAGTA; CYP24BisR1, 5′-ACTTCCAACCCCAAAAAACTCTAAC; CYP24BisF2, 5′-TGGTTTTAGTTAGATTTTAGAGGTTAGTTT; CYP24BisR2, 5′-TACTATTTCTTAAAACCCCCTTTCC; CYP24BisF3, 5′-GAGTATGTTTTGGGTGGTTAATGAG; CYP24BisR3, 5′-TCCAAACTAAAAATATCTAACTCCCC; CYP27BisF1, 5′-TTTTGGTTGTTTTATATTTGTTTTG; CYP27BisR1, 5′-AACCATAACCCAAACCCTCAAATA.
Marmoset amplification primers were marF1 (5′-GTATTTTAGGGAAGGTTTGGAG) and marR1 (5′-CCTCCTTCCTACAACAACAAC). Mouse primers were mF1 (5′-TGTTTAGTAGTGGTTAGTTGGTGGG) and mR1 (5′-ACATCCCTAAACATCTACCTCTTCC). Amplification conditions were as follows: 95 °C for 5 min, 56 °C for 1 min 30 s, and 72 °C for 1 min 30 s × 40 cycles, 72 °C for 7 min. Resulting amplicons were cloned into TOPO TA Cloning Vector (Invitrogen) for automated fluorescent sequencing as described in Ref. 45. Data were analyzed using BiQ Analyser software (46), and clones showing less than 80% conversion or that were identified as clonal in origin were not included in subsequent analysis. Mean methylation levels for each amplicon were determined using the formula (number of methylated CpG sites/total CpG sites examined × 100).
CYP24A1 Gene Reporter Assays—A 758-bp region of the promoter/exon 1 of the human CYP24A1 gene previously shown to confer vitamin D responsiveness (47) was amplified using primers; Sal_CYP24_–548F (5′-CTCTCTGTCGACTTGGAGCTCCGCAGAAAGCCAAACTTCCT) and SalI_CYP24_+209R (5′-CTCTCTGTCGACGTACTCGAGACAGCCTCAGAGCATTGGTG) with Phusion Hi-Fidelity DNA polymerase (Finnzymes). The SalI-digested amplicon was cloned into a promoterless luciferase expression vector pGL3:basic (Promega, Madison, WI) to produce the plasmid pGL3:–594/+209. Reverse orientation cloning was also carried out to produce pGL:+209/–594. Following sequence verification, DNA was in vitro methylated using either Sss1, HpaII, or HhaI methylases (New England Biolabs, Ipswich, MA) according to the manufacturer's instructions. 100 ng of pGL3:control vector (SV40 constitutive promoter), pGL3:basic (promoterless), or unmethylated or methylated CYP24A1 promoter constructs were then transfected into JAR and HIPEC65 cells. Co-transfection with 10 ng of pRn (HSV-TK promoter linked to the Renilla luciferase gene) was carried out in all cases to normalize for transfection efficiency.
Gene Induction Experiments—24 h prior to induction, CVS-derived cells, skin, and placental-derived fibroblasts were seeded in a 6-well plate at a density of 250,000 cells/well. Half of the wells were then treated with 100 nm calcitriol (Sigma), and the other half were left untreated. Cells were then incubated for 7 h prior to total RNA isolation using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA synthesis was performed with the SuperScript III kit (Invitrogen), from 1 μg of total RNA. Quantitative reverse transcription-PCR was performed in a 96-well optical plate (Applied Biosystems, Foster City, CA) on a 7300 ABi thermal cycler (Applied Biosystems). The master mix consisted of 15 μl of Sybr Green Master Mix (Sigma), 1 μl of forward primer, 1 μl of reverse primer (50 pm final concentration), 8 μl of water, 5 μl of cDNA template in a total volume of 30 μl. Nondiluted cDNA was used for CYP24A1, CYP27B1, and VDR assays, and a 1:10 dilution was used for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) assay. Primer sequences were as follows: CYP24A1_F, 5′-CCCAAAGGAACAGTGCTCATG; CYP24A1_R, 5′-TCTCCTGAAGCCAACGTTCAG; CYP27B1_F, 5′-ATGAGGAACGGAGGACAGCC; CYP27B1_R, 5′-GAGCGTGTTGGACACCGTGT; VDR_F, 5′-TTGCGCTCCAATGAGTCCTTC; VDR_R, 5′-CTGTGTCCGGCTTTGGTCAC; GAPDH_F, 5′-TTCGACAGTCAGCCGCATCTT; GAPDH_R, 5′-CCCAATACGACCAAATCCGTT.
RESULTS
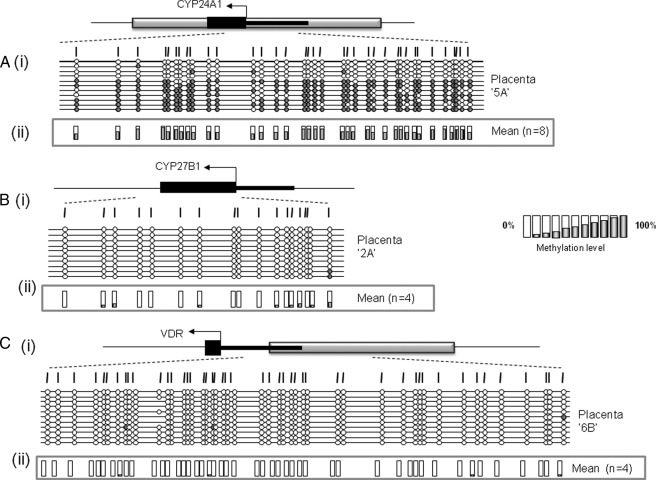
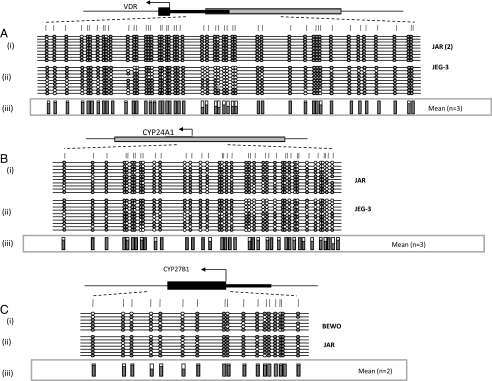
The CYP24A1 but Not the VDR or CYP27B1 Gene Promoter Is Methylated in Full Term Human Placental Tissue—As a first step in the identification of potential epigenetic regulation of vitamin D levels in the placenta, we designed specific methylation amplification assays for regulatory regions of the VDR, CYP24A1, and CYP27B1 genes. In the case of the VDR and CYP24A1 genes, assays were designed to span the transcription start site and exon 1 within the associated CpG island, whereas for CYP27B1, we analyzed a region of exon 1 previously shown to be methylated in some cancers (48, 49). Initial experiments on human term placenta revealed a complete lack of methylation at the VDR and CYP27B1 gene regulatory regions, with clear (but variable) methylation of the CYP24A1 promoter region (Fig. 1 and Table 1). This was not due to problems with specific assays, since methylation of both VDR and CYP27B1 were detected in choriocarcinoma cells (see below). Three independent assays spanning the CYP24A1-associated CpG island were then examined with each showing evidence of methylation in full term placenta (supplemental Fig. 1). A total of 10 term placentas were then examined using the CYP24A1-2 assay spanning a region of differential methylation recently identified in the placenta by whole genome methylation data (50). These results confirmed methylation of the CYP24A1 CpG island in all full term placental samples, with a mean methylation level of 56.5% (Table 1). Whereas some CYP24A1 alleles showed nearly complete methylation, others were almost completely unmethylated. This is reminiscent of monoallelic methylation, previously described for the APC tumor suppressor gene in the placenta (37), but may equally reflect differing methylation profiles in a mixed population of cells.
