Abstract
The troponin complex on the thin filament plays a crucial role in the regulation of muscle contraction. However, the precise location of troponin relative to actin and tropomyosin remains uncertain. We have developed a method of reconstructing thin filaments using single particle analysis that does not impose the helical symmetry of actin and is independent of a starting model. We present a single particle three-dimensional reconstruction of the thin filament. Atomic models of the F-actin filament were fitted into the electron density maps and troponin and tropomyosin located. The structure provides evidence that the globular head region of troponin labels the two strands of actin with a 27.5-Å axial stagger. The density attributed to troponin appears tapered with the widest point toward the barbed end. This leads us to interpret the polarity of the troponin complex in the thin filament as reversed with respect to the widely accepted model.
Regulation of actin filament function is a fundamental biological process with implications ranging from cell migration to muscle contraction. Skeletal and cardiac muscle thin filaments consist of actin and the regulatory proteins troponin and tropomyosin. Contraction is initiated by release of Ca2+ into the sarcomere and the consequent binding of Ca2+ to regulatory sites on troponin. Troponin is believed to undergo a conformational change leading to an azimuthal movement of tropomyosin, which allows myosin heads to interact with actin, hydrolyze ATP, and generate force. The molecular basis by which troponin acts to regulate muscle contraction is only partly understood. It is essential that the structure of troponin in the thin filament at high and low Ca2+ is determined to properly understand the mechanism of regulation.
The basic structure of the thin filament was described by Ebashi in 1972 (1). In this structure each tropomyosin molecule covers seven actin monomers, and there is a 27.5-Å stagger between troponin molecules. The 7-Å tropomyosin structure (2), the atomic model of F-actin (3), and the troponin “core domain” (4) have recently been used to generate atomic models of the thin filament in low and high Ca2+ states (5). While the position of troponin in these models was constrained by known distance measurements between filament components, the exact arrangement of the complex on the filament has not been determined a priori. Although recently published crystal structures of partial troponin complexes (4, 6) have provided valuable insights into the arrangement of the globular head or core domain, the complex in its entirety has not been crystallized.
Troponin is believed to consist of a globular core domain with an extended tail (7). The globular core contains the Ca2+-binding subunit (TnC),2 the inhibitory subunit (TnI), and the C-terminal part (residues 156–262) of the tropomyosin-binding subunit (TnT). The extended tail consists of the N-terminal part of TnT (residues 1–155). A structural rearrangement associated with Ca2+ dissociation from the troponin core has been observed (4) such that the helix connecting the two domains of TnC collapses, releasing the TnI inhibitory segment. It is postulated that the TnI inhibitory segment then becomes able to bind actin, in so doing biasing tropomyosin (8). To understand properly how Ca2+ binding to TnC leads to movement of tropomyosin, it is necessary to determine a high resolution structure of troponin attached to the thin filament, allowing unambiguous docking of the available crystal structures and direct observation of any changes at a molecular level caused by Ca2+ binding.
Direct visualization of the thin filament is possible using electron microscopy. Tropomyosin strands have been resolved in the low and high Ca2+ states confirming the movement of tropomyosin and the steric blocking model (9, 10). Until recently the actin helical repeat has been imposed in the majority of reconstructions of the thin filament causing artifacts. Helical averaging using the actin repeat spreads troponin density over every actin monomer, which prevents the detailed position and shape of the troponin complex from being found (11). It is possible to avoid this effect by applying a single particle approach. Individual filament images are divided into segments and each segment treated as a particle. Three-dimensional reconstruction may then be carried out by single particle techniques of alignment, classification (12, 13), Euler angle assignment (14–16) and exact filter back-projection (17, 18).
Two forms of single particle analysis have emerged: helical single particle analysis (19), where the determined helical symmetry is applied to the final reconstruction, and non-helical single particle analysis, which treats the complex as a truly asymmetric particle. Helical single particle analysis has been used to successfully reconstruct a myosin containing invertebrate thick filament to a resolution of 25 Å (20), and non-helical single particle analysis has been applied to the vertebrate skeletal muscle thick filament allowing azimuthal perturbations of the myosin heads to be observed (21).
Model-based single particle image processing methods have recently been applied to the structural analysis of the vertebrate (5, 22, 23) and the insect thin filament (24). We have deliberately avoided starting with a model and any potential model bias by using a reference-free alignment procedure. The adaptation of conventional procedures and their application to the structural study of the muscle thin filament has been documented (25).
EXPERIMENTAL PROCEDURES
Filament Isolation and Electron Microscopy—Thin filaments were isolated from goldfish (Carassius auratus) in relaxing solution as described in Kensler and Stewart (1989) (26). Electron micrographs were recorded of negatively stained filaments applied to a thin carbon film support over the holes of holey carbon grids. The images were collected on a JEOL 1200EX at a nominal magnification of ×20,000. The images were digitized on a Nikon Coolscan 8000ED with a step size of 6.35 μm. The focal level of the micrographs was such that the 1st minimum in the Thon rings was at substantially higher resolution than the final resolution of our analysis (∼35 Å, see below). From an initial dataset of 29 micrographs a selection of filaments from each micrograph were assessed. The Fourier transforms were computed and the strong 59 Å actin reflection used to calibrate the magnification and adjust the sampling of each filament to 3.28 Å/pixel.
