Abstract
Caprazamycins are potent anti-mycobacterial liponucleoside antibiotics isolated from Streptomyces sp. MK730-62F2 and belong to the translocase I inhibitor family. Their complex structure is derived from 5′-(β-O-aminoribosyl)-glycyluridine and comprises a unique N-methyldiazepanone ring. The biosynthetic gene cluster has been identified, cloned, and sequenced, representing the first gene cluster of a translocase I inhibitor. Sequence analysis revealed the presence of 23 open reading frames putatively involved in export, resistance, regulation, and biosynthesis of the caprazamycins. Heterologous expression of the gene cluster in Streptomyces coelicolor M512 led to the production of non-glycosylated bioactive caprazamycin derivatives. A set of gene deletions validated the boundaries of the cluster and inactivation of cpz21 resulted in the accumulation of novel simplified liponucleoside antibiotics that lack the 3-methylglutaryl moiety. Therefore, Cpz21 is assigned to act as an acyltransferase in caprazamycin biosynthesis. In vivo and in silico analysis of the caprazamycin biosynthetic gene cluster allows a first proposal of the biosynthetic pathway and provides insights into the biosynthesis of related uridyl-antibiotics.
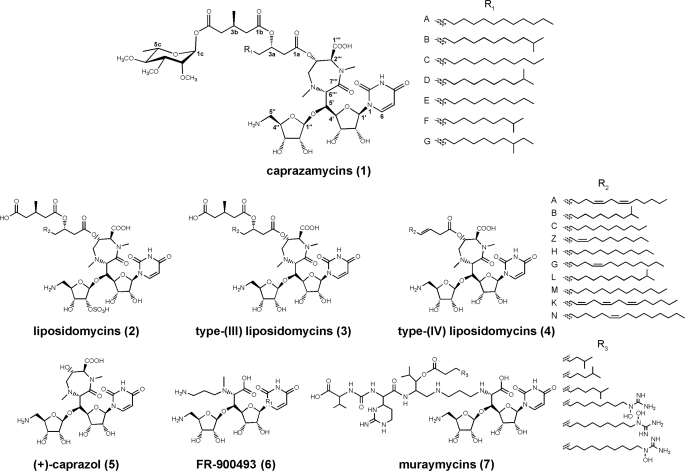
Caprazamycins (CPZs)2 (Fig. 1, 1) are liponucleoside antibiotics isolated from a fermentation broth of Streptomyces sp. MK730-62F2 (1, 2). They show excellent activity in vitro against Gram-positive bacteria, in particular against the genus Mycobacterium including Mycobacterium intracellulare, Mycobacterium avium, and Mycobacterium tuberculosis (3). In a pulmonary mouse model with M. tuberculosis H37Rv, administration of caprazamycin B exhibited a therapeutic effect but no significant toxicity (4). Structural elucidation (2) revealed a complex and unique composition of elements the CPZs share only with the closely related liposidomycins (LPMs, 2) (5). The core skeleton is the (+)-caprazol (5) composed of an N-alkylated 5′-(β-O-aminoribosyl)-glycyluridine, also known from FR-900493 (6) (6) and the muraymycins (7) (7), which is cyclized to form a rare diazepanone ring. Attached to the 3′″-OH are β-hydroxy fatty acids of different chain length resulting in CPZs A–G (1). They differ from the LPMs in the absence of a sulfate group at the 2″-position of the aminoribose and the presence of a permethylated l-rhamnose β-glycosidically linked to the 3-methylglutaryl (3-MG) moiety.
FIGURE 1.
Nucleoside antibiotics of the translocase I inhibitor family.
The LPMs have been shown to inhibit biosynthesis of the bacterial cell wall by targeting the formation of lipid I (8). The CPZs are expected to act in the same way and are assigned to the growing number of translocase I inhibitors that include other nucleoside antibiotics, like the tunicamycins and mureidomycins (9). During peptidoglycan formation, translocase I catalyzes the transfer of UDP-MurNAc-pentapeptide to the undecaprenyl phosphate carrier to generate lipid I (10). This reaction is considered an unexploited and promising target for new anti-infective drugs (11).
Recent investigations indicate that the 3″-OH group (12), the amino group of the aminoribosyl-glycyluridine, and an intact uracil moiety (13) are essential for the inhibition of the Escherichia coli translocase I MraY. The chemical synthesis of the (+)-caprazol (5) was recently accomplished (14), however, this compound only shows weak antibacterial activity. In contrast, the acylated compounds 3 and 4 exhibit strong growth inhibition of mycobacteria, suggesting a potential role of the fatty acid side chain in penetration of the bacterial cell (15, 16). Apparently, the acyl-caprazols (4) represent the most simplified antibiotically active liponucleosides and a good starting point for further optimization of this class of potential therapeutics.
Although chemical synthesis and biological activity of CPZs and LPMs has been studied in some detail, their biosynthesis remains speculative and only few data exists about the formation of other translocase I inhibitors (17, 18). Nevertheless, we assume that the CPZ biosynthetic pathway is partially similar to that of LPMs, FR-90043 (6), and muraymycins (7) and presents a model for the comprehension and manipulation of liponucleoside formation. Considering the unique structural features of the CPZs we also expect some unusual biotransformations to be involved in the formation of, e.g. the (+)-caprazol.
Here we report the identification and analysis of the CPZ gene cluster, the first cluster of a translocase I inhibitor. A set of gene disruption experiments provide insights into the biosynthetic origin of the CPZs and moreover, heterologous expression of the gene cluster allows the generation of novel bioactive derivatives by pathway engineering.
EXPERIMENTAL PROCEDURES
Bacterial Strains and General Methods—Chemicals, microbiological, and molecular biological agents were purchased from standard commercial sources. Streptomyces sp. MK730-62F2 and Streptomyces coelicolor M512 (SCP1-, SCP2-, ΔactIIorf4, ΔredD) and their respective derivatives were maintained and grown on either MS agar (2% soy flour, 2% mannitol, 2% agar; components purchased from Carl Roth, Karlsruhe, Germany) or TSB medium (BD Biosciences). E. coli strains were cultivated in LB medium (components purchased from Carl Roth) supplemented with appropriate antibiotics. Mycobacterium phlei was cultured in nutrient agar (BD Biosciences) and used as an indicator strain in agar diffusion assays for the detection of bioactivity in culture extracts of Streptomyces sp. MK730-62F2, S. coelicolor M512, and their derivatives. DNA isolation and manipulations were carried out according to standard methods for E. coli (19) and Streptomyces (20).
Production, Extraction, and Detection of Caprazamycin Derivatives—50 ml of TSB media was inoculated with spore suspension of Streptomyces sp. MK730-62F2, S. coelicolor M512, or a derivative thereof. The cultures were incubated for 2 days at 30 °C and 200 rpm. For the production of CPZs, 1 ml of the pre-cultures were inoculated into 100 ml of a medium containing 1% soytone, 1% soluble starch, and 2% d-maltose adjusted to pH 6.7 (components purchased from BD Biosciences). The cultures were incubated for 7 days at 30 °C and 200 rpm. For rapid identification of CPZs, cells were harvested and extracted with ice-cold methanol. The extract was directly applied to LC-MS and agar diffusion assay. Partial purification of CPZs was achieved by the adjustment of the culture supernatant to pH 4 and its subsequent extraction with an equal volume of butanol. The organic phase was evaporated and extracts were resolved in 500 μl of methanol. LC-MS/MS analysis was performed on a Surveyor HPLC system equipped with a Reprosil-Pur Basic C18 (5 μm, 250 × 2 mm) column (Dr. Maisch, Ammerbuch, Germany) coupled to a Thermo Finnigan TSQ Quantum triple quadrupole mass spectrometer (heated capillary temperature, 320 °C; sheath gas, nitrogen). For sample separation, a linear gradient from 2 to 40% acetonitrile in aqueous formic acid (0.1%) over 4 min followed by a linear gradient from 40 to 100% acetonitrile in aqueous formic acid (0.1%) over 31 min was used; the flow rate was 0.2 liters min-1 and detection at 262 nm. Positive electrospray ionization ((+)-ESI) was performed with electrospray voltage of 3.8 kV and collision-induced dissociation spectra were recorded with collision energy of 35 eV. Accordant parameters in negative mode ((-)-ESI) were 4.0 kV and 25 eV, respectively.