FIGURE 1.
Methylation status of CYP24A1, CYP27B1, and VDR in human term placenta. Schematic representation of methylation data generated for regulatory regions of genes coding for 24-hydroxylase (A), 1α-hydroxylase (B), and the vitamin D receptor (C). i, gene structure showing the location of methylation assays. Gray box, CpG island; black boxes, exon regions; arrow, translational start site. ii, mean methylation levels seen at each CpG site for each type of sample tested. Numbers of each type of tissue are listed in parentheses. Between 8 and 12 individual clones were sequenced for each sample. Circles, CpG sites denoted by vertical dashes. Closed circles, methylation; open circles, lack of methylation. Missing circles indicate CpG sites for which no information was obtained.
TABLE 1.
Mean promoter methylation levels of CYP24A1, CYP27B1, and VDR genes in human placental samples and cell lines
A summary of CpG island methylation status for genes regulating vitamin D homeostasis and activity was determined by bisulfite sequencing. Placenta, full term placental tissue; 1st trimester CT, purified cytotrophoblasts showing >95% CK-7 positive staining; Terminated pregnancy, placental tissue at 9–12 weeks gestation; Pre-eclampsia, diseased placental tissue at 9–12 weeks gestation; Choriocarcinoma, cell lines (BeWo, JEG-3, and/or JAR). Placental fibroblasts were isolated following serial passaging of Percoll gradient-separated cell fractions. Decidual cells, human endometrial stromal cells grown in culture. HIPEC65, an SV40-transformed trophoblast cell line. Normal tissue data consist of pooled data across a variety of different human tissues isolated from different subjects. The number of each type of cell/tissue is shown in parentheses.
| Gene | Tissue | Mean methylation | S. D. |
|---|---|---|---|
| % | |||
| CYP24A1 | Placenta (n = 8) | 56.53 | 10.49 |
| Uncultured CVS (n = 4) | 33.07 | 9.31 | |
| Cultured CVS (n = 4) | 33.79 | 19.02 | |
| First trimester CT (n = 4) | 48.17 | 24.49 | |
| Terminated pregnancy (n = 3) | 48.62 | 19.18 | |
| Pre-eclampsia (n = 3) | 39.85 | 6.32 | |
| Choriocarcinoma (n = 3) | 80.40 | 10.47 | |
| Placenta fibroblasts (n = 3) | 2.95 | 2.28 | |
| Decidual cells (n = 4) | 1.57 | 0.57 | |
| HIPEC65 (n = 2) | 2.03 | 0.96 | |
| Nonplacental tissue (n = 10) | 1.56 | 1.12 | |
| CYP27B1 | Placenta (n = 4) | 5.23 | 4.88 |
| Choriocarcinoma (n = 2) | 86.63 | 1.51 | |
| VDR | Placenta (n = 2) | 0.65 | 0.14 |
| Choriocarcinoma (n = 3) | 85.84 | 11.61 |
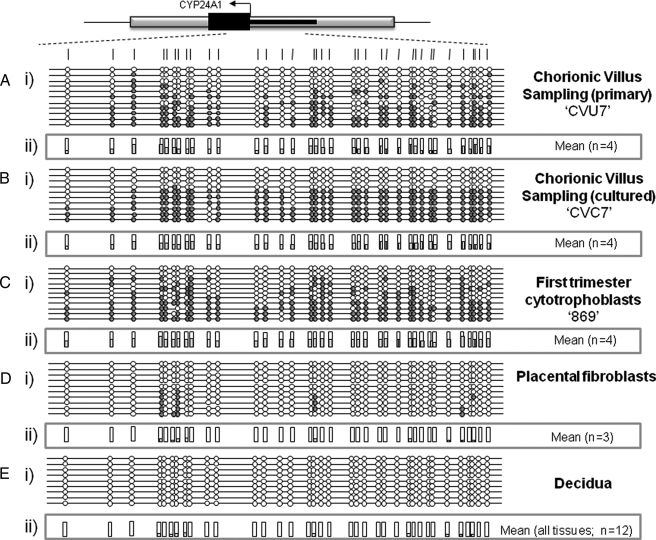
CYP24A1 Placental Methylation in Multiple First Trimester Placental Cell Types—In order to ascertain the cell types and timing of methylation of the CYP24A1 promoter within the placenta, we examined the methylation status of chorionic villous tissue (pre- and post-cell culturing), first trimester termination placental tissue, purified first trimester cytotrophoblasts (>90% cytokeratin-7-positive), and full term placental fibroblasts. Promoter methylation comparable with that seen in full term placental tissue was observed both in primary and cultured CVS tissue samples (Fig. 2 and Table 1) and in first trimester termination tissue (Table 1). Since the CVS culturing conditions do not favor trophoblast growth or division, the methylation of CYP24A1 seen in the cultured samples is probably mesodermal in origin. In addition, purified first trimester cytotrophoblasts also show methylation of the CYP24A1 gene (Fig. 2), demonstrating that this methylation is present in multiple cell types in first trimester placenta. No methylation was detected in purified full term placental fibroblasts or an SV40-transformed first trimester trophoblast cell line (Fig. 2). A summary of mean methylation levels obtained in this study is presented in Table 1.
FIGURE 2.
Methylation of CYP24A1 in different human placental tissues and cell types. Representative methylation data generated for the CYP24A1-2 assay in cytokeratin-7 positive cytotrophoblasts (A), uncultured CVS (B), cultured CVS (C), full term placental fibroblasts (D), and whole blood (E). i, between 8 and 12 individual clones were sequenced for each sample. Circles, CpG sites denoted by vertical dashes. Closed circles, methylation; open circles, lack of methylation. Missing circles indicate CpG sites for which no information was obtained. Gray boxes, to GpG island locations; arrows, start site of translation within exon 1 (black line). ii, mean methylation levels seen at each CpG site for each type of sample tested. Numbers of each type of tissue are listed in parentheses.
