Abstract
Remodeling of the actin cytoskeleton is a critical early step in skeletal muscle differentiation. Smooth muscle α-actin (SMA) is one of the earliest markers of myoblast differentiation and is important for the migration and cell shape changes that precede fusion. We have found that satellite cell-derived primary myoblasts from mice lacking the Barx2 homeobox gene show altered patterns of actin remodeling, reduced cell migration, and delayed differentiation. Consistent with the role of SMA in these processes, Barx2–/– myoblasts also show reduced expression of SMA mRNA and protein. The proximal SMA promoter contains binding sites for muscle regulatory factors and serum response factor as well as a conserved homeodomain binding site (HBS). We found that Barx2 binds to the HBS element and potentiates up-regulation of SMA promoter activity by MyoD. We also show that Barx2, MyoD, and serum response factor simultaneously occupy the SMA promoter in cells and that Barx2 interacts with MyoD. Overall these data indicate that Barx2 cooperates with other muscle-expressed transcription factors to regulate the early cytoskeletal remodeling events that underlie efficient myoblast differentiation.
A critical process in skeletal muscle development is the terminal differentiation and fusion of myoblasts and the concomitant activation of muscle-specific genes. These events are regulated by a suite of muscle-expressed transcription factors, including bHLH3 muscle regulatory factors (MRFs), such as MyoD and myogenin, and MADS domain proteins, such as myocyte enhancer factor (MEF) 2 family and serum response factor (SRF) (1–3). Muscle differentiation is also accompanied by up-regulation of muscle-specific actin isoforms and the down-regulation of nonmuscle actins (4). Although less than 10% different in sequence, actin isoforms are typically involved in different processes within the cell, and thus their regulation may differentially affect cell shape, motility, and fate (5).
Actin isoform nomenclature has not been used consistently in the literature, leading to potential confusion about which actins are under investigation. To clarify, mammalian genomes contain six actin genes: skeletal α-actin (ACTA1), cardiac α-actin (ACTC), β-actin (ACTB), γ-actin (ACTG1), enteric γ-actin (ACTG2), and vascular α-actin (ACTA2) (6). The vascular α-actin gene (ACTA2) is also known as smooth muscle alpha actin (SMA). In addition to its expression in smooth muscle cells (7), SMA is transiently expressed in skeletal and cardiac myoblasts during differentiation (8). In maturing skeletal myofibers, SMA is down-regulated and replaced by skeletal α (sarcomeric)-actin (8, 9). The exact role of transient SMA expression in skeletal myoblasts is unknown; however, a recent study has shown that down-regulation of SMA inhibits cardiomyocyte differentiation (10). In other cell types, including myofibroblasts, SMA has been shown to be involved in the cytoskeletal remodeling events that control cell spreading and migration. SMA is frequently enriched in stress fibers, where it plays key roles in cell shape change and force generation (11–14). In cultured skeletal myoblasts, serum withdrawal induces actin polymerization, and the conversion of G-actin to F-actin acts as a signal that increases the activity of SRF and thus its target genes, such as SMA (15–17). It is likely therefore that changes in the expression and distribution of SMA influence myoblast differentiation via mechanical effects on in cell shape and motility as well as by modulation of transcriptional pathways.
Studies of the chicken, mouse, and human SMA genes have shown that different DNA regulatory elements are required for transcriptional regulation in different cell types (18–20). The proximal region of the mouse SMA promoter (20) contains canonical CArG and E-box motifs that bind to SRF and ubiquitous class I bHLH factors, respectively, in smooth muscle cells (21). In skeletal muscle, myogenic bHLH factors, such as MyoD and myogenin, bind to the E-box motifs (22). Moreover, removing both E-box motifs was shown to abolish almost all expression in vivo (21), indicating the essential nature of these elements for both smooth and skeletal muscle expression of the SMA gene.
Hautmann and co-workers (23, 24) identified a conserved homeobox binding site (HBS) within the SMA promoter located between the previously characterized E-box and CArG motifs. In smooth muscle cells, this HBS is bound by the Mhox homeobox factor, which transactivates the promoter in response to angiotensin stimulation. Mhox also promotes binding of SRF to the adjacent CArG box elements, which activates the promoter further (23). Whether particular homeobox factors also regulate the transient expression of SMA in skeletal muscle is currently unknown.
Barx2 is a member of the Antennapedia family of homeobox proteins (25) that is expressed in both smooth and skeletal muscle (26–28). Barx2 has also been shown to interact directly with SRF and can increase the binding of SRF to DNA (26). We showed previously that Barx2 is expressed in skeletal myoblasts and that down-regulation of Barx2 expression in primary limb bud cell cultures delayed myotube formation, whereas its overexpression accelerated the appearance of myotubes (28). However, Barx2 could not induce myogenic differentiation in cells that do not express canonical MRFs, such as MyoD, suggesting that Barx2 may function in cooperation with these factors (28).
In this study, we used myoblasts from the Barx2 null mouse (29) to explore the role of Barx2 in regulation of SMA expression and myoblast differentiation. Our results indicate that Barx2 is a direct regulator of the SMA gene in cooperation with classical muscle regulatory factors and that reduced SMA expression in the absence of Barx2 may inhibit the cell shape change and migration events that are required for efficient differentiation.
EXPERIMENTAL PROCEDURES
Barx2 Null Mice—Mice were obtained from Dr. Geoff Rosenfeld and maintained by heterozygous crosses. The detailed muscle phenotype of these mice will be described elsewhere. Limb bud mesenchymal and satellite cell-derived primary myoblast cultures were prepared from sibling pairs as described below.
Cell Cultures and Staining—Two different types of primary cultures were used in these studies, primary myoblasts and limb bud mesenchymal cells. Primary myoblasts were generated as previously described (30) from postnatal day 4 (P4) Barx2+/+ and Barx2–/– mice. Cells were cultured on collagen-coated plates in Dulbecco's modified Eagle's medium/F10 with 20% fetal bovine serum and 5 ng/ml basic fibroblast growth factor. Differentiation was induced by transferring cells to Dulbecco's modified Eagle's medium supplemented with 2% horse serum. Cells were fixed at various time points after induction of differentiation with 2% paraformaldehyde and stained with monoclonal antibodies to SMA (clone 1A4; Sigma), myogenin (clone F5D; BD Bioscience Pharmingen or Santa Cruz Biotechnology, Inc., Santa Cruz, CA) with rhodamine-conjugated phalloidin or combinations thereof. Single optical sections or Z-series were obtained using a Bio-Rad (Zeiss) Radiance 2100 Rainbow laser-scanning confocal microscope. Images were analyzed with IMARIS imaging software. Limb bud mesenchymal cell (micro-mass) cultures were prepared and maintained as described previously in serum-free CMRL growth medium (27, 31, 32). COS1, C3H10T1/2, and C2C12 cells were obtained from ATCC and used variously in coimmunoprecipitation, reporter gene assays, gel shift, and ChIP experiments as described below. These cell lines were grown in basal medium or Dulbecco's modified Eagle's medium supplemented with 10–20% fetal bovine serum as appropriate.