Bioactivity of culture extracts was monitored using M. phlei as an indicator strain. 50 μl of a glycerol culture of M. phlei was spread out on a nutrient agar plate. 5 μl of the butanolic culture extracts were applied to filter paper discs (5 mm) and placed on the top of the agar. The assay was incubated at 30 °C for 48 h.
Generation and Screening of a Cosmid Library—A Streptomyces sp. MK730-62F2 genomic cosmid library was constructed. Chromosomal DNA was partially digested with Bsp1431 (Fermentas, Vilnius, Lithuania), dephosphorylated, and ligated with BamHI/XbaI-digested SuperCos1 vector. The ligation products were packed (Gigapack III XL, Stratagene, Heidelberg, Germany) and transferred into E. coli SURE®. A 0.45-kb fragment of cpz28 was amplified from genomic DNA using primer omt1CH_fw, CCGTCCGCTACGGCTCNSMNAARTGG and omt1CH_rv, GCGGTCCACAGGTCCTCNAYNACRTA. Perfect matching primers omt7218_fw, GGCTGCACTGGTTCACGGG and omt7218_rv, CCAGAGGTCCTCGATCACG amplifying a 0.39-kb fragment of cpz28 were applied in a PCR screening of the cosmid library.
DNA Sequencing and Computer-assisted Sequence Analysis—Double-stranded sequencing of the entire cosmid clone 31C2 (42,300-bp insert) was performed by GenoTech (Baejeon, Korea) by using a shotgun library with DNA fragments of ∼0.5–1.0 kb in length. The DNASIS software package (Hitachi Software Engineering, Tokyo, Japan) and Artemis (Wellcome Trust Genome Campus, Cambridge, UK) were used for sequence analysis and annotation. Data base comparisons were carried out in the GenBank™ data base by using the BLAST program (21). Alignment and comparison of sequences were performed using the ClustalX algorithm (22) and GeneDoc alignment editor.
Heterologous Expression of the Caprazamycin Gene Cluster—An integration cassette from pIJ787 was introduced into the bla sequence of cosmid 31C2 as described previously (23) generating cosmid cpzLK09. cpzLK09 was verified by restriction analysis and non-methylated DNA was used for protoplast transformation into S. coelicolor. In brief, for the preparation of protoplasts, S. coelicolor M512 was cultivated in 100 ml of S-YEME broth (34% sucrose, 1% glucose, 0.5% peptone, 0.3% yeast extract, 0.3% malt extract, 5 mm MgCl2; components purchased from Carl Roth) for 48 h at 30 °C. Cells were harvested and washed with an aqueous solution of 10.3% sucrose. The degradation of the cell wall was performed at 30 °C in P-buffer (10.3% sucrose, 0.025% K2SO4, 0.202% MgCl2·6H2O, 0.005% KH2PO4, 0.368% CaCl2·2H2O, and 0.573% TES) supplemented with 4 mg/ml lysozyme (Serva, Heidelberg, Germany) and monitored by microscopy. Subsequent polyethylene glycol-mediated protoplast transformation was performed as described by Kieser et al. (20). Kanamycin resistance mutants were selected and designated as S. coelicolor M512/cpzLK09(1–3).
Generation of Δcpz1–4, Δcpz1–5, Δcpz1–6, Δcpz33–34, and Δcpz32–34 in S. coelicolor M512—An apramycin resistance (aac(3)IV) cassette was amplified from plasmid pIJ773 (24) using primer pairs B4_rv/B5_fw (Δcpz1–4), B4_rv/B6_fw (Δcpz1–5), B4_rv/B7_fw (Δcpz1–6), B1_rv/B1_fw (Δcpz33–34), and B1_rv/B2_fw (Δcpz32–34) (supplemental Table S1). The genes were replaced in E. coli BW25113/pIJ790/cpzLK09 by using the PCR targeting system (24). Resulting cosmids were confirmed by restriction analysis. Excision of the cassette was performed in E. coli BT340 taking advantage of the FLP/FRT recognition sites adjacent to the resistance cassette (25). Positive cosmids were screened for their apramycin sensitivity and verified by restriction analysis and PCR using primers B1_test_rv, B1_test_fw, B2_test_fw, B4_test_fw, B5_test_rv, B6_test_rv, and B7_test_rv (supplemental Table S1). Cosmids cpzWP01 (Δcpz33–34), cpzWP02 (Δcpz32–34), cpzWP05 (Δcpz1–4), czWP06 (Δcpz1–5), and cpzWP07 (Δcpz1–6) were transferred into E. coli ET12567 (26) and introduced into S. coelicolor M512 by triparental intergeneric conjugation with the help of E. coli ET12567/pUB307 (27). Kanamycin resistance clones were selected, confirmed by PCR, and designated as S. coelicolor M512/cpzWP01(1–3), S. coelicolor M512/cpzWP02(1–3), S. coelicolor M512/cpzWP05(1–3), S. coelicolor M512/cpzWP06(1–3), and S. coelicolor M512/cpzWP07(1–3).
Generation of Δcpz21 and Δcpz23 Mutants in S. coelicolor M512—Deletion mutants were generated in accordance with the generation of the mutants for the detection of the cluster boundaries. Primer pairs cpz21_rv/cpz21_fw and cpz23_rv/cpz23_fw were used to amplify the apramycin resistance cassette (supplemental Table S1). The resulting mutants were designated S. coelicolor M512/cpzLL06(1–3) (Δcpz21) and S. coelicolor M512/cpzLL07(1–3) (Δcpz23).
To generate the expression plasmids for mutant complementation, cpz21 and cpz23 were amplified from cosmid cpzLK09 using primer pairs cpz21Eco_fw/cpz21Hind_rv and cpz23Eco_fw/cpz23Hind_rv and cloned into the vector pGEM®-T (Promega). The genes were subcloned into the EcoRI/SpeI sites of expression vector pUWL201 (28) under the control of the ermE* promoter. This resulted in plasmids pLL06 (cpz21) and pLL07 (cpz23), respectively. DNA sequencing of these plasmids confirmed the correct sequence of all constructs. For protoplast transformation, the two plasmids were transferred into the non-methylating E. coli strain ET12567 and DNA was isolated by standard procedures. Transformation of the S. coelicolor mutant strains by polyethylene glycol-mediated protoplast transformation (20) finally generated strains S. coelicolor M512/cpzLL06/pLL06 and S. coelicolor M512/cpzLL07/pLL07.