CYP24A1 Methylation Is Placenta-specific—Given the importance of the 24-hydroxylase enzyme in regulating levels of active vitamin D, it is surprising how little is known about the epigenetic regulation of this gene in human tissues. In order to address this, we assessed the methylation status of the CYP24A1 gene in a panel of human tissues consisting of kidney, skeletal muscle, skin fibroblasts, brain (prefrontal cortex), sperm, whole blood, buccal mucosa, endometrial stroma, decidualized stroma, bone marrow, and umbilical cord tissue. The results, (summarized in Fig. 2 and Table 1; detailed in supplemental Fig. 2), demonstrate the specificity of placental CYP24A1 methylation in humans, with no evidence for methylation in any human somatic tissue tested.
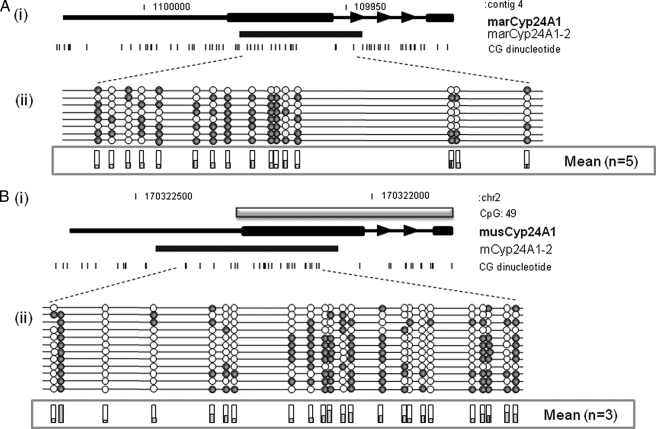
The CYP24A1 Gene Is Differentially Methylated in Full Term Placentas from Different Mammals—In previous studies, we have demonstrated that some of the specific promoter methylation recently documented in human term placental tissue is present in primates but absent in E18 (day 18 of pregnancy) mouse placenta. We therefore examined methylation of the CYP24A1 gene promoter regions in full term placenta from marmoset and mouse E18 and E11.5 (Fig. 3). Assays were designed to span the region corresponding to the human CYP24A1-2 assay. This revealed varying levels of CYP24A1 methylation in both marmoset (mean 27 ± 18.7%) and mouse (mean 35.1 ± 4.5%) placental tissue with generally lower levels relative to that seen in human full term placentas (mean 56.5 ± 10.5%; Fig. 3) Interestingly, less methylation was seen in E18 mouse placental tissue compared with E11.5 tissue (data not shown).
FIGURE 3.
Variable levels of CYP24A1 methylation in different mammalian full term placentas. Representative methylation data were generated for homologous CYP24A1 promoter region in marmoset (A) and mouse (B). i, gray boxes, GpG island locations; arrows, orientation of transcription. Black boxes, assay location. ii, eight individual clones were sequenced for each sample. Circles, CpG sites denoted by black dashes. Closed circles, methylation; open circles, lack of methylation. Shaded bars, mean methylation levels seen at each CpG site for each type of sample tested.
Methylation of the Core CYP24A1 Gene Promoter Directly Attenuates Basal Promoter Activity in Human Trophoblast-derived Cells—Despite circumstantial evidence of an inverse correlation of CYP24A1 mRNA levels with promoter gene methylation (above), no direct evidence for DNA methylation-mediated silencing of this gene promoter has been reported. To investigate this further, we PCR-amplified the previously described vitamin D-responsive region of the human CYP24A1 gene promoter (47) and generated a luciferase reporter construct to directly test the effects of promoter methylation on both basal and vitamin D-induced promoter activity.
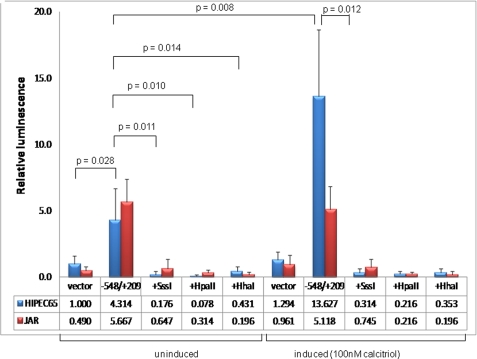
The SV40-transformed human trophoblast cell line, HIPEC65, and choriocarcinoma-derived JAR cells were transfected with two different reporter constructs (plus and minus orientation) with or without in vitro methylation with SssI or HpaII methylases. SssI methylase methylates all CpGs; HpaII methylase methylates only the CpG within the sequence CCGG. Results, summarized in Fig. 4, reveal a low level of promoter activity of this promoter region in the plus orientation in both cell lines, ∼6.5-fold (HIPEC65; p = 0.028) and 12-fold (JAR; p < 0.001) higher than that of the promoterless luciferase vector. Promoter activity in the reverse (minus) orientation was negligible (1.3-fold), demonstrating a directionality of the basal promoter element (data not shown). Basal activity was completely attenuated by prior methylation with any of SssI (p = 0.011), HpaII (p = 0.01), or HhaI (p = 0.014) methylases in HIPEC65 cells and inhibited in the JAR cell line (p < 0.001 for each of SssI and HpaII; supplemental Table 1). The 768-bp region of this gene used for reporter activity assays contains a total of 5 HhaI and 15 HpaII methylase target sites, suggesting that even low level methylation of this promoter is sufficient to abrogate gene expression activity.
FIGURE 4.
CYP24A1 promoter activity in human trophoblast cell lines. SV40-transformed cytotrophoblast (HIPEC-65) or choriocarcinoma (JAR) cells were transfected (n = 8 for each cell line) with a promoterless luciferase reporter (vector) with or without the cloned CYP24A1 gene promoter (–548/+209 relative to the start site of transcription). This region has previously been demonstrated to contain vitamin D-responsive elements (47). Basal promoter activity was detected in both cell lines and was abolished with in vitro methylation of the promoter prior to transfection (Sss1, HpaII, and HhaI methylases). 1,25-(OH)2D (active vitamin D) induction of the promoter was observed only in HIPEC-65 cells (JAR cells are predicted to lack the vitamin D receptor, as evidenced by complete methylation of the associated CpG island), but this induction was attenuated with prior methylation of the promoter region. Luminescence values were normalized to Renilla luciferase to correct for transfection efficiency, and all data are displayed relative to the mean promoterless control vector in HIPEC-65 cells. Error bars denote 95% confidence intervals. Statistical analysis for comparing between transfected groups was carried out using a two-tailed Student's t test with unequal variance.