Image Processing—Image processing software packages were used as follows: Imagic (28), the MRC suite of programs (60), Spider (34), and ImageJ (27). The straightening of filament images, was achieved using the straighten plugin for ImageJ (27). Helical reconstruction was carried out using adapted programs from the MRC Image package.
Single Particle Analysis—The image processing package Imagic provided the framework for the single particle analysis (28). Adaptations to the standard procedures, specific to filamentous particles, were implemented including the modified back-projection algorithm (25).
Particle Selection, Alignment, and Classification—Individual filaments were interactively selected into a large box (∼3000–4000 Å2). The filaments were straightened and subsequently segmented into smaller boxes ∼850 Å2 (128 × 128 pixels). These filament segments made up our 2638 selected particles. Particles were translationally aligned through the straightening procedure. Refinement was carried out using Spider alignment routines. Particles originating from the same filament should clearly be assigned the same polarity. The occurrence of erroneously oriented filaments was checked by monitoring the alignment procedure allowing particles that were assigned polarity inconsistent to their originating filament to be identified and subsequently removed from the data.
The consistency of alignment in the final data set was found to be 87%: those segments, which were inconsistently aligned were eliminated prior to calculating the final three-dimensional map. The relatively high level of consistency is characteristic of a strongly polar structure and effective alignment. Multivariate statistical analysis was performed on the data. The hierarchical ascendant classification algorithm was then used to generate 225 classes. The images in each of these classes were summed together to create class averages with an increased signal to noise ratio.
Angle Assignment and Three-dimensional Reconstruction—A helical reconstruction was calculated using the MRC helical processing routines from twelve straightened filaments. The helical reconstruction was then used to produce an “anchor set” of reference images which were evenly spaced over the Euler sphere. The “anchor set” of reference images were used to create sinograms, sets of one-dimensional line projections of each image as a function of projection angle, for use in angular reconstitution. The alternate angle assignment methods of projection matching using reference images generated from the helical reconstruction rotated about the filament axis and analysis of the central section overlap (29) were tested using the first set of class averages (generated before any multi-reference alignment was performed). Consistency between the three angle assignment methods was a criterion in angle selection to generate the first three-dimensional reconstruction. In subsequent rounds of angle assignment the angular reconstitution method was employed with an anchor set of 288 references again evenly distributed over the Euler sphere. The class averages were assigned angles with good coverage around the equator of the Euler sphere (Fig. 1). The modified exact filter back projection algorithm (25) was applied to create a three-dimensional electron density map from two-dimensional class averages.
FIGURE 1.
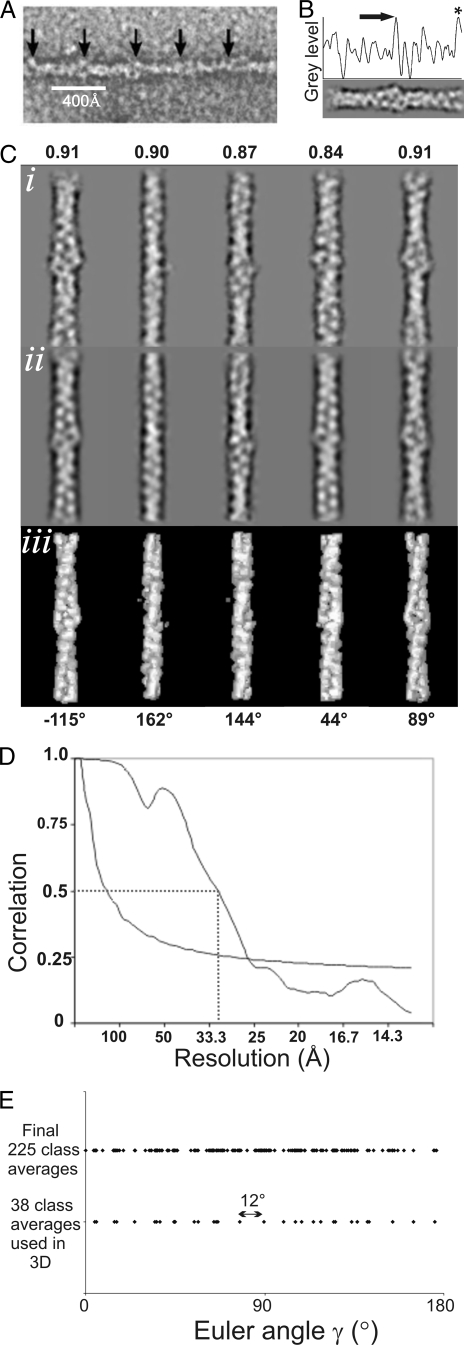
Single particle analysis of the thin filament. A, typical goldfish thin filament negatively stained with regularly spaced “bumps” of density attributed to troponin. B, two-dimensional class average and the corresponding one-dimensional density profile. The arrow indicates a large peak locating the troponin complex in the center of the segment. The asterisk highlights a second peak due to contributions from the neighboring troponin complex. C, five columns represent different viewing angles of the filament described by the Euler angles (α = 0, β = 90 throughout and γ =-115°, 162°, 144°, 44°, and 89°. (i) Class averages, (ii) reprojections, and (iii) surface views of the three-dimensional reconstruction are displayed at the corresponding angles. The cross-correlation coefficient between each of the class averages and the corresponding reprojection are given at the top of each column. D, fourier shell correlation curve of the single particle reconstruction prior to averaging. The dotted line gives the 0.5 threshold, and the exponential decay (dashed) shows the ½ bit criteria. Where the curve crosses these two thresholds an estimate of the highest significant spatial frequency present can be made and hence the resolution of the data. E, distribution of Euler angles assigned to the 225 class averages generated in the final round of processing and the 38 class averages back-projected to create the final three-dimensional single particle reconstruction. The distribution of angles demonstrates a random orientation about the filament axis. The largest gap in the angular coverage in Euler angle γ in the final three-dimensional single particle reconstruction is 12° (indicated). A gap of 12° taken as the sampling step for an evenly distributed data set tilted about a single axis theoretically could produce a reconstruction of ∼15 Å (59). Using this calculation we can rule out the possibility that angular distribution and the number of class averages used in our reconstruction has limited the resolution.