RESULTS AND DISCUSSION
Identification and Cloning of the Caprazamycin Gene Cluster—The unusual structure of CPZs and the lack of information about the biosynthetic origin make it difficult to select genetic probes for the identification of the gene cluster. However, the formation of the permethylated l-rhamnose moiety is known from other antibiotics like elloramycin (29) and spinosyn (30). Oligonucleotides deduced from a multiple sequence alignment based on the elloramycin methyltransferase elmM1 led to the amplification of a partial sequence of cpz28 with high similarity to sugar O-methyltransferases. Primer walking revealed two adjacent genes cpz29 and cpz30 to be homologous to other O-methyltransferases suggesting the presence of the CPZ gene cluster. To our knowledge, this is the first study demonstrating the successful application of degenerated primers for O-methyltransferases for probe development. Commonly, methyltransferases are considered to be too diverse on the nucleotide sequence level and too widely distributed in bacterial metabolism to be useful in the identification of a specific gene cluster.
Perfect matching primers were applied to a genomic library of Streptomyces sp. MK730-62F2 constructed in a SuperCos1 vector. Eight positive cosmids of 3000 could be identified and proven to contain overlapping DNA by restriction mapping. Cosmid 31C2 was finally selected for complete shotgun sequencing (nucleotide sequence of the gene cluster has been deposited at GenBank, accession number FJ490409).
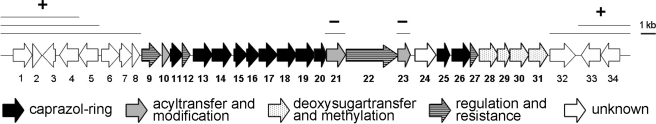
Sequence Analysis of the Caprazamycin Gene Cluster—A contiguous 42.3-kb region could be assembled with an average GC content of 70.2%, a typical value for Streptomyces DNA. In silico sequence analysis guided by BLAST homology searches (21), conserved protein domain searches (31), and the GC frame plot method (32) revealed 34 candidate genes. A total of 23 open reading frames, designated cpz9–31, were assigned to the CPZ gene cluster putatively encoding for biosynthesis, resistance, transport, and regulatory functions (Fig. 2). Table 1 summarizes the orthologous and proposed functions of the annotated genes. Notably, most of the putative gene products did not show homology to proteins found previously in other secondary metabolite gene clusters that reflects the unusual structure of the CPZs.
FIGURE 2.
Organization of the caprazamycin gene cluster. The putative assignment of the genes to different steps in the biosynthesis is indicated. Bars above the cluster mark the gene deletions performed in this study. - indicates that the deletion of the respective region led to an abolishment of CPZ production. + indicates that CPZ production was not influenced.
TABLE 1.
Deduced functions of genes in the caprazamycin gene cluster
| Gene | AA | Protein homolog | Accession number | Identity/similaritya | Proposed function |
|---|---|---|---|---|---|
| cpz1 | 477b | SchA30, Streptomyces sp. SCC 2136 | CAH10130 | 84/90 | Feruloyl-CoA synthase |
| cpz2 | 151 | SSEG_02365, Streptomyces sviceus ATCC 29083 | YP002207296 | 90/95 | Acyl dehydratase |
| cpz3 | 278 | SchA31, Streptomyces sp. SCC 2136 | CAH10131 | 76/84 | Transcriptional regulator |
| cpz4 | 513 | SACE_7046, Saccharopolyspora erythraea | CAM06207 | 44/58 | Hypothetical protein |
| cpz5 | 419 | hmgs, Streptomyces sp. CL190 | BAB07795 | 70/79 | HMG-CoA synthase |
| cpz6 | 349 | SaRppA, Streptomyces antibioticus | BAD89289 | 30/46 | Type III polyketide synthase |
| cpz7 | 260 | SACE_3529, S. erythraea | YP001105278 | 57/65 | PAPSc 3′-phosphatase |
| cpz8 | 212 | SACE_5947, S. erythraea | YP001108055 | 43/60 | Hypothetical protein |
| cpz9 | 348 | SSEG_03332, S. sviceus ATCC 29083 | YP002206529 | 27/41 | Transcriptional regulator |
| cpz10 | 182 | RSK20926_04892, Roseobacter sp. SK209-2-6 | ZP01755256 | 25/40 | β-Hydroxylase |
| cpz11 | 210 | SCO_1731, S. coelicolor A3(2) | NP626004 | 29/40 | Methyltransferase |
| cpz12 | 189 | SSEG_10045, S. sviceus ATCC 29083 | YP002202959 | 23/34 | Kinase |
| cpz13 | 441 | SACE_4299, S. erythraea | YP001106493 | 16/30 | Aminotransferase |
| cpz14 | 424 | Orf-4, Streptomyces atroolivaceus | AAN85510 | 36/53 | Hydroxymethyltransferase |
| cpz15 | 274 | Xccb100_2413, Xanthomonas campestris | YP001903823 | 26/40 | Dioxygenase |
| cpz16 | 233 | Smed_4814, Sinorhizobium medicae | YP001313540 | 20/30 | Nucleotidyltransferase |
| cpz17 | 377 | AprG2, Streptomyces tenebrarius | AAN06006 | 19/30 | Glycosyltransferase |
| cpz18 | 424 | CetH, Actinomyces sp. LU 9419 | ACH85568 | 34/49 | Aminotransferase |
| cpz19 | 460 | GK_2312, Geobacillus kaustophilus | YP148165 | 39/54 | Pyrimidine phosphorylase |
| cpz20 | 354 | TMCL4, Streptomyces sp. CK 4412 | ABI94375 | 44/58 | Acyl-CoA synthase |
| cpz21 | 500 | TMCL1, Streptomyces sp. CK 4412 | ABI94378 | 35/46 | Carboxyesterase |
| cpz22 | 1238 | SAV_5299, Streptomyces avermitilis MA 4680 | NP826476 | 36/53 | ABC transporter |
| cpz23 | 344 | AviX9, Streptomyces viridochromogenes | AAK83171 | 31/46 | Lipase |
| cpz24 | 598 | Cja_1569, Cellvibrio japonicus | YP001982049 | 26/40 | Hypothetical protein |
| cpz25 | 341 | SAV_2980, S. avermitilis MA 4680 | NP824156 | 45/63 | Dehydrogenase |
| cpz26 | 417 | SSDG_05270, S. pristinaespiralis ATCC 25486 | YP002195839 | 15/24 | Methyltransferase |
| cpz27 | 192 | SSEG_10045, S. sviceus ATCC 29083 | YP002202959 | 39/54 | Kinase |
| cpz28 | 396 | spnK, Saccharopolyspora spinosa | AAG23272 | 50/66 | O-Methyltransferase |
| cpz29 | 265 | elmM3, Streptomyces olivaceus | CAD57141 | 46/60 | O-Methyltransferase |
| cpz30 | 419 | elmM1, S. olivaceus | CAD57139 | 43/53 | O-Methyltransferase |
| cpz31 | 396 | elmGT, S. olivaceus | CAC16413 | 40/53 | Deoxysugar transferase |
| cpz32 | 543 | Tfu_2432, Thermobifida fusca | YP290488 | 22/28 | Nucleotidyltransferase |
| cpz33 | 556 | SchA32, Streptomyces sp. SCC 2136 | CAH10132 | 81/88 | Hypothetical protein |
| cpz34 | 544 | SchA33, Streptomyces sp. SCC 2136 | CAH10133 | 85/93 | Metallophosphoesterase |
Overall homology (%)
Gene lacks an appropriate stop codon and is considered to be uncomplete on cosmid 31C2 cpz. Bold genes are proposed to be essential for caprazamycin production
PAPS, adenosine 3′-phosphate,5′-phosphosulfate
As proposed, the CPZ gene cluster would start with cpz9, which encodes for a putative regulator of the AraC family. Most members of this family are positive transcriptional activators containing a helix-turn-helix motif. They are known from sugar degradation and other pathways but are rarely found in gene clusters of secondary metabolism (33). The predicted gene product of cpz22 shows homology to ABC-transporters. Similar proteins can be found in many antibiotic gene clusters and are usually involved in self-resistance and export (34). cpz12 and cpz27 are two putative sugar kinase genes similar to tunicamycin resistance proteins, e.g. TmrD from Deinococcus radiodurans, which structure has been reported recently (35). The 2′-, 3′-, and 5′-hydroxy groups of the uridine have been suggested as potential targets for phosphorylation by TmrD, resulting in inactivation of the nucleoside antibiotic tunicamycin.