Cell Migration Assays—Barx2+/– and Barx2–/– primary myoblasts were seeded into collagen-coated Oris cell migration chambers as described by the manufacturer. Well inserts were removed, and the cells were allowed to migrate into the clear field over 24 h. Cells were fixed with paraformaldehyde, photographed, and then counted manually. Data were averaged from at least six fields.
Time Lapse Imaging—Barx2+/– and Barx2–/– primary myoblasts were plated onto 3-cm glass bottom dishes and maintained for 48 h in growth medium. They were then transferred to differentiation medium and imaged using an Olympus IX70 inverted microscope equipped with phase optics and a cooled CCD CH350 Roper camera (DeltaVision System, Applied Precision, LLC Issaquah, Washington). Cells were maintained while filming at 37 °C under a CO2- and humidity-controlled environment chamber unit (Pathology Devices Inc., Westminster, MD). Images were collected using the DeltaVision software, SoftWoRx, and imported into Image Pro Plus (Media Cybernetics, Bethesda, MD) or Image J (National Institutes of Health, Bethesda, MD) for further processing. Cells were observed for 24–48 h, and the time for fusion events to occur between cells was determined using the Image Pro Plus software and Image J software (National Institutes of Health).
Co-immunoprecipitation—Co-immunoprecipitation of Barx2 with MyoD, CREB-binding protein (CBP), and PGC-1 from cells was performed as essentially as described in Ref. 33. Briefly, C3H10T1/2 or COS1 cells were co-transfected with the Myc-tagged Barx2:pcDNA3 expression plasmid (28) and either Myc-tagged MyoD:pcDNA3, hemagglutinin (HA)-tagged-CBP, or HA-tagged PGC-1 in the pCMV5 expression vector using Lipofectamine 2000 (Invitrogen). COS1 cells were used for co-activator experiments because of their high transfection efficiency. The CBP and PGC-1 expression plasmids were gifts of Dr. Michael Downes. Cell lysates were prepared 48 h after transfection, precleared with either Protein A- or G-Sepharose, and then incubated overnight at 4 °C with 5 μg of the appropriate antibodies or rabbit IgG (Jackson ImmunoResearch Laboratories). Complexes were precipitated with Protein A- or G-Sepharose and washed four times with an Nonidet P-40-containing buffer (33). All complexes were resolved by SDS-PAGE (Novex) and immunoblotted with appropriate antibodies as described below.
Co-immunoprecipitation of MyoD with Barx2 was performed using a custom made Barx2 anti-peptide polyclonal antibody or rabbit IgG (Covance), and gels were immunoblotted with mouse monoclonal MyoD antibodies (clone MoAb5.8A; BD PharMingen). Coimmunoprecipitation of Barx2 with HA-tagged CBP and PGC-1 was performed using monoclonal HA antibodies (F-7; Santa Cruz Biotechnology) and immunoblotted with polyclonal Barx2 antibody (M-186; Santa Cruz Biotechnologies). Additional experiments were performed in which Barx2 was immunoprecipitated with chicken (59268, ICN Pharmaceuticals, Inc.) or mouse (clone 9E10; Sigma) anti-c-Myc antibodies and probed with HA antibodies, yielding similar results.
For in vitro co-immunoprecipitation, Barx2 and MyoD proteins were generated using the Promega TNT Quick Coupled transcription/translation kit. Co-immunoprecipitation was performed using 5 μg of custom Barx2 polyclonal antibody or rabbit IgG as described previously (34). Gels were immunoblotted with monoclonal MyoD antibody (clone D7F2; Developmental Studies Hybridoma Bank).
Transfections and Promoter Assays—The wild type (WT) SMA promoter construct was generated by cloning a 264-bp segment of the murine SMA promoter into the pGL3Basic firefly luciferase reporter vector (Promega). In addition, a variant of this construct was generated in which the HBS motif was mutated (ΔHBS-SMA). Limb bud mesenchymal cells or C2C12 cells were plated in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum and glutamine or in 24-well tissue culture plates at an initial density of 2 × 104 cells/well and co-transfected with SMA promoter constructs and either Barx2:pcDNA3, MyoD:pcDNA3, or empty pcDNA3 expression plasmids alone or in combination, as described under “Results” (600 ng of DNA/well total) using Lipofectamine 2000. Either the LacZ reporter CMVβgal (Clontech) or the Renilla luciferase reporter pRLCMV (Promega) was included at one-tenth of the total DNA amount to provide an internal reference for transfection efficiency. All experiments were performed in duplicate, and the data shown were derived from at least three independent experiments. The statistical significance of the differences in promoter activity was assessed using the nonparametric Wilcoxon signed rank test (35). p < 0.01 was considered to reflect a statistically significant difference.
Gel Shift Assay—Oligonucleotide probes were designed that correspond to the HBS and flanking CArG boxes from the murine SMA gene as well as mutant versions of these sequences in which the core motifs were disrupted. Sequences were as follows: wild type SMA probe, 5′-TGCAGTGGAAGAGACCCACGCTCTGGCCACCCAGATTAGAGAGTTTTGTGCTGAGGTCCCTATATGGTTGTGT-3′; ΔCArG probe, 5′-TGCAGTATAAGAGATTCACGCTCTGGCCACCCAGATTAGAGAGTTTTGTGCTGAGGTCATTATATTTTTGTGT-3′; ΔHBS probe, 5′-TGCAGTGGAAGAGACCCACGCTCTGGCCACCCAGACGAGAGAGTTTTGTGCTGAGGTCCCTATATGGTTGTGT-3′ (Fig. 1B). Probes were end-labeled with polynucleotide kinase and [γ-32P]ATP, as described previously (36). Nuclear extracts were prepared as described previously (37) from E12.5 mouse limbs. Equal amounts of nuclear protein were used in each binding reaction, as described previously (36). For antibody blockade experiments, 2 μg of polyclonal rabbit Barx2 antibody (M-186; Santa Cruz Biotechnology), polyclonal rabbit SRF antibody (G-20; Santa Cruz Biotechnologies), or preimmune rabbit serum was added to the binding reaction, as indicated under “Results.”
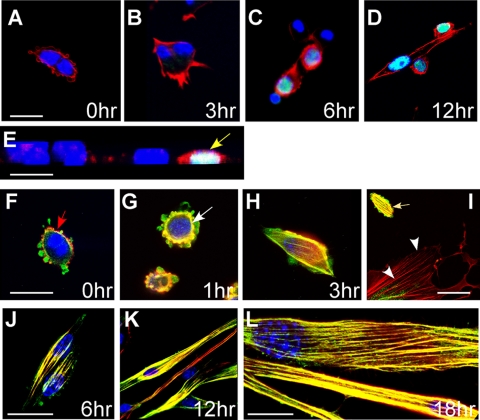
FIGURE 1.