CpG Island Methylation Abrogates 1,25-(OH)2D-mediated Up-regulation of CYP24A1 Transcription—In addition to examining the effects of promoter methylation on the basal CYP24A1 promoter activity, we also set out to investigate the consequences of this methylation on the previously demonstrated inducibility of this gene promoter by 1,25-(OH)2D (47). JAR and HIPEC65 cells were transfected with the 768-bp core promoter with or without previous methylation with Sss1 methylase as above. 10 h post-transfection, one-half of cells were supplemented with 100 nm 1,25-(OH)2D, and reporter (luciferase) activity was then monitored. Although 1,25-(OH)2D treatment resulted in a clear induction (3.5-fold) of basal promoter activity in HIPEC65 cells in the absence of previous promoter methylation (p = 0.008), this response was abolished with in vitro methylation of the reporter construct prior to transfection (Fig. 4). 1,25-(OH)2D treatment had no effect on promoter activity in JAR cells (p = 0.663), most likely reflecting the methylation of the VDR gene in this cell line (see below) that silences the gene and inhibits exogenous 1,25-(OH)2D signaling through the VDR.
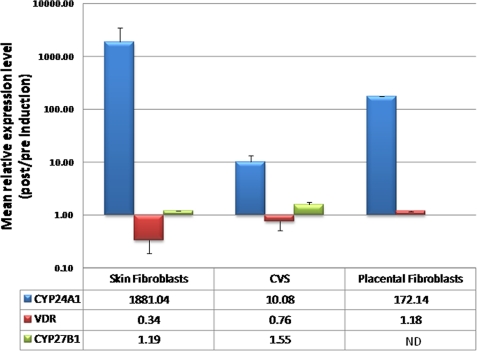
We also examined the effect of promoter methylation on the induction of endogenous CYP24A1, CYP27B1, and VDR gene expression in primary cells derived from CVS tissue (average methylation level of 33.4%) and placental and skin fibroblasts (both lacking CYP24A1 methylation). Cultured skin fibroblasts showed a mean CYP24A1 induction level approaching 2000-fold (n = 2; Fig. 5), consistent with previous data (47, 51, 52). Placental fibroblasts also showed a significant level of induction (mean 172-fold; n = 2). In contrast, cultured CVS cells showed a reduced capacity for gene induction (mean ∼10 fold; n = 4). In contrast to the major effects of vitamin D induction on CYP24A1 expression in skin and placental fibroblasts, VDR expression levels were slightly reduced (mean reduction of ∼3-fold) in skin fibroblasts, with little change apparent in placental fibroblasts (mean 1.18-fold increase) and CVS-derived cells (mean 0.76-fold change; Fig. 5). Similarly, no major effects on CYP27B1 expression level were seen in skin fibroblasts (mean 1.19-fold increase) or CVS-derived cells (mean 1.55-fold increase) following vitamin D induction (Fig. 5).
FIGURE 5.
Induction of endogenous CYP24A1 mRNA in cultured primary cells. Cells were cultured in multiple wells, and 100 nm 1,25-(OH)2D3 (active vitamin D) was added to half of the cultures for 7 h, followed by total RNA isolation and quantitative real time reverse transcription-PCR. Mean levels of induction (n = 2–4) are listed in the corresponding table. Whereas cultured skin fibroblasts show a nearly 2000-fold induction of CYP24A1 expression, consistent with previous data (47, 51, 52), and placental fibroblasts show a mean 172-fold induction, cultured CVS biopsies (containing cells with CYP24A1 methylation) show a greatly reduced level of gene induction (∼10-fold). No major effects on VDR or CYP27B1 expression were observed. Error bars denote 95% confidence interval. ND, no data available.
Altered CYP24A1 Methylation in Placental Tissue from Pre-eclampsia Pregnancies?—We carried out a preliminary analysis of methylation levels of the CYP24A1-2 assay region in a small set of available preeclampsia tissue (Table 1). These samples showed a mean methylation level of only 39.5 ± 6% (n = 3), compared with the 56.53 ± 10% level seen in nonpre-eclampsia placental tissue (n = 8). The potential significance (p = 0.017) of these data will require further investigation in a larger sample size in the future.
Complete Methylation of Vitamin D Regulatory Genes in Choriocarcinoma Cell Lines—Trophoblast hyperplasia is a common feature of complete and partial hydatidiform mole with the potential for malignant transformation to choriocarcinoma (CCA) (53). Trophoblasts that comprise CCA resemble the primitive trophoblast of the previllous stage during placental development, arrested in specific stages of differentiation (54).
We analyzed the methylation status of CYP24A1 in three widely studied CCA cell lines, JEG-3, BeWo, and JAR. In all three lines, methylation at the CYP24A1 promoter was increased to nearly complete methylation (Fig. 6). These results correlate with the very low levels of CYP24A1 mRNA seen in each of these cell types (data not shown). We also assessed methylation at the VDR and CYP27B1 CpG islands, which also revealed nearly complete methylation of each of these genes in all cell lines tested. This contrasts with the complete lack of methylation seen in full term placental tissue and first trimester cytotrophoblast (described above).
FIGURE 6.
Methylation analysis of vitamin D-associated genes in CCA-derived trophoblast cell lines. High levels of methylation were detected for both VDR (A) and CYP27B1 (C) in choriocarcinoma cell lines (n = 2). All choriocarcinoma cell lines (n = 3; BeWo not shown) examined showed hypermethylation of the CYP24A1 gene relative to full term placental tissue or purified first trimester trophoblasts. Numbers of each type of tissue are listed in parentheses. Between 8 and 12 individual clones were sequenced for each sample. Circles, CpG sites within the assayed region. Closed circles, methylation; open circles, lack of methylation. Missing circles indicate CpG sites for which no information was obtained. Gray boxes correspond to GpG island locations, and arrows denote start site of translation within exon 1 (black line). iii, mean methylation levels seen at each CpG site for each type of sample tested.
DISCUSSION
Placental insufficiency, due to inadequate remodeling of the maternal vasculature in early pregnancy, is thought to play a major role in several adverse pregnancy outcomes, including the early stages of pre-eclampsia (55). Lowered 1,25-(OH)2D and altered calcium metabolism have been implicated in this process (10, 11). For example, a recent Cochrane review of randomized control trials demonstrated a role for calcium supplementation in reducing pre-eclampsia risk.5 Altered expression levels of 1-hydroxylase, VDR, and 24-hydroxylase genes have all been reported in different placental cells and tissue from preeclamptic pregnancies, possibly in response to decreasing maternal 1,25-(OH)2D (19, 57, 58). In addition, levels of CYP27B1 mRNA are reduced in cultured syncytial trophoblasts derived from pre-eclamptic patients compared with controls (57). These data highlight the likely functional importance of 1,25-(OH)2D in placental function and implicate disruption of vitamin D homeostasis in adverse pregnancy outcomes.