Cpz10 exhibits similarity to the Fe(II)/2-oxoglutarate-dependent oxygenase family (36). Cpz11 and Cpz26 are two putative methyltransferases that contain conserved S-adenosylmethionine-binding domains (cd02440) but share low overall homology to each other (10% identity/18% similarity). Interestingly, both genes are translationally coupled to possible resistance genes cpz12 and cpz27 by overlap of start and stop codons. cpz13 shows weak homology to aminotransferase genes and is most likely translationally coupled to the predicted serine hydroxymethyltransferase gene cpz14.
The genes cpz15–23 seem to be co-translated as indicated by the overlap of start and stop codons. This subcluster would encode for Cpz15, another hypothetical Fe(II)/2-oxoglutarate-dependent oxygenase, a putative nucleotidyltransferase Cpz16 and Cpz17, which shows similarity to the glycosyltransferases. Cpz18 seems to belong to the class III aminotransferases, whereas Cpz19 resembles pyrimidine-nucleoside phosphorylases. Cpz20 and Cpz21 are similar to a putative acyl-CoA synthase (TMCL4) and a carboxyesterase (TMCL1) from the tautomycetin gene cluster (37). Another possible esterase is encoded by cpz23 the last gene in the proposed subcluster. The deduced gene product of cpz25 is a hypothetical alcohol dehydrogenase.
cpz28, cpz29, cpz30, and cpz31 apparently constitute an operon for the attachment and methylation of a deoxysugar as indicated by probable translational coupling of these genes. They show strong similarity to O-methyltransferases and glycosyltransferases from known antibiotic gene clusters in particular to proteins participating in the formation of elloramycin (29, 38) and spinosyn (39). Both compounds contain the same permethylated l-rhamnose moiety as found in the CPZs.
Interestingly, we could not identify genes for the dTDP-l-rhamnose biosynthesis on the cosmid. This was initially surprising, because all genes for the production of a bacterial secondary metabolite are usually clustered. However, neither the gene cluster of elloramycin (40) nor of spinosyn (41), steffimycin (42), or arranciamycin (43) contain genes for dTDP-l-rhamnose formation. We therefore suggest the genes for the CPZs deoxysugar biosynthesis to be located elsewhere on the genome of the natural producer.
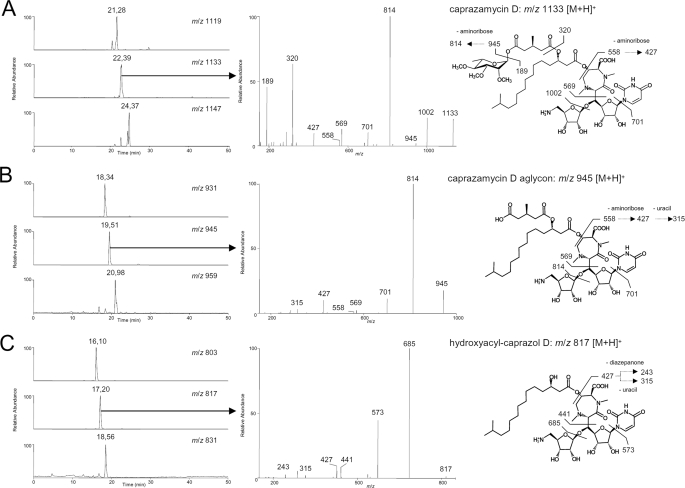
Heterologous Expression of the Caprazamycin Gene Cluster—To investigate whether the genes found on cosmid 31C2 were sufficient for biosynthesis of CPZs we intended to express the cosmid heterologously. For this purpose, the betalactamase (bla) gene on the backbone of 31C2 was replaced with an integration cassette of pIJ787 (23, 44) containing the attP attachment site and the integrase gene (int) of phage ΦC31, a tetracycline resistance gene (tet) and an origin of transfer (oriT) using λ-Red recombination. The generated cosmid cpzLK09 was introduced into S. coelicolor M512 by polyethylene glycol-mediated protoplast transformation (20) and three kanamycin resistance clones were selected, referred to as S. coelicolor M512/cpzLK09(1), -(2), and -(3). Extracts of cultures of the wild-type and mutant strains were applied to HPLC and ESI-MS/MS. In the wild-type strain the known CPZs A-G, whose different fatty acid side chains result in three different masses were detected readily as depicted in the selected ion monitoring chromatograms of Fig. 3A (m/z 1119 [M + H]+ for CPZ E and FatRt = 21.28 min, m/z 1133 [M + H]+ for CPZ C, D, and G at Rt = 22.39 min, and m/z 1147 [M + H]+ for CPZ A and B at Rt = 24.37 min). Characteristic MS/MS fragmentation patterns were observed by collision-induced dissociation of the methylated rhamnosyl group, aminoribose, uracil, and fatty acid groups. Similar fragmentations have been reported for liposidomycins in fast atom bombardment mass spectroscopy (45). For all CPZs, product ions of m/z 189, 320, 427, and 558 could be found correlating with the deacyl-components (Fig. 3 and supplemental Fig. S1). For the parent ion m/z 1133 [M + H]+ fragments of m/z 1002, 945, 814, 701, and 569 were detected. Corresponding fragments with a mass shift of -14 Da and +14 Da were observed for the parent ions m/z 1119 [M + H]+ and m/z 1147 [M + H]+, respectively (supplemental Fig. S1). Additional evidence was obtained by product ion scans in negative mode using the same extracts (supplemental Fig. S2).
FIGURE 3.
Identification of caprazamycin derivatives produced by wild-type and recombinant strains. Selected ion monitoring chromatograms obtained from LC-ESI-MS mass scans in positive mode. n-Butanolic culture extracts from A, wild-type Streptomyces sp. MK730-62F2 with m/z 1119 (CPZ E/F), 1133 (CPZ C/D/G), and 1147 (CPZ A/B); B, S. coelicolor M512/cpzLK09 with m/z 931 (CPZ E/F aglyca), 945 (CPZ C/D/G aglyca), and 959 (CPZ A/B aglyca); C, S. coelicolor M512/cpzLL06 with m/z 803 (hydroxyacylcaprazol E/F), 817 (hydroxyacylcaprazol C/D/G), and 831 (hydroxyacylcaprazol A/B). Mass spectrometric fragmentation pattern in the collision-induced dissociation experiment are shown for CPZ C/D/G from A, for CPZ C/D/G aglyca from B, and for hydroxyacylcaprazol C/D/G from C. The suggested fragmentation schemes are depicted for CPZ D (A), CPZ D aglycone (B), and hydroxyacylcaprazol D (C).