Assembly of SMA into actin filaments is an early event in the differentiation of primary postanatal myoblasts. A–D, cells were activated by serum withdrawal and fixed after 0, 3, 6, or 12 h for staining with rhodamine-phalloidin to visualize F-actin (red), antibodies to myogenin (green), and 4′,6-diamidino-2-phenylindole (blue). Cells begin to show increased F-actin assembly and onset of myogenin expression within 3–6 h of activation (C). Optical section through the nuclei of fusing myoblasts showing strong expression of myogenin (D). E, transverse view of activated myoblasts at 6 h. Cells with strong phalloidin staining indicating F-actin assembly also show myogenin expression (yellow arrow), suggesting that actin remodeling is involved in myoblast differentiation. F–L, cells were co-stained with rhodamine-phalloidin (red), antibodies to SMA (green), and 4′,6-diamidino-2-phenylindole (blue). There is no overlap of SMA and phalloidin staining in proliferating myoblasts (F); however, F-actin is colocalized with SMA and visible as a ring (white arrow) around the nuclei at 1 h after activation by serum withdrawal; from 3 to 18 h after activation, the majority of SMA becomes incorporated into actin filaments of differentiating myotubes (H–L). Interestingly, a fibroblast found within the myoblast culture does not show incorporation of SMA into F-actin stress-fibers (I, white arrowheads), whereas myoblasts do (I, yellow arrow). Scale bars (A–K), 25 μm; L, 10 μm.
ChIP—Primary limb bud mesenchymal cultures (27) and C3H10T1/2 mesenchymal cells were used for ChIP. C3H10T1/2 cells were transfected with Barx2:pcDNA3 and MyoD:pcDNAs plasmids using Lipofectamine 2000 (Invitrogen) and then grown for an additional 48 h. ChIP was performed as previously described (27, 38). Briefly, equivalent amounts of precleared, cross-linked chromatin were precipitated with 5 μg of polyclonal rabbit Barx2 antibody (M-186; Santa Cruz Biotechnology), polyclonal rabbit MyoD antibody (C20; Santa Cruz Biotechnology), polyclonal rabbit SRF antibody (G-20; Santa Cruz Biotechnology), or preimmune rabbit serum and captured with Protein A beads. After precipitation, the cross-links were reversed, and the DNA was recovered. Enrichment of genomic DNA corresponding to the SMA promoter region was determined by PCR amplification of equal aliquots of ChIP-derived DNA using SMA promoter-specific primers (forward, 5′-ccagtgtctgggcatttgag-3′; reverse, 5′-gatcccctcccactcgcttcc-3′). Quantification was performed using gel densitometry. To test whether Barx2, SRF, and MyoD bind to the SMA promoter in vivo, we performed double ChIP as follows. After the first precipitation, the eluted protein-DNA complexes were divided into two aliquots and reprecipitated with either a second antibody or with preimmune serum. The remaining steps were as for single ChIP.
Immunoblotting—Total protein was prepared from Barx2+/– and Barx2–/– primary myoblast cultures using Nonidet P-40 lysis buffer and sonicated. Equal aliquots of protein were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with antibodies to SMA or to β-actin, which acted as a reference. The bands were quantified by densitometry, and the ratio of SMA/β-actin was calculated. Two experiments were performed, and the results were normalized to the values from the Barx2+/– cultures.
Quantitative Reverse Transcription-PCR—RNA was prepared from Barx2+/– and Barx2–/– primary myoblast cultures using TRIzol, and DNA was removed using the Ambion DNA-free kit. Five micrograms of RNA were used for reverse transcription using Moloney murine leukemia virus reverse transcriptase (New England Biolabs) and random primers. Real time reverse transcription-PCR was performed on an ABL 7400 gene detection system using the PowerSYBR green reagent and primers to SMA: forward primer, 5′-CGTGCCTATCTATGAGGGCTATG-3′; reverse primer, 5′-GTGGCCATCTCATTTTCAAAGTC-3′. β-Actin was used a reference for normalization. Three independent experiments were performed in duplicate.
RESULTS
Incorporation of SMA into Filamentous Actin Is Coincident with Differentiation—When maintained in medium with 20% serum and basic fibroblast growth factor, primary skeletal myoblasts retained a high proliferation rate and undifferentiated phenotype for several passages (Fig. 1A). Activation of differentiation by serum withdrawal induced rapid changes in cell shape and remodeling of the actin cytoskeleton as observed using rhodamine-phalloidin staining for F-actin (red) (Fig. 1, B and C). Moreover, formation of prominent actin filaments correlated with the expression of the myoblast differentiation marker myogenin (green) between 3 and 12 h after serum withdrawal (Fig. 1, C–E).
Previous studies have indicated that SMA is rapidly assembled into actin filaments in differentiating smooth muscle cells (11); however, its distribution with respect to the F-actin network during differentiation of skeletal myoblasts has not been described. To examine this distribution, we co-stained differentiating primary myoblasts with rhodamine-phalloidin and SMA antibodies and collected confocal optical sections (Fig. 1, F–L). Before activation by serum withdrawal, SMA expression (green) was found throughout the cell, whereas F-actin (phalloidin staining; red) was found predominantly at the cell periphery with little or no overlap between the two signals, suggesting that most SMA was not yet assembled into a filamentous network (Fig. 1F). Overlap of SMA and phalloidin staining (appearing yellow/orange) increased between 1 and 18 h after activation and as cells elongated and fused. Interestingly, a fibroblast in the culture shown in Fig. 1I does not show SMA staining and F-actin overlap, and its stress fibers are exclusively red. These data suggest that assembly of SMA into actin filaments in skeletal myoblasts is strongly correlated with differentiation, as indicated by elongation, fusion, and myogenin expression.
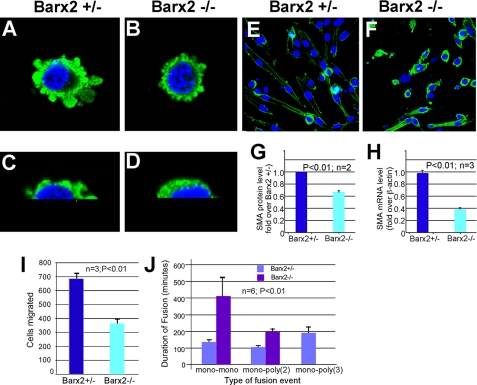
Loss of Barx2 Function Alters SMA Expression and Distribution in Myoblasts—We previously found that overexpression of the Barx2 homeobox gene can increase SMA expression in C2C12 cells (27). To better understand the role of Barx2 in SMA expression and remodeling, primary myoblast cultures were established from P4 Barx2+/– and Barx2–/– mice. Cells were stained with SMA antibodies and examined using serial optical sections that were reconstructed into three-dimensional images. In undifferentiated Barx2+/– myoblasts, SMA was expressed throughout the cytoplasm and at the cell periphery in processes that were particularly abundant on the upper surface of the cells (Fig. 2, A and C). In contrast, Barx2–/– myoblasts had fewer and smaller processes (Fig. 2, compare A and C with B and D). Consistent with the appearance of reduced SMA expression in Barx2–/– myoblasts, immunoblotting and quantitative reverse transcription-PCR revealed 30% less SMA protein and 50% less SMA mRNA in undifferentiated Barx2–/– myoblasts compared with Barx2+/– cells (Fig. 2, G and H). After 12–24 h of differentiation, Barx2+/– cells elongated and fused, and SMA expression was reduced in longer myotubes and redistributed to the periphery of the cells (Fig. 2E). In contrast, elongation and fusion of Barx2–/– cells was delayed, and SMA appeared to persist throughout the cytoplasm (Fig. 2F). At 48–72 h, Barx2–/– myoblasts did form myotubes (not shown), indicating that loss of Barx2 leads to a delay, rather than a block, in differentiation.