Epigenetic regulation of steroid signaling is an emerging field of research and hints at a complex interplay between environmental or dietary influences with underlying genetic variation, epigenetic variation, steroid metabolism, and subsequent downstream effects (59). However, at present, very little is known about the consequences of altered environmental, genetic, or epigenetic regulation of vitamin D homeostasis at the fetomaternal interface. In this study, we found no evidence for methylation of the VDR or CYP27B1 genes in human full term or first trimester placental tissue. Since methylation is generally associated with gene silencing, this supports previous data demonstrating high levels of expression of 1α-hydroxylase and VDR in the placenta (18, 19, 60). In contrast, we have identified methylation of the CYP24A1 gene, encoding 24α-hydroxylase, the major catabolic enzyme of 1,25-(OH)2D and 25-(OH)2D in placental tissue and cell subtypes. We have demonstrated that this methylation is present in both the trophoblast compartment and cells of mesodermal origin and is present (albeit at variable levels) in primate and rodent placentas. This contrasts with the recently described placenta-specific methylation of the APC tumor suppressor gene that is primate-specific and localizes solely to the trophoblast compartment (37) and the SFRP2 tumor suppressor methylation that is primate-specific but localizes to multiple placental cell subtypes (38).
Importantly, we have demonstrated (by in vitro reporter studies) that methylation of the CYP24A1 gene promoter directly down-regulates basal gene activity and abolishes vitamin D induction of the proximal promoter region. This is further supported by data demonstrating a reduced inducibility (by 2–3 orders of magnitude) of the methylated endogenous CYP24A1 gene in cultured primary human CVS-derived cells relative to unmethylated fibroblast cells. We observed much higher levels of induction of the endogenous gene in cultured primary fibroblast cells in comparison with that seen with a transfected reporter construct (containing only the proximal promoter region) in transformed trophoblast cells. This is consistent with previous data demonstrating a requirement for distal upstream sequence elements, in addition to the two previously described vitamin D-responsive elements, contained within the proximal promoter region, for maximal induction of the CYP24A1 gene (47). Interestingly, we did not find any major effects of exogenous 1,25-(OH)2D on the levels of VDR or CYP27B1 expression in skin or placental fibroblast cells. This contrasts with previous studies showing down-regulation of each of these genes in some cancer cell lines (61) in response to 1,25-(OH)2D treatment. In addition, the CYP27B1 gene was recently reported to be down-regulated in response to 1,25-(OH)2D in cultured trophoblasts undergoing syncitialization (62). The proximal human CYP27B1 gene promoter contains putative vitamin D-responsive elements and has been shown to mediate 1,25-(OH)2d-mediated down-regulation of this gene in in vitro reporter experiments in mouse proximal tubule cells (63). However, in primary human keratinocytes, 1,25-(OH)2D does not regulate 1α-hydroxylase mRNA or protein expression (64), supporting our findings in this study.
Methylation of the proximal promoter region of the mouse Cyp24a1 gene has been previously reported in tumor-derived endothelial cells, where it contributes to selective growth inhibition by 1,25-(OH)2D (30). Conversely, a lack of methylation in corresponding non-tumor-derived mouse endothelial cells is associated with a lack of 1,25-(OH)2D-mediated growth inhibition (30). No equivalent data have previously been reported for the human CYP24A1 gene.
It is well documented that maternal circulating 1,25-(OH)2D levels show a dramatic increase during pregnancy (reviewed in Ref. 12), with ∼50% of maternal circulating 1,25-(OH)2D derived from maternal decidua and placenta (13). This observation demonstrates that the tight regulation of 1,25-(OH)2D levels is uncoupled during pregnancy. There are two potential mechanisms by which this can be achieved. First, this can be achieved through an increase in expression of the CYP27B1 gene, which results in increasing 1α-hydroxylase and elevated production of 1,25-(OH)2D from less active precursors. The second potential mechanism involves a decrease in the catabolism of 1,25-(OH)2D by the 24-hydroxylase enzyme encoded by CYP24A1. Increasing levels of maternal circulating 1,25-(OH)2D during pregnancy have been linked to increased synthesis rather than decreasing catabolic activity in animal studies (65–67). In addition, CYP27B1 expression is increased in early pregnancy decidua in comparison with nonpregnant endometrium (19, 68). In this study, we have demonstrated that the increasing synthesis of 1,25-(OH)2D occurs in concert with a placenta-specific epigenetic down-regulation of the CYP24A1 (24-hydroxylase) gene. Thus, both mechanisms for elevating 1,25-(OH)2D levels are present during pregnancy.
We are currently investigating the specific role of 1,25-(OH)2D in mediating various aspects of placental functioning. 1,25-(OH)2D has widely diverse roles in regulating cell division, apoptosis, and immune modulation. It also shows antiproliferative effects in many different cell types, is proapoptotic, and is inhibitory to cell migration and invasion (5, 6, 69–71). Each of these processes plays an important role in the development and function of the mammalian placenta.
The interesting finding of simultaneous methylation of the VDR, CYP24A1, and CYP27B1 genes in choriocarcinoma-derived trophoblast cell lines is unprecedented in cancer. Methylation of the VDR gene is consistent with recent data demonstrating a lack of expression in BeWo and JEG-3 choriocarcinoma cell lines (72), and dysregulation of all three genes has been reported in cancers of different origin (73). Although abolition of VDR and CYP27B1 expression is anticipated to attenuate vitamin D-mediated growth inhibition in tumor cells, silencing of CYP24A1 would have the effect of maximizing active vitamin D bioavailability. It is possible that the precursor cells from which CCA developed already had the CYP24A1 methylation described in this study and that acquisition of epigenetic silencing of the VDR and CYP27B1 genes was associated with the development of this tumor type. It is interesting to speculate that a sequential series of methylation events may occur in tumor development and progression. Future studies examining the epigenetic regulation of this and other pathways in hydatidiform mole, recurring mole, and choriocarcinoma, may shed valuable light on the acquisition of methylation changes in gestational tumors.
Confirmation of epigenetic regulation of active vitamin D levels at the fetomaternal interface provides compelling circumstantial evidence for the intersection of two metabolic pathways in regulating pregnancy outcome, namely vitamin D and 1-carbon (folate or methyl donor) metabolism. One-carbon metabolism is a process by which methyl groups are passed from one donor molecule to the next (74, 75). This cycle produces S-adenosylmethionine, which donates its methyl group to cytosine to produce methylated CpG. Each of these pathways relies on external inputs for the production of precursor molecules, and enzymes involved in each pathway (including VDR and CYP27B1) have known functional polymorphic variants implicated in moderating disease risk (76–83), including adverse pregnancy outcomes (84–89). There are numerous polymorphisms throughout the CYP24A1 exonic, intronic, and regulatory gene regions (56), but as yet there is little information regarding the functionality or clinical relevance of such variants. The convergence/intersection of 1-carbon metabolism with other metabolic and signaling pathways is potentially very important in modulating disease risk and warrants further investigation.
The placenta-specific epigenetic regulation of several genes and metabolic pathways, independent of well characterized imprinted regions, is now a firmly established phenomenon. Uncovering the mechanisms leading to the establishment of such markings, their timing, and sensitivity to environmental and genetic disruption has the potential to reveal valuable insights into the evolution of different placental functions and may reveal novel risk pathways associated with adverse pregnancy outcomes and tumor development.