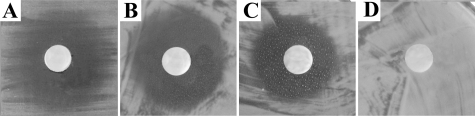
Although the masses for the CPZs could only be detected in the natural producer Streptomyces sp. MK730-62F2 (Fig. 3A and supplemental Fig. S4), prominent mass peaks for the CPZ aglyca were observed in S. coelicolor M512/cpzLK09 (selected ion monitoring chromatograms in Fig. 3B; for the CPZ E and F aglyca m/z 931 [M + H]+ at Rt = 18.34 min; for CPZ C, D, and G aglyca m/z 945 [M + H]+ at Rt = 19.51 min; and for CPZ A and B aglyca m/z 959 [M + H]+ at Rt = 20.98 min). Displaying the more hydrophilic character of the free carboxylic acid group, the aglyca elute 3 min earlier from the reversed phase HPLC column than the corresponding intact CPZs. Positive ESI-collision-induced dissociation fragmentation of the new compounds was identical to CPZs except for the absence of the l-rhamnose moiety. Fragments of m/z 814, 701, and 569 were obtained from the parent ion m/z 945 [M + H]+ (Fig. 3B). Analogous fragments with a mass shift of -14 Da and +14 Da resulted from m/z 931 [M + H]+ and m/z 959 [M + H]+ representing the CPZ derivatives with fatty acids of different chain length (supplemental Fig. S1). Molecular ions of m/z 558, 427, and 315, assigned to components of the caprazol structure, were found in all three spectra. S. coelicolor M512 without the gene cluster did not produce any of these new substances (supplemental Figs. S3 and S4). The analytical data strongly implicates the production of non-glycosylated CPZs, which are similar to the type-(III) LPMs (Fig. 1, 3) isolated previously (46). These compounds have been reported to show excellent activity against Mycobacteria. Thus, culture extracts from the heterologous producer were applied to an agar diffusion assay against M. phlei (Fig. 4). An inhibition zone of similar size could be observed with extracts from the wild-type and mutant strains, whereas extracts from S. coelicolor M512 without the gene cluster showed no bioactivity in this assay.
FIGURE 4.
Bioactivity of culture extracts against M. phlei. Butanolic culture extracts of: A, Streptomyces sp. MK730-62F2; B, S. coelicolor M512/cpzLK09; C, S. coelicolor M512/cpzLL06; and D, S. coelicolor M512 were applied to an agar diffusion assay against M. phlei.
The production of the non-glycosylated CPZ derivatives correlates with the absence of genes for the dTDP-l-rhamnose biosynthesis on the cosmid. Apparently, S. coelicolor M512 is unable to provide the dTDP-l-rhamnose in compensation as the corresponding enzymes are not encoded on the genome. Both, analytical and biological data verify that the genes identified on cosmid 31C2 indeed represent the CPZ biosynthetic gene cluster.
Validation of the Cluster Boundaries—A set of gene disruption experiments was carried out to determine the cluster boundaries. Sequence analysis of overlapping cosmids suggested the CPZ gene cluster to be inserted into a genomic region apparently conserved in several Streptomyces strains. cpz1 and cpz3 at the one end of the cluster and cpz33 and cpz34 at the other are almost identical with a continuous part of sequence from Streptomyces sp. SCC 2136 (47). Primer walking and terminal sequencing of overlapping cosmids showed that this similarity extends further upstream of cpz1.
Although cpz4 encodes for a hypothetical protein with unknown function, cpz5 showed homology to 3-hydroxymethylglutaryl (HMG)-CoA synthases. 3-Hydroxymethylglutaryl-CoA synthases catalyze the aldol addition of acetyl-CoA onto acetoacetyl-CoA and usually participate in the mevalonate pathway (48). A biosynthetic route to the uncommon 3-MG moiety was proposed involving a 3-hydroxymethylglutaryl-CoA synthase, a dehydratase, and a hydrogenase. No function in CPZ formation could be assigned to a putative type III polyketide synthase encoded by cpz6 and the possibly co-transcribed genes cpz7 and cpz8.
To validate the left border of the cluster cpz1, cpz2, cpz3, and cpz4 were deleted in cpzLK09 to generate cosmid cpzWP05 and cpz5 was additionally deleted to generate cpzWP06. By inactivation of the suggested biosynthetic pathway to 3-MG in cpzWP06 we hoped to produce compounds similar to the highly bioactive type (IV) LPMs (Fig. 1, 4). cpz6 was inactivated in addition to cpz1–cpz5 in cosmid cpzWP07. At the right end of the cluster cpz33 and cpz34 encoding for a hypothetical protein and a metallophosphoesterase were deleted in cosmid cpzWP01. A possible nucleotidyltransferases encoded by cpz32 was additionally deleted in cosmid cpzWP02.
After introducing the modified cosmids into S. coelicolor M512, positive candidates were selected by their kanamycin resistance and verified by PCR. Cultivation and analysis by HPLC and ESI-MS/MS revealed production of CPZ aglyca in all mutants (data not shown). In addition, bioassays of the culture extracts against M. phlei did not show any difference in inhibitory activity compared with S. coelicolor M512/cpzLK09 containing the intact gene cluster. In the case of Δcpz4, Δcpz5, and Δcpz32 complementation by host genes seems unlikely as the S. coelicolor genome contains no homologues. Therefore we concluded cpz1–6 and cpz32–34 to be non-essential in CPZ biosynthesis. Given that cpz6 is most likely co-transcribed with its downstream positioned genes, a functional knock-out of cpz7 and cpz8 can be assumed in cosmid cpzWP07. Consequently, the biosynthetic gene cluster for CPZs is predicted to span from cpz9 to cpz31 (Fig. 2).
Deletion of cpz21 and cpz23 and Production of Hydroxyacylcaprazols—Because cpz5 seems not to be required for CPZ formation and the corresponding mutant S. coelicolor M512/cpzWP06 did not accumulate the desired β-hydroxyacylcaprazols (Fig. 5, 4), we searched for possible acyltransferases within the gene cluster. Two acyl moieties, the 3-MG and β-hydroxy fatty acids have to be attached during CPZ biosynthesis and the two putative hydrolases Cpz21 and Cpz23 could be considered for these transfer reactions. Cpz21 is predicted to contain a typical α/β-hydrolase fold, the catalytic triad Ser208–Glu326–His409, and a GXSXG motif (49). The overall homology of Cpz21 is strongest with TMCL1 from Streptomyces sp. CK4412 (37). TMCL1, also named TmcC, is assigned to the esterification of a dialkymaleic anhydrid moiety to the linear polyketide during tautomycetin formation.
FIGURE 5.
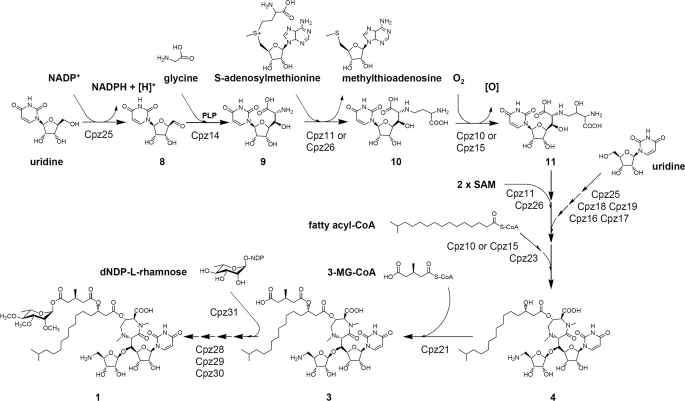
A hypothetical pathway for caprazamycin biosynthesis.
The amino acid sequence deduced from cpz23 shows highest overall homology to several hypothetical proteins from Streptomyces including AviX9 of the avilamycin gene cluster from Streptomyces viridochromogenes (50). According to the conserved protein domain search the C terminus of Cpz23 is similar to SGNH-hydrolases, a diverse family of lipases and carboxyesterases (51).