FIGURE 2.
Barx2 null myoblasts show altered SMA expression and distribution and delayed migration and fusion after serum withdrawal. A–D, undifferentiated primary myoblast cultures from Barx2+/– (A and C) and Barx2–/– (B and D) mice were stained with antibodies to SMA (green) and 4′,6-diamidino-2-phenylindole (blue) and examined using serial optical sections that were reconstructed into three-dimensional images. Higher magnification images are shown in C and D. G and H, undifferentiated Barx2–/– myoblasts show lower SMA expression than Barx2+/– myoblasts. SMA expression was examined by immunoblotting (G) and by quantitative reverse transcription-PCR (H). Results are the average of 2–3 experiments. E and F, Barx2–/– myoblasts differentiate more slowly than Barx2+/– myoblasts. Cells were induced to differentiate by serum withdrawal. By 24 h, Barx2+/– cultures have differentiated into myotubes and show reduced SMA expression (E) whereas differentiation of Barx2–/– cultures is delayed and strong SMA expression persists (F). I and J, Barx2–/– myoblasts show slower migration and fusion relative to Barx2+/– myoblasts. I, cells were seeded in Oris migration chambers and counted after 24 h (n = 3). J, time lapse analysis was performed as described under “Experimental Procedures.” The time until fusion occurred between two mononucleated cells (mono-mono) or between a mononucleate cell and a myotube containing either 2 or 3 nuclei (mono-poly(2) or mono-poly(3)) was recorded. Average fusion times were derived from six different experiments per condition and observing 20–30 cells/field/experiment.
Barx2 Null Cells Show Delayed Migration and Fusion—Alterations in SMA expression and distribution in Barx2–/– myoblasts could delay the cell spreading and migration events that are required for myoblast fusion. To examine whether Barx2–/– myoblasts have impaired migration, we measured migration using collagen-coated cell migration chambers (Oris) as described under “Experimental Procedures.” Nearly 2-fold fewer Barx2–/– myoblasts migrated into the clear detection zone of the chamber than Barx2+/– myoblasts within 48 h of seeding (Fig. 2I), indicating that Barx2–/– myoblasts are indeed slower to migrate.
To investigate myoblast migration and fusion in more detail, we used time lapse microscopy to measure the time for fusion events to occur after removing serum. We observed and measured fusion of mononucleated cells to each other (mono-mono fusion), fusion of mononucleated cells to nascent myotubes containing two nuclei (mono-poly(2) fusion), and fusion of mononucleated cells to myotubes with three or more nuclei (mono-poly(3) fusion). Mononucleated Barx2–/– cells fused to each other nearly 3 times more slowly than Barx2+/– cells (Fig. 2J). Consistent with this delay in fusion, we observed no fusion events involving cells with three or more nuclei in Barx2–/– cultures within the observation period (Fig. 2J). Overall, these results suggest that regulation of SMA expression and redistribution by Barx2 is required for efficient cell migration and cell fusion during myoblast differentiation.
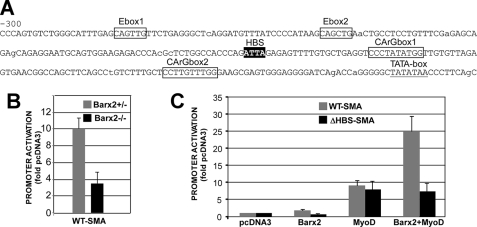
SMA Promoter Activity Is Reduced in Barx2 Null Cells—The observation that SMA mRNA levels are reduced in Barx2 null myoblasts prompted us to investigate whether Barx2 directly regulates the SMA gene. This possibility was suggested by analysis of the SMA promoter sequence. The proximal SMA promoter contains recognition motifs for MRFs and SRF (39) as well as a potential HBS. The HBS is conserved in the mouse, rat, and human genes and is flanked by two conserved SRF binding sites (CArG-boxes) (Fig. 3A). We generated a luciferase reporter construct that contains a 264-bp segment of the SMA promoter upstream of the predicted transcription start site. This construct was transfected into mesenchymal progenitor cell cultures prepared from Barx2+/– and Barx2–/– embryonic limbs (E11.5) (27, 40). The promoter construct was ∼3-fold less active in Barx2–/– cells than in Barx2+/– cells (Fig. 3B), strongly suggesting that Barx2 regulates the SMA promoter.
FIGURE 3.
Barx2 and the HBS potentiate activation of the SMA promoter by MyoD. A, sequence of the 300-bp proximal SMA promoter region used in SMA promoter-luciferase reporter constructs. B, luciferase assay of the SMA promoter construct transfected into primary limb bud cells from Barx2+/– and Barx2–/– mice shows that Barx2 is required for full promoter activity. C, co-transfection of the SMA promoter construct with Barx2:pcDNA3, MyoD:pcDNA3, or both MyoD:pcDNA3 and Barx2:pcDNA3 plasmids in C2C12 cells shows that Barx2 cooperates with MyoD to increase SMA promoter activity. Co-transfection of the ΔHBS-SMA promoter construct with the same effector plasmids shows that mutation of the HBS element prevents cooperative activation by Barx2 and MyoD. Promoter activity is in arbitrary units, and experiments were performed at least four times. All values are significant at p < 0.05.
To determine whether Barx2 works in cooperation with other myogenic regulatory factors in activation of the SMA promoter, we performed co-transfection experiments in which C2C12 cells were co-transfected with the SMA reporter construct and with either a Barx2 or MyoD expression plasmid or with both MyoD and Barx2 plasmids. Barx2 alone had little effect on the promoter, increasing its activity less than 2-fold. MyoD activated the promoter ∼9-fold, and the combination of both factors increased activity nearly 25-fold (Fig. 3C). This suggests that Barx2 cooperates with MyoD in regulation of the SMA expression.
To verify the role of the HBS element in regulating SMA activity, we generated a mutated version of the reporter, which contained a GC for TT nucleotide substitution within the ATTA core motif (ΔHBS). The activity of the ΔHBS-SMA reporter plasmid was compared with the WT SMA promoter in cotransfection experiments in C2C12 cells The ΔHBS-SMA promoter had ∼3-fold lower activity than the WT SMA promoter (not shown), and although MyoD activated the ΔHBS-SMA promoter construct ∼8-fold, there was no further increase when MyoD and Barx2 were co-expressed (Fig. 3C). Thus the HBS is required for the synergistic response to MyoD and Barx2.
Barx2 Binds to the HBS Element in the SMA Gene Promoter—To determine whether Barx2 binds directly to the SMA promoter HBS, we prepared a 73-bp probe spanning the HBS and downstream SRF binding site and a mutant version of this probe in which the HBS was mutated as shown in Fig. 4A. Binding of embryonic limb nuclear extracts to these elements was tested using gel mobility shift assays, and the specificity of the interactions was examined by antibody blockade (Fig. 4B).