Acknowledgments
We thank Dr. Nicole Brooks and Tina Vaino (Mercy Hospital for Women, Australia) and Sarah Healy (Royal Women's Hospital) for help with collection of placental tissue, Professor Lois Salamonson (Prince Henry's Institute of Medical Research, Australia) and Professor Samuel Breit (University of NSW, Australia) for CCA cancer cell lines, Drs. Patrick Western and Craig Smith (Murdoch Childrens Research Institute) for mouse placental tissue, and Dr. Mark Pertile for human CVS samples. We especially thank Lavinia Gordon for bioinformatics help in this and previous studies.
Author's Choice—Final version full access.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: 1,25-(OH)2D, biologically active form of vitamin D; 25-(OH)2D, immediate precursor of 1,25-(OH)2D; CVS, chorionic villus sampling; CCA, choriocarcinoma; En, embryonic day n.
G. J. Hofmeyr, A. N. Atallah, and L. Duley (2006) Cochrane Database of Systematic Reviews 3, CD001059, available on the World Wide Web.
References
- 1.von Rango, U. (2008) Immunol. Lett. 115 21–32 [DOI] [PubMed] [Google Scholar]
- 2.Carter, A. M. (2007) Placenta 28 Suppl. A, 41–47 [Google Scholar]
- 3.Soundararajan, R., and Rao, A. J. (2004) Reprod. Biol. Endocrinol. 2 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, M. G., Nakane, M., Ruan, X., Kroeger, P. E., and Wu-Wong, J. R. (2006) Cancer Chemother. Pharmacol. 57 234–240 [DOI] [PubMed] [Google Scholar]
- 5.DeLuca, H. F. (2004) Am. J. Clin. Nutr. 80 (suppl.) 1689–1696 [Google Scholar]
- 6.Dusso, A. S., Brown, A. J., and Slatopolsky, E. (2005) Am. J. Physiol. 289 F8–F28 [DOI] [PubMed] [Google Scholar]
- 7.Masuda, S., and Jones, G. (2006) Mol. Cancer Ther. 5 797–808 [DOI] [PubMed] [Google Scholar]
- 8.Muller, K., and Bendtzen, K. (1996) J. Investig. Dermatol. Symp. Proc. 1 68–71 [PubMed] [Google Scholar]
- 9.Peehl, D. M., Skowronski, R. J., Leung, G. K., Wong, S. T., Stamey, T. A., and Feldman, D. (1994) Cancer Res. 54 805–810 [PubMed] [Google Scholar]
- 10.Bodnar, L. M., Catov, J. M., Simhan, H. N., Holick, M. F., Powers, R. W., and Roberts, J. M. (2007) J. Clin. Endocrinol. Metab. 92 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seely, E. W. (2007) J. Clin. Endocrinol. Metab. 92 3402–3403 [DOI] [PubMed] [Google Scholar]
- 12.Kovacs, C. S., and Kronenberg, H. M. (1997) Endocr. Rev. 18 832–872 [DOI] [PubMed] [Google Scholar]
- 13.Delvin, E. E., Arabian, A., Glorieux, F. H., and Mamer, O. A. (1985) J. Clin. Endocrinol. Metab. 60 880–885 [DOI] [PubMed] [Google Scholar]
- 14.Delvin, E. E., Glorieux, F. H., Salle, B. L., David, L., and Varenne, J. P. (1982) Arch. Dis. Child 57 754–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salle, B. L., Delvin, E. E., Lapillonne, A., Bishop, N. J., and Glorieux, F. H. (2000) Am. J. Clin. Nutr. 71 (suppl.) 1317–1324 [DOI] [PubMed] [Google Scholar]
- 16.Ron, M., Levitz, M., Chuba, J., and Dancis, J. (1984) Am. J. Obstet. Gynecol. 148 370–374 [DOI] [PubMed] [Google Scholar]
- 17.Tsang, R. C., Greer, F., and Steichen, J. J. (1981) Clin. Perinatol. 8 287–306 [PubMed] [Google Scholar]
- 18.Barrera, D., Avila, E., Hernandez, G., Mendez, I., Gonzalez, L., Halhali, A., Larrea, F., Morales, A., and Diaz, L. (2008) Reprod. Biol. Endocrinol. 6 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, K. N., Bulmer, J. N., Kilby, M. D., and Hewison, M. (2004) J. Soc. Gynecol. Investig. 11 263–271 [DOI] [PubMed] [Google Scholar]
- 20.Du, H., Daftary, G. S., Lalwani, S. I., and Taylor, H. S. (2005) Mol. Endocrinol. 19 2222–2233 [DOI] [PubMed] [Google Scholar]
- 21.Barrera, D., Avila, E., Hernandez, G., Halhali, A., Biruete, B., Larrea, F., and Diaz, L. (2007) J. Steroid Biochem. Mol. Biol. 103 529–532 [DOI] [PubMed] [Google Scholar]
- 22.Stephanou, A., Ross, R., and Handwerger, S. (1994) Endocrinology 135 2651–2656 [DOI] [PubMed] [Google Scholar]
- 23.Liu, N., Kaplan, A. T., Low, J., Nguyen, L., Liu, G. Y., Equils, O., and Hewison, M. (2009) Biol. Reprod. 80 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans, K. N., Nguyen, L., Chan, J., Innes, B. A., Bulmer, J. N., Kilby, M. D., and Hewison, M. (2006) Biol. Reprod. 75 816–822 [DOI] [PubMed] [Google Scholar]
- 25.Kumar, R. (1984) Physiol. Rev. 64 478–504 [DOI] [PubMed] [Google Scholar]
- 26.Lohnes, D., and Jones, G. (1987) J. Biol. Chem. 262 14394–14401 [PubMed] [Google Scholar]
- 27.Makin, G., Lohnes, D., Byford, V., Ray, R., and Jones, G. (1989) Biochem. J. 262 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siu-Caldera, M. L., Zou, L., Ehrlich, M. G., Schwartz, E. R., Ishizuka, S., and Reddy, G. S. (1995) Endocrinology 136 4195–4203 [DOI] [PubMed] [Google Scholar]
- 29.Albertson, D. G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S. H., Kowbel, D., Kuo, W. L., Gray, J. W., and Pinkel, D. (2000) Nat. Genet. 25 144–146 [DOI] [PubMed] [Google Scholar]
- 30.Chung, I., Karpf, A. R., Muindi, J. R., Conroy, J. M., Nowak, N. J., Johnson, C. S., and Trump, D. L. (2007) J. Biol. Chem. 282 8704–8714 [DOI] [PubMed] [Google Scholar]