Both genes were individually deleted from cosmid cpzLK09 using λ-Red-mediated recombination. To create in-frame deletions, the disruption cassette from pIJ773 was subsequently removed by the use of FLP-recombinase (25) generating cosmids cpzLL06 (Δcpz21) and cpzLL07 (Δcpz23). After introduction of the cosmids in S. coelicolor M512 kanamycin-resistant mutants were cultivated and extracts were analyzed by LC-MS/MS. Production of the CPZ aglyca was abolished in S. coelicolor M512/cpzLL06 and S. coelicolor M512/cpzLL07 (data not shown). This proves that both Cpz21 and Cpz23 play an essential role in CPZ biosynthesis. Metabolites were only identified in extracts of S. coelicolor M512/cpzLL06 (Δcpz21) with m/z 803 [M + H]+ at Rt = 16.10 min; m/z 817 [M + H]+ at Rt = 17.20 min; and m/z 831 [M + H]+ at Rt = 18.56 min by LC-ESI-MS mass scan in positive mode (selected ion monitoring chromatograms in Figs. 3C and supplemental S4, and UV chromatograms in supplemental Fig. S3). Collision-induced fragmentation corresponds to the CPZ aglyca by sequential loss of the aminoribose (-131 Da), uracil (-111 Da), and ribose (-132 Da) but m/z values of product ions indicate the absence of the 3-methylglutarate (129 Da) (see also supplemental Fig. S1). Overall, fragments matched exactly the predicted characteristics of β-hydroxyacylcaprazols, structurally minimized liponucleosides antibiotics. In a bioassay against M. phlei (Fig. 4) extracts of a S. coelicolor M512/cpzLK09 and S. coelicolor M512/cpzLL06 (Δcpz21) cultivation broths showed both similar inhibiting activities, whereas no growth inhibition could be observed with S. coelicolor M512 and S. coelicolor M512/cp-zLL07 (data not shown) extracts. Co-expression of intact copies of the deleted genes under the constitutive ermE* promotor in the respective mutants restored the production of CPZ aglyca (data not shown).
Although similar to the type (IV) LPMs (4) the β-hydroxyacylcaprazols are expected to be slightly more hydrophilic due to the additional OH-group at the 3a-C position. Thus, they represent interesting novel compounds for further investigations, e.g. in structure/activity relationships. Moreover, the data indicated that Cpz21 is most likely involved in attachment of the 3-MG moiety. Beside Cpz21 two other enzymes with homology to para-nitrobenzyl esterases are known from bacterial secondary metabolism. Both of these enzymes, TmcC and TtmK from the tautomycin gene cluster (52), were proposed to catalyze the attachment of an acyl group. However, inactivation of the corresponding genes in the gene cluster did not lead to the identification of an accumulated intermediate. Therefore the data presented here provides the first functional evidence that these family of enzymes indeed act as acyltransferases.
A Model for Caprazamycin Biosynthesis—Sequence analysis of the gene cluster combined with analytical data from heterologous expression and gene inactivation experiments may allow a first proposal of the CPZ biosynthetic pathway (Fig. 5) although many of the suggested reactions remain speculative at present.
A key question in the biosynthesis of CPZs and translocase I inhibitors of the same class is the origin of the glycyluridine (Fig. 5, 9). Metabolic labeling studies have shown that uridine is incorporated directly into related uridyl antibiotics such as tunicamycins (53). A pathway to the tunicamycins has been proposed to start with the oxidation of uridine to form uridine 5′-aldehyde (8) (17). We suggest a similar reaction for CPZ biosynthesis, which may be catalyzed by the putative alcohol dehydrogenase Cpz25. The resulting product (8) could undergo a subsequent aldol addition with a pyridoxal phosphate-glycine adduct to generate 9. This mechanism would be very similar to that of the well studied serine hydroxymethyltransferases, which are known to produce β-hydroxy α-amino acids from glycine and various aldehydes (54). Cpz14, with significant sequence similarity to serine hydroxymethyltransferases, is an obvious candidate for the catalysis of this reaction. The next step would be the transfer of a 3-amino-3-carboxypropyl group to the 5′″-amino group of 9 to form 10. A corresponding reaction occurs in the nocardicin biosynthesis (55). In this pathway, the gene product Nat utilizes S-adenosylmethionine to transfer the 3-amino-3-carboxypropyl moiety to a nucleophilic acceptor (56). Nat shows conserved domains of S-adenosylmethionine-dependent methyltransferases, which are also found in Cpz11 and Cpz26, but overall sequence similarity is low with 16% to Cpz26 and 15% to Cpz11. Hence, both genes may be candidates for a 3-amino-3-carboxypropyl transfer in CPZ biosynthesis, although we rather consider them to be involved in the two N-methylation steps discussed below. We further speculate that 10 could be a common intermediate in the caprazamycin (1), liposidomycin (2), FR900493 (6), and the muraymycin (7) biosynthesis. β-Hydroxylation of the 3-amino-3-carboxypropyl group of 10 would lead to 11 and could be catalyzed by either Cpz10 or Cpz15. Both proteins show homology to oxygenases.
Subsequent biosynthetic steps, including formation and transfer of the aminoribose, cyclization, and N-methylation of the diazepanone ring and attachment of the fatty acid would finally lead to 4 (Fig. 5). Compounds of this structure were accumulated in the Δcpz21 mutant strain and are probable intermediates of the CPZ pathway. Reasonable candidate genes for these biosynthetic steps can be found in the cluster. However, the sequence of these reactions, described in the following paragraphs, is speculative at present.
Cyclization of 11 by amide bond formation between the carboxyl group and the secondary amino group would immediately result in the characteristic diazepanone ring. For this reaction, a previous activation of the carboxyl group, e.g. in the form of an acyl adenylate, a coenzyme A ester, or an acyl phosphate would be required. The hypothetical acyl-CoA synthase Cpz20 or the putative kinases Cpz12 and Cpz27 may be involved in this reaction.
Interestingly, a contiguous set of genes, cpz16–19, was found in the CPZ cluster, which can be assigned to all steps required for the generation and attachment of the aminoribosyl moiety. This reaction sequence may start from a second molecule of 8, derived from uridine by a Cpz25-mediated oxidation as described above. Subsequently, the 5-aldehyde group could undergo an aminotransfer reaction, yielding a 5-aminated nucleoside possibly catalyzed by the hypothetical aminotransferase Cpz18. CetH, an orthologoue of Cpz18, has recently been assigned to the aminotransfer reaction in biosynthesis of the aminocyclitol cetoniacytone (57). 5-Amino-ribose-1-phosphate and uracil would be generated from the aminated nucleoside by Cpz19, a putative pyrimidine-nucleoside phosphorylase. A similar reaction has been shown in fluorothreonine biosynthesis where a 1-phosphoribosyl derivative is formed under catalysis of the pyrimidine phosphorylase FlB (58). Subsequently, the potential nucleotidyltransferase Cpz16 may convert the 5-amino-ribose-1-phosphate to dNDP-5-aminoribose. Then, the putative glycosyltransferase Cpz17 could transfer the aminoribose moiety forming a glycosidic bond.
Generally, ribosyl moieties are attached by phosphoribosyl-transferases (59) using 5-phosphoribosyl-1-diphosphate as a donor to generate a 5′-phosphoribosylated product. Then, the 5′-phospho group is removed by a phosphatase. Similar reactions have recently been shown to lead to the ribosyl moiety in butirosin biosynthesis involving BtrL and BtrP (60). However, no orthologoues to BtrL and BtrP were found in the CPZ gene cluster, making the pathway described above a more likely alternative.