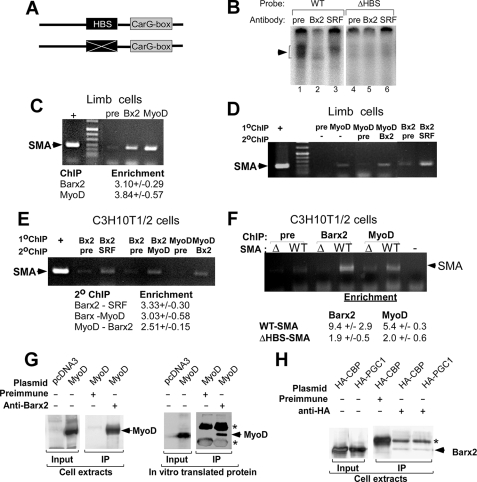
FIGURE 4.
Barx2 binds to the SMA promoter in cooperation with MyoD and SRF. A, schematic of 73-bp wild type and mutated probes containing the SMA CArG box and HBS motifs. B, gel mobility shift analysis of complexes formed between wild type and mutated 32P-labeled probes and nuclear extracts from embryonic limbs. Antibody blockade was performed by the addition of 2 μg of polyclonal antibodies to Barx2 or SRF or rabbit preimmune serum (pre). C, single ChIP in limb bud cell cultures. Sheared cross-linked chromatin was immunoprecipitated with antibodies to Barx2 (Bx2) or MyoD or with preimmune serum. Equal aliquots of each precipitated DNA sample were used in PCR assays with primers corresponding to the SMA promoter region. +, positive control (input chromatin). D, double ChIP analysis in limb bud cell cultures. Complexes that were immunoprecipitated as in D were eluted and reprecipitated with antibodies as described in each lane. E, double ChIP analysis in C3H10T1/2 cells transfected with Barx2 and MyoD expression vectors. As a control for nonspecific enrichment during double ChIP, we performed PCR amplification of each sample using primers to a nontarget sequence (tRNA gene), which consistently gave no detectable product (not shown). F, ChIP analysis of wild type and ΔHBS-SMA promoter plasmids in C3H10T1/2 cells using Barx2 and MyoD antibodies. To ensure that we measured enrichment of the plasmid rather than the endogenous gene, PCR assays used a forward primer that binds in the SMA promoter and a reverse primer (GL2) that binds in the pGL3basic vector sequence downstream of the cloning site. In C, E, and F, amplification of SMA promoter DNA was quantified by densitometry of PCR products, and the enrichment using each specific antibody was calculated by normalizing to the background level of amplification in samples that were mock precipitated. n = 3. G, left, immunoblot analysis of co-immunoprecipitation (IP) from COS1 cells transfected with plasmids expressing Barx2 and MyoD. Right, immunoblot analysis of co-immunoprecipitation using in vitro transcribed/translated Barx2 and MyoD proteins. In both panels, complexes were immunoprecipitated with either Barx2 antibodies or preimmune serum and immunoblotted with MyoD antibodies. Expressed MyoD is slightly larger than native MyoD due to the presence of an epitope tag and additional amino acids encoded in the vector sequence. The asterisk indicates cross-reacting IgG. H, immunoblot analysis of co-immunoprecipitation from COS1 cells transfected with plasmids expressing Barx2 and either HA-tagged human CBP or PGC-1. Complexes were immunoprecipitated using either HA antibodies or preimmune serum and immunoblotted with Barx2 antibody. The asterisk indicates cross-reacting IgG.
The wild type probe formed an intense complex with the nuclear extract that migrated as a broad band (Fig. 4B, lane 1, arrow). The addition of antibodies to Barx2 or SRF reduced formation of the complex (Fig. 4B, lanes 2 and 3), whereas a preimmune serum did not (lane 1). Moreover, we found that Barx2 antibodies inhibited mainly the upper part of the complex, whereas SRF antibodies mainly inhibited the lower part of the band, indicating that the complex is heterogeneous. Mutation of the HBS motif (ΔHBS) essentially abolished binding to the probe (Fig. 4B, lanes 4–6), suggesting that the HBS element may be important to recruit or stabilize a complex containing both Barx2 and SRF.
Barx2, SRF, and MyoD Simultaneously Occupy the SMA Promoter—To investigate whether Barx2 binds to the endogenous SMA promoter, we performed ChIP using limb bud cell cultures (Fig. 4C). Chromatin was immunoprecipitated using antibodies to Barx2 or MyoD or mock precipitated using a preimmune rabbit IgG (27, 38). The recovered DNA was subjected to PCR amplification using primers that span the region of the SMA promoter shown in Fig. 1A. ChIP using either the Barx2 or MyoD antibody significantly enriched the SMA sequence, whereas the preimmune serum did not. Thus, both MyoD and Barx2 can bind to the endogenous SMA promoter (Fig. 4C).
To test whether Barx2, MyoD, and SRF simultaneously occupy the SMA promoter, we performed sequential or double ChIP either in limb bud cell cultures (Fig. 4D) or in C3H10T1/2 cells that were transiently transfected with Barx2 and MyoD expression plasmids (Fig. 4E). Chromatin was immunoprecipitated with the first antibody, eluted, and then reprecipitated with the second antibody or with preimmune rabbit serum as indicated in each lane of the gel. Double ChIP using combinations of MyoD and Barx2 antibodies or Barx2 and SRF antibodies enriched the SMA promoter in both cellular contexts (Fig. 4, D and E), suggesting that Barx2 binds to the promoter together with MyoD and SRF. The enrichment of the SMA promoter by double ChIP (calculated by densitometry and presented as a ratio between the PCR amplification from the antibody sample and the preimmune serum sample) was between 2.5- and 3.3-fold (Fig. 4E). Primers to a control nontarget genomic sequence (tRNA gene) did not generate a product from any sample, indicating that the precipitation was specific (not shown). Overall, the results suggest that Barx2, MyoD, and SRF simultaneously occupy the SMA promoter in both limb bud and C3H10T1/2 mesenchymal cells. These data together with the gel shift data shown in Fig. 4B suggest that Barx2 binds to the HBS element within the SMA promoter and is associated with a complex of factors that can regulate SMA expression.
The HBS Controls Binding of Barx2 and MyoD to the SMA Promoter—To test the role of the HBS element in recruiting MyoD to the SMA promoter, C2C12 cells were transfected with either the WT or mutated (ΔHBS) SMA promoter reporter constructs, and ChIP was performed using Barx2 or MyoD antibodies. Control samples were mock-precipitated with preimmune serum. Precipitated chromatin was analyzed by PCR using primers that are anchored in the reporter plasmid as described in the legend to Fig. 4. Mutation of the HBS reduced binding of Barx2 to essentially background levels, as might be expected; moreover, it also significantly reduced MyoD binding (Fig. 4F). Thus, the HBS not only binds to Barx2 but may also help to recruit MyoD to the SMA promoter.
Barx2 Interacts with MyoD—The ChIP studies show that Barx2, SRF, and MyoD bind simultaneous to the SMA promoter (Fig. 4, D and E). It has been previously shown that Barx2 and SRF interact directly (26). To test whether Barx2 and MyoD can also interact, we performed co-immunoprecipitation from C3H10T1/2 cells transfected with Barx2 and MyoD expression plasmids. Barx2 antibodies co-immunoprecipitated MyoD, whereas preimmune rabbit serum did not (Fig. 4G), suggesting that Barx2 can form a complex with MyoD. To assess whether this interaction is direct, we generated proteins by in vitro transcription/translation and performed co-immunoprecipitation with Barx2 antibodies. Similarly to the results obtained in cell lysates, Barx2 antibodies co-immunoprecipitated MyoD, whereas preimmune rabbit serum did not (Fig. 4G). Thus, the interaction of Barx2 and MyoD appears to be direct and not mediated solely by co-interacting proteins, such as SRF.