- 31.Ohyama, Y., Kusada, T., Yamasaki, T., and Ide, H. (2002) Nucleic Acids Symp. Series 2 249–250 [DOI] [PubMed] [Google Scholar]
- 32.Rahnama, F., Shafiei, F., Gluckman, P. D., Mitchell, M. D., and Lobie, P. E. (2006) Endocrinology 147 5275–5283 [DOI] [PubMed] [Google Scholar]
- 33.Serman, L., Vlahovic, M., Sijan, M., Bulic-Jakus, F., Serman, A., Sincic, N., Matijevic, R., Juric-Lekic, G., and Katusic, A. (2007) Placenta 28 803–811 [DOI] [PubMed] [Google Scholar]
- 34.Coan, P. M., Burton, G. J., and Ferguson-Smith, A. C. (2005) Placenta 26 Suppl. A, 10–20 [DOI] [PubMed] [Google Scholar]
- 35.Chiu, R. W., Chim, S. S., Wong, I. H., Wong, C. S., Lee, W. S., To, K. F., Tong, J. H., Yuen, R. K., Shum, A. S., Chan, J. K., Chan, L. Y., Yuen, J. W., Tong, Y. K., Weier, J. F., Ferlatte, C., Leung, T. N., Lau, T. K., Lo, K. W., and Lo, Y. M. (2007) Am. J. Pathol. 170 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, X., Shiu, S., Cal, A., and Borevitz, J. O. (2008) PLoS Genet. 4 e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong, N. C., Novakovic, B., Weinrich, B., Dewi, C., Andronikos, R., Sibson, M., Macrae, F., Morley, R., Pertile, M. D., Craig, J. M., and Saffery, R. (2008) Cancer Lett. 268 56–62 [DOI] [PubMed] [Google Scholar]
- 38.Novakovic, B., Rakyan, V., Ng, H. K., Manuelpillai, U., Dewi, C., Wong, N. C., Morley, R., Down, T., Beck, S., Craig, J. M., and Saffery, R. (2008) Mol. Hum. Reprod. 14 547–554 [DOI] [PubMed] [Google Scholar]
- 39.Tapia, A., Salamonsen, L. A., Manuelpillai, U., and Dimitriadis, E. (2008) Hum. Reprod. 23 1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler, P. O., and Bridson, W. E. (1971) J. Clin. Endocrinol. Metab. 32 683–687 [DOI] [PubMed] [Google Scholar]
- 41.Pattillo, R. A., and Gey, G. O. (1968) Cancer Res. 28 1231–1236 [PubMed] [Google Scholar]
- 42.Pattillo, R. A., Gey, G. O., Delfs, E., Huang, W. Y., Hause, L., Garancis, D. J., Knoth, M., Amatruda, J., Bertino, J., Friesen, H. G., and Mattingly, R. F. (1971) Ann. N. Y. Acad. Sci. 172 288–298 [DOI] [PubMed] [Google Scholar]
- 43.Pavan, L., Tarrade, A., Hermouet, A., Delouis, C., Titeux, M., Vidaud, M., Therond, P., Evain-Brion, D., and Fournier, T. (2003) Carcinogenesis 24 1325–1336 [DOI] [PubMed] [Google Scholar]
- 44.Dimitriadis, E., Robb, L., and Salamonsen, L. A. (2002) Mol. Hum. Reprod. 8 636–643 [DOI] [PubMed] [Google Scholar]
- 45.Wong, N. C., Wong, L. H., Quach, J. M., Canham, P., Craig, J. M., Song, J. Z., Clark, S. J., and Choo, K. H. (2006) PLoS Genet. 2 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock, C., Reither, S., Mikeska, T., Paulsen, M., Walter, J., and Lengauer, T. (2005) Bioinformatics 21 4067–4068 [DOI] [PubMed] [Google Scholar]
- 47.Tashiro, K., Ishii, C., and Ryoji, M. (2007) Biochem. Biophys. Res. Commun. 358 259–265 [DOI] [PubMed] [Google Scholar]
- 48.Kim, S., Li, M., Paik, H., Nephew, K., Shi, H., Kramer, R., Xu, D., and Huang, T. H. (2008) Pacific Symposium on Biocomputing, Fairmont Orchid, Hawaii, January 4–8, 2008, pp. 315–326, PSB, Stanford, CA [DOI] [PubMed]
- 49.Taylor, K. H., Kramer, R. S., Davis, J. W., Guo, J., Duff, D. J., Xu, D., Caldwell, C. W., and Shi, H. (2007) Cancer Res. 67 8511–8518 [DOI] [PubMed] [Google Scholar]
- 50.Rakyan, V., Down, T., Thorne, N., Flicek, P., Kulesha, E., Graf, S., Tomazou, E., Backdahl, L., Johnson, N., Herberth, M., Howe, K., Jackson, D., Miretti, M., Fiegler, H., Marioni, J., Birney, E., Hubbard, T., Carter, N., Tavare, S., and Beck, S. (2008) Genome Res. 18 1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malloy, P. J., Wang, J., Peng, L., Nayak, S., Sisk, J. M., Thompson, C. C., and Feldman, D. (2007) Arch. Biochem. Biophys. 460 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Y., Wang, J., Malloy, P. J., Dolezel, Z., and Feldman, D. (2009) J. Bone Miner. Res. 24 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, H. W., Tsao, S. W., and Cheung, A. N. (2002) Placenta 23 20–31 [DOI] [PubMed] [Google Scholar]
- 54.Shih Ie, M., and Kurman, R. J. (2002) Curr. Mol. Med. 2 1–12 [DOI] [PubMed] [Google Scholar]
- 55.Chaddha, V., Viero, S., Huppertz, B., and Kingdom, J. (2004) Semin. Fetal Neonatal Med. 9 357–369 [DOI] [PubMed] [Google Scholar]
- 56.Holick, C. N., Stanford, J. L., Kwon, E. M., Ostrander, E. A., Nejentsev, S., and Peters, U. (2007) Cancer Epidemiol. Biomarkers Prev. 16 1990–1999 [DOI] [PubMed] [Google Scholar]
- 57.Diaz, L., Arranz, C., Avila, E., Halhali, A., Vilchis, F., and Larrea, F. (2002) J. Clin. Endocrinol. Metab. 87 3876–3882 [DOI] [PubMed] [Google Scholar]
- 58.Fischer, D., Schroer, A., Ludders, D., Cordes, T., Bucker, B., Reichrath, J., and Friedrich, M. (2007) Clin. Exp. Obstet. Gynecol. 34 80–84 [PubMed] [Google Scholar]
- 59.Leader, J. E., Wang, C., Fu, M., and Pestell, R. G. (2006) Biochem. Pharmacol. 72 1589–1596 [DOI] [PubMed] [Google Scholar]