The fatty acid moieties of LPMs and CPZs are probably derived from primary metabolism, as feeding studies with labeled palmitic acid in Streptomyces griseosporeus showed the direct incorporation into LPMs (61). Hydroxylation of the fatty acids could either occur within primary metabolism or by oxygenases Cpz10 or Cpz15. Cpz23 may be involved in the attachment of the hydroxy fatty acids, due to its homology to lipases.
In the diazepanone ring, both nitrogens are methylated. The N-methylation reactions are likely to be catalyzed by Cpz11 and/or Cpz26. Notably, Cpz11 shows sequence similarity (55%) to one of the few characterized N-methyltransferases AtM1 from the gene cluster of AT2433 (62).
The biosynthetic origin of the 3-MG moiety remains elusive. By our inactivation experiments we could exclude an involvement of the putative 3-hydroxymethylglutaryl-CoA synthase Cpz5. Therefore, this moiety is likely generated by enzymes encoded outside the cluster, probably in the form of a coenzyme A ester. We assign the catalysis of the subsequent acyltransfer to Cpz21 as indicated by functional investigations in this study.
Analogous to the biosynthesis of elloramycin, l-rhamnose would be synthesized from enzymes encoded elsewhere on the genome (40). The dNDP-l-rhamnose probably constitutes the substrate for a transfer reaction to the CPZ aglycon catalyzed by the putative rhamnosyltransferase Cpz31. Sequential methylation of the deoxysugar moiety is likely catalyzed by the hypothetical sugar O-methyltransferases Cpz28, Cpz29, and Cpz30.
The identification and analysis of the caprazamycin gene cluster provides the first molecular basis for the proposal of a translocase I inhibitor biosynthetic pathway. Because the formation of intermediate 10 can be speculated to be similar for other structurally related compounds, this work may help in the development of probes for the discovery of gene clusters of other uridyl antibiotics. As proposed, several biosynthetic steps to the caprazamycins seem to be distinctive and unique in bacterial secondary metabolism. Apparently they represent intriguing subjects for further functional investigations. A detailed understanding of the caprazamycins biosynthetic pathway combined with the successful establishment of a heterologous expression system sets the basis for genetic and metabolic engineering toward the production of new liponucleoside antibiotics with improved properties.
Supplementary Material
Acknowledgments
We are grateful to Prof. Lutz Heide (Tübingen University, Germany) for comments on the manuscript and Prof. Tim Bugg (University of Warwick, UK) for valuable discussions. We thank the students of the “Wahlpflichtfach Pharmazeutische Bio und Gentechnologie 2007” of Tübingen University for the generation of mutant cosmids cpzWP01-07 and Dr. Manuel Wolpert for kind supervision. We also thank Stefanie Khartulyari for help with the ESI-MS.
This work was supported European Commission Grant IP005224 ActinoGEN.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
Footnotes
The abbreviations used are: CPZ, caprazamycin; LPM, liposidomycin; 3-MG, 3-methylglutaryl; MS, mass spectrometry; HPLC, high pressure liquid chromatography; ESI, electrospray ionization; TES, 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
References
- 1.Igarashi, M., Nakagawa, N., Doi, N., Hattori, S., Naganawa, H., and Hamada, M. (2003) J. Antibiot. 56 580-583 [DOI] [PubMed] [Google Scholar]
- 2.Igarashi, M., Takahashi, Y., Shitara, T., Nakamura, H., Naganawa, H., Miyake, T., and Akamatsu, Y. (2005) J. Antibiot. 58 327-337 [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi, T., Igarashi, M., Naganawa, H., and Hamada, M. (May 9, 2002) U. S. Patent 6,780,616
- 4.Igarashi, M., Nakagawa, S., Hattori, S., Doi, T., Nasuda, T., Mijake, M., Ishizuka, H., Naganawa, H., Shomura, T., and Hamada, M. (2002) Caprazamycins A–F, Novel Anti-TB Antibiotics, from Streptomyces sp., September 22, 2002, P. 232 (Abstr. F-2031), 42nd Interscience, Conference on Antimicrobial Agents and Chemotherapy, San Diego, CA
- 5.Isono, K., Uramoto, M., Kusakabe, H., Kimura, K., Isaki, K., Nelson, C. C., and McCloskey, J. A. (1985) J. Antibiot. 38 1617-1621 [DOI] [PubMed] [Google Scholar]
- 6.Ochi, K., Ezaki, M., Iwani, M., Komori, T., and Kohsaka, M. (March 6, 1989) U. S. Patent 4,950,605
- 7.McDonald, L. A., Barbieri, L. R., Carter, G. T., Lenoy, E., Lotvin, J., Petersen, P. J., Siegel, M. M., Singh, G., and Williamson, R. T. (2002) J. Am. Chem. Soc. 124 10260-10261 [DOI] [PubMed] [Google Scholar]
- 8.Brandish, P. E., Kimura, K. I., Inukai, M., Southgate, R., Lonsdale, J. T., and Bugg, T. D. (1996) Antimicrob. Agents Chemother. 40 1640-1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura, K., and Bugg, T. D. (2003) Nat. Prod. Rep. 20 252-273 [DOI] [PubMed] [Google Scholar]
- 10.Struve, W. G., Sinha, R. K., and Neuhaus, F. C. (1966) Biochemistry 5 82-93 [DOI] [PubMed] [Google Scholar]
- 11.Dini, C. (2005) Curr. Top. Med. Chem. 5 1221-1236 [DOI] [PubMed] [Google Scholar]
- 12.Dini, C., Drochon, N., Guillot, J. C., Mauvais, P., Walter, P., and Aszodi, J. (2001) Bioorg. Med. Chem. Lett. 11 533-536 [DOI] [PubMed] [Google Scholar]
- 13.Dini, C., Drochon, N., Feteanu, S., Guillot, J. C., Peixoto, C., and Aszodi, J. (2001) Bioorg. Med. Chem. Lett. 11 529-531 [DOI] [PubMed] [Google Scholar]
- 14.Hirano, S., Ichikawa, S., and Matsuda, A. (2005) Angew. Chem. Int. Ed. Engl. 44 1854-1856 [DOI] [PubMed] [Google Scholar]
- 15.Hirano, S., Ichikawa, S., and Matsuda, A. (2008) J. Org. Chem. 73 569-577 [DOI] [PubMed] [Google Scholar]
- 16.Kimura, K., Ikeda, Y., Kagami, S., Yoshihama, M., Suzuki, K., Osada, H., and Isono, K. (1998) J. Antibiot. 51 1099-1104 [DOI] [PubMed] [Google Scholar]
- 17.Price, N. P., and Tsvetanova, B. (2007) J. Antibiot. 60 485-491 [DOI] [PubMed] [Google Scholar]
- 18.Ohnuki, T., Muramatsu, Y., Miyakoshi, S., Takatsu, T., and Inukai, M. (2003) J. Antibiot. 56 268-279 [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 20.Kieser, T., Bibb, M., Buttner, M., Chater, K., and Hopwood, D. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK
- 21.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998) Trends Biochem. Sci. 23 403-405 [DOI] [PubMed] [Google Scholar]