Barx2 contains a C-terminal activation domain (36) that may recruit a variety of co-activators. To determine whether Barx2 can interact with known coactivators of myogenesis (41–45), we performed co-immunoprecipitation from COS1 cells that expressed Myc-tagged Barx2 and either HA-tagged CBP or proliferator-activated receptor γ-coactivator 1 (PGC-1) proteins. In both cases, antibodies to the HA tag co-immunoprecipitated Barx2 (Fig. 4H), suggesting that it interacts with both coactivators; however, the low intensity of the band may indicate that only a portion of expressed Barx2 interacts with these proteins. A similar result was obtained by immunoprecipitating with anti-Myc antibodies and immunoblotting with anti-HA antibodies (not shown). Overall, these data suggest that Barx2 could interact directly with MyoD and coactivators to promote SMA gene activation.
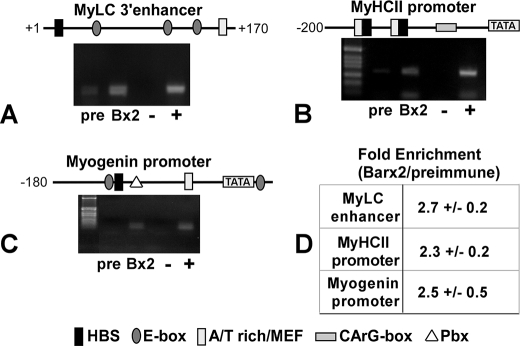
Many Muscle-specific Genes Contain HBS Binding Sites That Are Occupied by Barx2—MRFs cooperate with SRF and MEF proteins to regulate many muscle-specific genes, and clusters of their cognate binding motifs (E-box, CArG-box, and the MEF-binding AT-rich box) can be identified within such gene promoters. For several of these genes, potential HBSs have also been observed within these motif clusters (see Fig. 5). We used the ChIP assay to test whether Barx2 might also bind to these muscle-specific promoters. C3H10T1/2 cells were co-transfected with MyoD and Barx2 expression plasmids and allowed to differentiate. ChIP was then performed using Barx2 antibodies as shown in Fig. 4. Barx2 antibodies enriched the HBS-containing promoter regions of three of the genes tested: myogenin, myosin heavy chain II (MyHCII), and myosin light chain I (MyLCI) (Fig. 5). Thus, the binding of Barx2 and possibly other homeodomain transcription factors to HBS elements that are proximal to binding sites for other muscle-expressed transcription factors may occur in many muscle-specific genes. This could represent an important general mechanism for coordinating the activities of the various homeobox, bHLH, and MADS family proteins that control muscle development.
FIGURE 5.
Many muscle-specific gene promoters contain HBS motifs and can bind to Barx2. The promoter regions of several muscle-specific genes were examined for consensus recognition sites for homeodomain proteins (ATTA). Information about binding sites for MRFs, MEFs, and SRF was obtained from published sources or from the Catalogue of Muscle Specific Regulatory Elements (available on the World Wide Web). A–C, ChIP analysis of Barx2 binding to the MyLC1 (A), fast MyHCII (B), and myogenin (C) promoters in C3H10T1/2 cells that were induced to differentiate after transfection with Barx2 and MyoD expression plasmids. Chromatin was immunoprecipitated with either Barx2 antibodies or preimmune serum (pre) and analyzed by PCR using promoter-specific primers. +, input chromatin control. D, the average amplification of each promoter DNA sequence was measured as described in the legend to Fig. 4 (n = 2).
DISCUSSION
SMA is one of the earliest genes to be induced when skeletal myoblasts begin to differentiate; however, its role in differentiation is not well understood. Our data suggest that in the early steps of myoblast differentiation, SMA is rapidly assembled into the F-actin network at the cell periphery and into stress fibers. This remodeled cytoskeleton may promote cell spreading and migration, which are important for cells to find, contact, and eventually fuse with their neighbors. Consistent with this notion, Barx2–/– myoblasts, which have reduced expression of SMA, migrate more slowly and show delayed fusion after serum withdrawal. Interestingly, we have previously shown a role for Barx2 in migration of mammary epithelial cells (46), indicating that this may be a conserved function for Barx2 in different contexts.
It is not clear whether delayed differentiation of Barx2–/– myoblasts is due to reduced expression of SMA alone, particularly since SMA null mice (47) were not reported to have an overt skeletal muscle defect, although, given that down-regulation of SMA can inhibit cardiomyocyte differentiation (10), this remains a possibility. It is likely, however, that differentiation of Barx2–/– myoblasts is delayed due to the misregulation of multiple muscle-expressed Barx2 targets. For example, our earlier work has shown that Barx2 regulates the expression of the actin binding protein filamin (38), which mediates actin cross-linking and is associated with cortical F-actin and stress fibers (48). Moreover, in addition to cytoskeletal targets, such as actin and actin remodeling factors, Barx2 may regulate other muscle structural and regulatory genes either directly or indirectly, as discussed below.
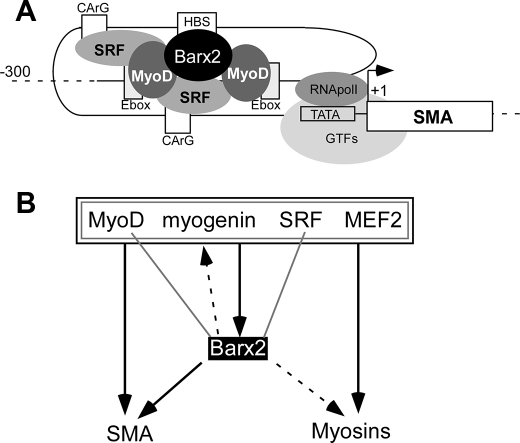
The SMA proximal promoter contains an HBS that is centrally located within a cluster of binding sites for MRFs and SRF (23), and we found that mutation of the HBS prevented synergistic activation by Barx2 and MyoD. Our ChIP data indicate that Barx2, MyoD, and SRF simultaneously occupy the SMA promoter. In this context, mutation of the HBS strongly inhibited binding of Barx2 to the promoter and also modestly reduced binding of MyoD. One interpretation of this result is that binding of Barx2 to the HBS helps to recruit or stabilize a complex that includes MyoD. We also found that Barx2 coimmunoprecipitates with MyoD from cells and in vitro. Given that Barx2 can also bind directly to SRF (26), and MyoD binds to SRF (49), we propose the existence of a Barx2-MyoD-SRF complex at the SMA promoter that is important for full activation (see Fig. 6A).
FIGURE 6.