- 60.Zehnder, D., Evans, K. N., Kilby, M. D., Bulmer, J. N., Innes, B. A., Stewart, P. M., and Hewison, M. (2002) Am. J. Pathol. 161 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lechner, D., Kallay, E., and Cross, H. S. (2007) Mol. Cell Endocrinol. 263 55–64 [DOI] [PubMed] [Google Scholar]
- 62.Avila, E., Diaz, L., Barrera, D., Halhali, A., Mendez, I., Gonzalez, L., Zuegel, U., Steinmeyer, A., and Larrea, F. (2007) J. Steroid Biochem. Mol. Biol. 103 90–96 [DOI] [PubMed] [Google Scholar]
- 63.Murayama, A., Takeyama, K., Kitanaka, S., Kodera, Y., Hosoya, T., and Kato, S. (1998) Biochem. Biophys. Res. Commun. 249 11–16 [DOI] [PubMed] [Google Scholar]
- 64.Xie, Z., Munson, S. J., Huang, N., Portale, A. A., Miller, W. L., and Bikle, D. D. (2002) J. Biol. Chem. 277 36987–36990 [DOI] [PubMed] [Google Scholar]
- 65.Paulson, S. K., Ford, K. K., and Langman, C. B. (1990) Am. J. Physiol. 258 E158–E162 [DOI] [PubMed] [Google Scholar]
- 66.Ross, R., Dorsey, J., and Ellis, K. (1990) Pediatr. Res. 27 192 (abstr.) [Google Scholar]
- 67.Ross, R., Halbert, K., and Tsang, R. C. (1989) Pediatr. Res. 26 633–638 [DOI] [PubMed] [Google Scholar]
- 68.Vigano, P., Lattuada, D., Mangioni, S., Ermellino, L., Vignali, M., Caporizzo, E., Panina-Bordignon, P., Besozzi, M., and Di Blasio, A. M. (2006) J. Mol. Endocrinol. 36 415–424 [DOI] [PubMed] [Google Scholar]
- 69.Pendas-Franco, N., Garcia, J. M., Pena, C., Valle, N., Palmer, H. G., Heinaniemi, M., Carlberg, C., Jimenez, B., Bonilla, F., Munoz, A., and Gonzalez-Sancho, J. M. (2008) Oncogene 27 4467–4477 [DOI] [PubMed] [Google Scholar]
- 70.Sung, V., and Feldman, D. (2000) Mol. Cell Endocrinol. 164 133–143 [DOI] [PubMed] [Google Scholar]
- 71.Wietrzyk, J., Filip, B., Milczarek, M., Klopotowska, D., Maciejewska, M., Dabrowska, K., Kurzepa, A., Dzimira, S., Madej, J., and Kutner, A. (2008) Oncol. Rep. 20 941–952 [PubMed] [Google Scholar]
- 72.Pospechova, K., Rozehnal, V., Stejskalova, L., Vrzal, R., Pospisilova, N., Jamborova, G., May, K., Siegmund, W., Dvorak, Z., Nachtigal, P., Semecky, V., and Pavek, P. (2009) Mol. Cell Endocrinol. 299 178–187 [DOI] [PubMed] [Google Scholar]
- 73.Deeb, K. K., Trump, D. L., and Johnson, C. S. (2007) Nat. Rev. Cancer 7 684–700 [DOI] [PubMed] [Google Scholar]
- 74.Davis, C. D., and Uthus, E. O. (2004) Exp. Biol. Med. (Maywood) 229 988–995 [DOI] [PubMed] [Google Scholar]
- 75.Friso, S., and Choi, S. W. (2005) Curr. Drug Metab. 6 37–46 [DOI] [PubMed] [Google Scholar]
- 76.Lopez, E. R., Regulla, K., Pani, M. A., Krause, M., Usadel, K. H., and Badenhoop, K. (2004) J. Steroid Biochem. Mol. Biol. 89- 90 155–157 [DOI] [PubMed] [Google Scholar]
- 77.Lopez, E. R., Zwermann, O., Segni, M., Meyer, G., Reincke, M., Seissler, J., Herwig, J., Usadel, K. H., and Badenhoop, K. (2004) Eur. J. Endocrinol. 151 193–197 [DOI] [PubMed] [Google Scholar]
- 78.Jennings, C. E., Owen, C. J., Wilson, V., and Pearce, S. H. (2005) J. Mol. Endocrinol. 34 859–863 [DOI] [PubMed] [Google Scholar]
- 79.Lauridsen, A. L., Vestergaard, P., Hermann, A. P., Brot, C., Heickendorff, L., Mosekilde, L., and Nexo, E. (2005) Calcif. Tissue Int. 77 15–22 [DOI] [PubMed] [Google Scholar]
- 80.Valdivielso, J. M., and Fernandez, E. (2006) Clin. Chim. Acta 371 1–12 [DOI] [PubMed] [Google Scholar]
- 81.Whitfield, G. K., Remus, L. S., Jurutka, P. W., Zitzer, H., Oza, A. K., Dang, H. T., Haussler, C. A., Galligan, M. A., Thatcher, M. L., Encinas Dominguez, C., and Haussler, M. R. (2001) Mol. Cell Endocrinol. 177 145–159 [DOI] [PubMed] [Google Scholar]
- 82.Arai, H., Miyamoto, K. I., Yoshida, M., Yamamoto, H., Taketani, Y., Morita, K., Kubota, M., Yoshida, S., Ikeda, M., Watabe, F., Kanemasa, Y., and Takeda, E. (2001) J. Bone Miner Res. 16 1256–1264 [DOI] [PubMed] [Google Scholar]
- 83.d'Alesio, A., Garabedian, M., Sabatier, J. P., Guaydier-Souquieres, G., Marcelli, C., Lemacon, A., Walrant-Debray, O., and Jehan, F. (2005) Hum. Mol. Genet. 14 3539–3548 [DOI] [PubMed] [Google Scholar]
- 84.van der Molen, E. F., Arends, G. E., Nelen, W. L., van der Put, N. J., Heil, S. G., Eskes, T. K., and Blom, H. J. (2000) Am. J. Obstet. Gynecol. 182 1258–1263 [DOI] [PubMed] [Google Scholar]
- 85.Nelen, W. L., Blom, H. J., Steegers, E. A., den Heijer, M., and Eskes, T. K. (2000) Fertil. Steril. 74 1196–1199 [DOI] [PubMed] [Google Scholar]
- 86.Nelen, W. L., Blom, H. J., Thomas, C. M., Steegers, E. A., Boers, G. H., and Eskes, T. K. (1998) J. Nutr. 128 1336–1341 [DOI] [PubMed] [Google Scholar]
- 87.Nelen, W. L., Steegers, E. A., Eskes, T. K., and Blom, H. J. (1997) Lancet 350 861. [DOI] [PubMed] [Google Scholar]
- 88.Sikora, J., Magnucki, J., Zietek, J., Kobielska, L., Partyka, R., Kokocinska, D., and Bialas, A. (2007) Neuroendocrinol. Lett. 28 507–512 [PubMed] [Google Scholar]
- 89.Dodds, L., Fell, D. B., Dooley, K. C., Armson, B. A., Allen, A. C., Nassar, B. A., Perkins, S., and Joseph, K. S. (2008) Clin. Chem. 54 326–334 [DOI] [PubMed] [Google Scholar]