- 23.Eustáquio, A. S., Gust, B., Galm, U., Li, S. M., Chater, K. F., and Heide, L. (2005) Appl. Environ. Microbiol. 71 2452-2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1541-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherepanov, P. P., and Wackernagel, W. (1995) Gene 158 9-14 [DOI] [PubMed] [Google Scholar]
- 26.MacNeil, D. J. (1988) J. Bacteriol. 170 5607-5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flett, F., Mersinias, V., and Smith, C. P. (1997) FEMS Microbiol. Lett. 155 223-229 [DOI] [PubMed] [Google Scholar]
- 28.Doumith, M., Weingarten, P., Wehmeier, U. F., Salah-Bey, K., Benhamou, B., Capdevila, C., Michel, J. M., Piepersberg, W., and Raynal, M. C. (2000) Mol. Gen. Genet. 264 477-485 [DOI] [PubMed] [Google Scholar]
- 29.Patallo, E. P., Blanco, G., Fischer, C., Brana, A. F., Rohr, J., Mendez, C., and Salas, J. A. (2001) J. Biol. Chem. 276 18765-18774 [DOI] [PubMed] [Google Scholar]
- 30.Waldron, C., Matsushima, P., Rosteck, P. R., Jr., Broughton, M. C., Turner, J., Madduri, K., Crawford, K. P., Merlo, D. J., and Baltz, R. H. (2001) Chem. Biol. 8 487-499 [DOI] [PubMed] [Google Scholar]
- 31.Marchler-Bauer, A., Anderson, J. B., Derbyshire, M. K., DeWeese-Scott, C., Gonzales, N. R., Gwadz, M., Hao, L., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z., Krylov, D., Lanczycki, C. J., Liebert, C. A., Liu, C., Lu, F., Lu, S., Marchler, G. H., Mullokandov, M., Song, J. S., Thanki, N., Yamashita, R. A., Yin, J. J., Zhang, D., and Bryant, S. H. (2007) Nucleic Acids Res. 35 237-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibb, M. J., Findlay, P. R., and Johnson, M. W. (1984) Gene 30 157-166 [DOI] [PubMed] [Google Scholar]
- 33.Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997) Microbiol. Mol. Biol. Rev. 61 393-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodríguez, A. M., Olano, C., Vilches, C., Méndez, C., and Salas, J. A. (1993) Mol. Microbiol. 8 571-582 [DOI] [PubMed] [Google Scholar]
- 35.Kapp, U., Macedo, S., Hall, D. R., Leiros, I., McSweeney, S. M., and Mitchell, E. (2008) Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 64 479-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stenflo, J., Holme, E., Lindstedt, S., Chandramouli, N., Huang, L. H., Tam, J. P., and Merrifield, R. B. (1989) Proc. Natl. Acad. Sci. U.S.A. 86 444-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi, S. S., Hur, Y. A., Sherman, D. H., and Kim, E. S. (2007) Microbiology 153 1095-1102 [DOI] [PubMed] [Google Scholar]
- 38.Blanco, G., Patallo, E. P., Braña, A. F., Trefzer, A., Bechthold, A., Rohr, J., Méndez, C., and Salas, J. A. (2001) Chem. Biol. 8 253-263 [DOI] [PubMed] [Google Scholar]
- 39.Waldron, C., Madduri, K., Crawford, K., Merlo, D. J., Treadway, P., Broughton, M. C., and Baltz, R. H. (2000) Antonie Leeuwenhoek 78 385-390 [DOI] [PubMed] [Google Scholar]
- 40.Ramos, A., Lombó, F., Braña, A. F., Rohr, J., Méndez, C., and Salas, J. A. (2008) Microbiology 154 781-788 [DOI] [PubMed] [Google Scholar]
- 41.Madduri, K., Waldron, C., and Merlo, D. J. (2001) J. Bacteriol. 183 5632-5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gullón, S., Olano, C., Abdelfattah, M. S., Braña, A. F., Rohr, J., Méndez, C., and Salas, J. A. (2006) Appl. Environ. Microbiol. 72 4172-4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luzhetskyy, A., Mayer, A., Hoffmann, J., Pelzer, S., Holzenkämper, M., Schmitt, B., Wohlert, S. E., Vente, A., and Bechthold, A. (2007) ChemBioChem 8 599-602 [DOI] [PubMed] [Google Scholar]
- 44.Gust, B., Chandra, G., Jakimowicz, D., Yuqing, T., Bruton, C. J., and Chater, K. F. (2004) Adv. Appl. Microbiol. 54 107-128 [DOI] [PubMed] [Google Scholar]
- 45.Ubukata, M., Kimura, K., Isono, K., Nelson, C. C., Gregson, J. M., and McCloskey, J. A. (1992) J. Org. Chem. 57 6392-6403 [Google Scholar]
- 46.Kimura, K., Ikeda, Y., Kagami, S., Yoshihama, M., Ubukata, M., Esumi, Y., Osada, H., and Isono, K. (1998) J. Antibiot. 51 647-654 [DOI] [PubMed] [Google Scholar]
- 47.Basnet, D. B., Oh, T. J., Vu, T. T., Sthapit, B., Liou, K., Lee, H. C., Yoo, J. C., and Sohng, J. K. (2006) Mol. Cells 22 154-162 [PubMed] [Google Scholar]
- 48.Dairi, T. (2005) J. Antibiot. 58 227-243 [DOI] [PubMed] [Google Scholar]
- 49.Holmquist, M. (2000) Curr. Protein Pept. Sci. 1 209-235 [DOI] [PubMed] [Google Scholar]
- 50.Weitnauer, G., Mühlenweg, A., Trefzer, A., Hoffmeister, D., Süssmuth, R. D., Jung, G., Welzel, K., Vente, A., Girreser, U., and Bechthold, A. (2001) Chem. Biol. 8 569-581 [DOI] [PubMed] [Google Scholar]
- 51.Akoh, C. C., Lee, G. C., Liaw, Y. C., Huang, T. H., and Shaw, J. F. (2004) Prog. Lipid Res. 43 534-552 [DOI] [PubMed] [Google Scholar]
- 52.Li, W., Ju, J., Rajski, S. R., Osada, H., and Shen, B. (2008) J. Biol. Chem. 283 28607-28617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsvetanova, B. C., Kiemle, D. J., and Price, N. P. (2002) J. Biol. Chem. 277 35289-35296 [DOI] [PubMed] [Google Scholar]
- 54.Makart, S., Bechtold, M., and Panke, S. (2007) J. Biotechnol. 130 402-410 [DOI] [PubMed] [Google Scholar]
- 55.Gunsior, M., Breazeale, S. D., Lind, A. J., Ravel, J., Janc, J. W., and Townsend, C. A. (2004) Chem. Biol. 11 927-938 [DOI] [PubMed] [Google Scholar]
- 56.Reeve, A. M., Breazeale, S. D., and Townsend, C. A. (1998) J. Biol. Chem. 273 30695-30703 [DOI] [PubMed] [Google Scholar]
- 57.Wu, X., Flatt, P. M., Xu, H., and Mahmud, T. (2009) ChemBioChem 10 304-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang, F., Haydock, S. F., Spiteller, D., Mironenko, T., Li, T. L., O'Hagan, D., Leadlay, P. F., and Spencer, J. B. (2006) Chem. Biol. 13 475-484 [DOI] [PubMed] [Google Scholar]
- 59.Sinha, S. C., and Smith, J. L. (2001) Curr. Opin. Struct. Biol. 11 733-739 [DOI] [PubMed] [Google Scholar]
- 60.Kudo, F., Fujii, T., Kinoshita, S., and Eguchi, T. (2007) Bioorg. Med. Chem. 15 4360-4368 [DOI] [PubMed] [Google Scholar]
- 61.Kagami, S., Esumi, Y., Nakakoshi, M., Yoshihama, M., and Kimura, K. (2003) J. Antibiot. 56 552-556 [DOI] [PubMed] [Google Scholar]
- 62.Gao, Q., Zhang, C., Blanchard, S., and Thorson, J. S. (2006) Chem. Biol. 13 733-743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.