A, the SMA gene acts as an exemplar for how Barx2 may interact with other muscle regulatory factors at target genes. The proximal SMA promoter region is modeled as a looped structure that brings the binding sites for Barx2, MyoD, and SRF into close proximity, consistent with the ability of these factors to physically interact. B, summary of interactions between Barx2 and muscle regulatory factors. Gray lines denote physical interactions, and black lines/arrows denote regulatory interactions; interactions demonstrated here or in previous studies are indicated by solid lines, whereas hypothesized interactions are indicated by dashed lines. Thus, MyoD, myogenin, SRF, and MEF2 factors have physical and/or regulatory interactions with one another, as indicated by the nested gray and black boxes. These factors also have binding sites in the Barx2 promoter and MyoD and myogenin were previously shown to induce Barx2 promoter activity. In this study, we showed that Barx2, MyoD, and SRF bind simultaneously to the SMA promoter and hypothesize that similar complexes could regulate other target genes, such as muscle myosins and myogenin.
There are similarities between our findings and those of previous studies on the Pbx homeodomain protein (50). The Pbx-Meis heterodimer was shown to form a constitutive complex adjacent to the E-box in the myogenin promoter that could act as a nucleus for assembly of an active transcription complex that includes MyoD (50). A mechanism was proposed in which Pbx marks the promoter with specific chromatin modifications for later activation by MyoD (50). Barx2 may have a function analogous to that of Pbx, although determination of whether they share similar mechanisms will require further studies of chromatin states at the SMA promoter in the presence and absence of Barx2.
Interestingly, Berkes et al. (50) reported putative Pbx recognition motifs adjacent to E-boxes in several muscle-specific genes, suggesting a conserved mechanism for marking genes for later activation by MRFs. Our bioinformatic searches also identified ATTA-containing HBS elements flanked by binding sites for bHLH factors, MEF2, and/or SRF in various muscle-specific genes, and we found that Barx2 could bind to at least three of these genes: two muscle myosins and myogenin (see Fig. 5). However, the Pbx recognition motif (TTGAT) is different from the antennapedia family homeodomain recognition motif (ATTA), and thus the HBS-containing motif clusters that we have studied appear to be different from those involving Pbx binding sites. However, in the myogenin gene, the Pbx recognition motif (50) is only 14 bp downstream from the E-box and ATTA motifs (see Fig. 5), raising the possibility that both Barx2 and Pbx could influence MyoD recruitment to this particular promoter. Mechanistically, it is unclear how Barx2, Pbx, and MyoD might interact. Pbx is known to form complexes with antennapedia family Hox proteins (51); however, it does so via the same domain that binds to bHLH proteins (52), which would seem to preclude the interaction of Pbx-Hox heterodimers with MyoD. Thus, a Pbx-Barx2-MyoD complex would be unlikely unless Barx2 interacts with a different domain of Pbx than the Hox proteins. It is also possible that Pbx-MyoD and Barx2-MyoD complexes are exclusive and have different functions.
In our earlier work, we showed that myogenin can regulate the Barx2 gene promoter (27), suggesting the possibility of a regulatory loop between these genes. We have summarized the known and predicted interactions of Barx2 with other regulators of myogenesis and their shared target genes in Fig. 6B. MyoD, myogenin, SRF, and MEF2 are known to participate in a complex network of regulatory interactions, and these factors have been variously shown to have binding motifs in the Barx2 promoter and/or regulate the promoter (27). Barx2 can interact with MyoD and SRF, and formation of a regulatory complex involving these three proteins was shown here for the SMA promoter and hypothesized for other muscle-specific genes, including muscle myosins and myogenin. In addition, such complexes may recruit co-activators, such as CBP/p300 and PGC-1, which are known to cooperate with MRFs and MADS domain factors (44, 45).
Recently the aristaless-related homeobox protein Arx was reported to have cooperative activities with classical muscle regulatory factors during myoblast differentiation similar to those described here for Barx2 (53). Specifically, Arx interacts with MEF2C and indirectly with MyoD. It also binds and regulates the myogenin promoter and is itself regulated by myogenin. Thus, interactions among homeobox, bHLH, and MADS domain proteins may be a general phenomenon mediating the cooperative and reciprocal regulation of a diverse set of genes that are required for skeletal muscle differentiation and function.
Acknowledgments
We are grateful to Maren Spillane for excellent technical assistance, and we appreciate critical readings of the manuscript by Drs. Kathryn Crossin, Bruce Cunningham, and Vince Mauro.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R03AR051131 (to R. M.) and 5R01AR053163-02 (to H. P. M.). This work was also supported by a grant from the Association Française contre les Myopathies (to H. P. M.).
Footnotes
The abbreviations used are: bHLH, basic helix-loop-helix; MRF, muscle regulatory factor; SMA, smooth muscle actin; HBS, homeobox binding site; SRF, serum response factor; ChIP, chromatin immunoprecipitation; HA, hemagglutinin; CBP, CREB-binding protein; CREB, cAMP-response element-binding protein; WT, wild type; MEF, myocyte enhancer factor.
References
- 1.Edmondson, D. G., and Olson, E. N. (1993) J. Biol. Chem. 268 755–758 [PubMed] [Google Scholar]
- 2.Duprey, P., and Lesens, C. (1994) Int. J. Dev. Biol. 38 591–604 [PubMed] [Google Scholar]
- 3.Molkentin, J. D., and Olson, E. N. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9366–9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, R. D., Garner, I., and Buckingham, M. E. (1990) Differentiation 43 183–191 [DOI] [PubMed] [Google Scholar]
- 5.Schevzov, G., Lloyd, C., and Gunning, P. (1992) J. Cell Biol. 117 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandekerckhove, J., and Weber, K. (1978) J. Mol. Biol. 126 783–802 [DOI] [PubMed] [Google Scholar]
- 7.Shimizu, R. T., Blank, R. S., Jervis, R., Lawrenz-Smith, S. C., and Owens, G. K. (1995) J. Biol. Chem. 270 7631–7643 [DOI] [PubMed] [Google Scholar]
- 8.Woodcock-Mitchell, J., Mitchell, J. J., Low, R. B., Kieny, M., Sengel, P., Rubbia, L., Skalli, O., Jackson, B., and Gabbiani, G. (1988) Differentiation 39 161–166 [DOI] [PubMed] [Google Scholar]
- 9.Springer, M. L., Ozawa, C. R., and Blau, H. M. (2002) Cell Motil. Cytoskeleton 51 177–186 [DOI] [PubMed] [Google Scholar]
- 10.Clement, S., Stouffs, M., Bettiol, E., Kampf, S., Krause, K.-H., Chaponnier, C., and Jaconi, M. (2007) J. Cell Sci. 120 229–238 [DOI] [PubMed] [Google Scholar]
- 11.Qu, G., Yan, H., and Strauch, A. R. (1997) J. Cell. Biochem. 67 514–527 [PubMed] [Google Scholar]
- 12.Hinz, B., Gabbiani, G., and Chaponnier, C. (2002) J. Cell Biol. 157 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinz, B., Dugina, V., Ballestrem, C., Wehrle-Haller, B., and Chaponnier, C. (2003) Mol. Biol. Cell 14 2508–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinz, B., and Gabbiani, G. (2003) Thromb. Haemost. 90 993–1002 [DOI] [PubMed] [Google Scholar]
- 15.Copeland, J. W., and Treisman, R. (2002) Mol. Biol. Cell 13 4088–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posern, G., Sotiropoulos, A., and Treisman, R. (2002) Mol. Biol. Cell 13 4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara, K., Barrientos, T., Pipes, G. C. T., Li, S., and Olson, E. N. (2005) Mol. Cell. Biol. 25 3173–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank, R. S., McQuinn, T. C., Yin, K. C., Thompson, M. M., Takeyasu, K., Schwartz, R. J., and Owens, G. K. (1992) J. Biol. Chem. 267 984–989 [PubMed] [Google Scholar]
- 19.Foster, D. N., Min, B., Foster, L. K., Stoflet, E. S., Sun, S., Getz, M. J., and Strauch, A. R. (1992) J. Biol. Chem. 267 11995–12003 [PubMed] [Google Scholar]
- 20.Min, B. H., Foster, D. N., and Strauch, A. R. (1990) J. Biol. Chem. 265 16667–16675 [PubMed] [Google Scholar]
- 21.Kumar, A., Velloso, C. P., Imokawa, Y., and Brockes, J. P. (2004) PLoS Biol. 2 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, A. D., and Owens, G. K. (1999) Am. J. Physiol. 276 C1420–C1431 [DOI] [PubMed] [Google Scholar]
- 23.Hautmann, M. B., Thompson, M. M., Swartz, E. A., Olson, E. N., and Owens, G. K. (1997) Circ. Res. 81 600–610 [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, T., Hoofnagle, M. H., and Owens, G. K. (2004) Circ. Res. 94 1075–1082 [DOI] [PubMed] [Google Scholar]
- 25.Jones, F. S., Kioussi, C., Copertino, D. W., Kallunki, P., Holst, B. D., and Edelman, G. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2632–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herring, B. P., Kriegel, A. M., and Hoggatt, A. M. (2001) J. Biol. Chem. 276 14482–14489 [DOI] [PubMed] [Google Scholar]
- 27.Meech, R., Edelman, D. B., Jones, F. S., and Makarenkova, H. P. (2005) Development 132 2135–2146 [DOI] [PubMed] [Google Scholar]
- 28.Meech, R., Makarenkova, H., Edelman, D. B., and Jones, F. S. (2003) J. Biol. Chem. 278 8269–8278 [DOI] [PubMed] [Google Scholar]
- 29.Olson, L. E., Zhang, J., Taylor, H., Rose, D. W., and Rosenfeld, M. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rando, T. A., and Blau, H. M. (1997) Methods Cell Biol. 52 261–272 [DOI] [PubMed] [Google Scholar]
- 31.Makarenkova, H., Becker, D. L., Tickle, C., and Warner, A. E. (1997) J. Cell Biol. 138 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel, A., and Tickle, C. (1993) Development 119 199–206 [DOI] [PubMed] [Google Scholar]
- 33.Zorn, A. M., Barish, G. D., Williams, B. O., Lavender, P., Klymkowsky, M. W., and Varmus, H. E. (1999) Mol. Cell 4 487–498 [DOI] [PubMed] [Google Scholar]
- 34.Busson, M., Daury, L., Seyer, P., Grandemange, S., Pessemesse, L., Casas, F., Wrutniak-Cabello, C., and Cabello, G. (2006) Endocrinology 147 3408–3418 [DOI] [PubMed] [Google Scholar]
- 35.Ostle, B., and Mensing, R. W. (1975) Statistics in Research, p. 596, Iowa State University Press
- 36.Edelman, D. B., Meech, R., and Jones, F. S. (2000) J. Biol. Chem. 275 21737–21745 [DOI] [PubMed] [Google Scholar]
- 37.Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K., Jaenisch, R., Wagschal, A., Feil, R., Schreiber, S. L., and Lander, E. S. (2006) Cell 125 315–326 [DOI] [PubMed] [Google Scholar]
- 38.Stevens, T. A., Iacovoni, J. S., Edelman, D. B., and Meech, R. (2004) J. Biol. Chem. 279 14520–14530 [DOI] [PubMed] [Google Scholar]
- 39.Sartorelli, V., Webster, K. A., and Kedes, L. (1990) Genes Dev. 4 1811–1822 [DOI] [PubMed] [Google Scholar]
- 40.Amthor, H., Nicholas, G., McKinnell, I., Kemp, C. F., Sharma, M., Kambadur, R., and Patel, K. (2004) Dev. Biol. 270 19–30 [DOI] [PubMed] [Google Scholar]
- 41.Phiel, C. J., Gabbeta, V., Parsons, L. M., Rothblat, D., Harvey, R. P., and McHugh, K. M. (2001) J. Biol. Chem. 276 34637–34650 [DOI] [PubMed] [Google Scholar]
- 42.Lin, J., Wu, H., Tarr, P. T., Zhang, C.-Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., Lowell, B. B., Bassel-Duby, R., and Spiegelman, B. M. (2002) Nature 418 797–801 [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Calvo, R., Jove, M., Coll, T., Camins, A., Sanchez, R. M., Alegret, M., Merlos, M., Pallas, M., Laguna, J. C., and Vazquez-Carrera, M. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61 773–780 [DOI] [PubMed] [Google Scholar]
- 44.Sartorelli, V., Huang, J., Hamamori, Y., and Kedes, L. (1997) Mol. Cell. Biol. 17 1010–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez, S., Ali, S. A. S., Robin, P., Trouche, D., and Harel-Bellan, A. (1997) J. Biol. Chem. 272 31016–31021 [DOI] [PubMed] [Google Scholar]
- 46.Stevens, T. A., and Meech, R. (2006) Oncogene 25 5426–5435 [DOI] [PubMed] [Google Scholar]
- 47.Schildmeyer, L. A., Braun, R., Taffet, G., Debiasi, M., Burns, A. E., Bradley, A., and Schwartz, R. J. (2000) FASEB J. 14 2213–2220 [DOI] [PubMed] [Google Scholar]
- 48.Stossel, T. P., Condeelis, J., Cooley, L., Hartwig, J. H., Noegel, A., Schleicher, M., and Shapiro, S. S. (2001) Nat. Rev. Mol. Cell Biol. 2 138–145 [DOI] [PubMed] [Google Scholar]
- 49.Groisman, R., Masutani, H., Leibovitch, M.-P., Robin, P., Soudant, I., Trouche, D., and Harel-Bellan, A. (1996) J. Biol. Chem. 271 5258–5264 [DOI] [PubMed] [Google Scholar]
- 50.Berkes, C. A., Bergstrom, D. A., Penn, B. H., Seaver, K. J., Knoepfler, P. S., and Tapscott, S. J. (2004) Mol. Cell 14 465–477 [DOI] [PubMed] [Google Scholar]
- 51.Mann, R. S., and Affolter, M. (1998) Curr. Opin. Genet. Dev. 8 423–429 [DOI] [PubMed] [Google Scholar]
- 52.Knoepfler, P. S., Bergstrom, D. A., Uetsuki, T., Dac-Korytko, I., Sun, Y. H., Wright, W. E., Tapscott, S. J., and Kamps, M. P. (1999) Nucleic Acids Res. 27 3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biressi, S., Messina, G., Collombat, P., Tagliafico, E., Monteverde, S., Benedetti, L., Cusella De Angelis, M. G., Mansouri, A., Ferrari, S., Tajbakhsh, S., Broccoli, V., and Cossu, G. (2007) Cell Death Differ. 15 94–104 [DOI] [PubMed] [Google Scholar]