Imposing Screw Symmetry—A novel method has been implemented to computationally sum the two strands of the thin filament. Once converted to a set of cylindrical sections (cylindrical surfaces flattened out at a fixed radius) Imagic was used to align, add, cut, and translate the images. After processing, the sections were used to create the desired averaged structure. The three-dimensional map consisting of cylindrical sections was then converted back into Cartesian coordinates.
Rendering in PyMol—The three-dimensional model, electron density maps, and Protein Data Bank (PDB) structures were rendered using PyMol (61). A hybrid method (using Imagic and PyMol) has been established that allows EM density maps to be displayed in PyMol at a theoretical volume corresponding to a specific molecular weight.
Docking Atomic Models and Difference Analysis—Situs version 2.2 (30) and URO (31) were used to dock the atomic model of actin into our electron density maps. To evaluate the fit, docked PDB files were viewed in stereo in PyMol using the Nvidia Quadro Stereo System and NuVision stereoscopic glasses. Electron density models were created from the aligned PDB files within the URO software package. The PDB models and the experimental data were converted into the Imagic format and low pass filtered to 35 Å. The density models of docked F-actin and docked F-actin with docked tropomyosin were used to create the difference datasets.
RESULTS
Two-dimensional Class Averages of the Thin Filament—A dataset of 2638 individual particles was selected from straightened thin filament images (see “Experimental Procedures”). The samples were prepared in the absence of Ca2+ and were negatively stained for electron microscopy. A typical filament is shown in Fig. 1A: periodic regions of extra density at a spacing of ∼385 Å are clearly visible, allowing particle selection centered on troponin. After a reference-free alignment procedure, which centered the particles, multivariate statistical analysis, and hierarchical ascendant classification were used to create classes of filament segments (13). The members of each class were averaged to form a class average with increased signal to noise ratio. Two strands of actin subunits twisting around each other, with a crossover repeat of ∼360 Å were clearly visible in the two-dimensional class averages (Fig. 1C). These features appeared clear and well-defined from the early stages of the analysis.
Each class average is representative of a particular viewing angle of the filament. 225 classes were generated in the final cycle of image processing. Euler angles were assigned to the class averages using angular reconstitution initially with a reference set of images from the helical reconstruction calculated from the same data (see below) and subsequently using the three-dimensional map generated in the previous cycle of processing. As part of the refinement procedure a careful selection of class averages was made at the stage of three-dimensional reconstruction so as only to include those with a good level of detail which agreed well with two-dimensional reprojections of the three-dimensional map. This was achieved by visual and statistical comparison between classes and reprojections of the three-dimensional map, taking into account the distribution of assigned angles. This served to ensure that only well preserved and properly aligned filament segments were used in the analysis. Accordingly, in the final round of refinement 38 classes were selected out of the 225 calculated from the full data set. The distribution of angles assigned to the final 225 class averages, the entire data set, and the 38 used in the map provided a good coverage around the equator of the Euler sphere (Fig. 1). Model tests have shown that angular reconstitution can be successfully applied using a starting model that deviates from the desired structure: a model of F-actin provides enough information to accurately assign Euler angles to projections of a model of the full thin filament (25). Alternative angle assignment methods of projection matching using projection images of the helical reconstruction as references and a method based on the extent of correlation between corresponding central sections (29) provided consistent results. Once Euler angles had been assigned, an initial three-dimensional map was calculated using the exact filter back projection algorithm adapted for filamentous macromolecular complexes (25).
The consistency of Euler angle assignment within filaments was monitored. The Euler angles of adjacent segments assigned via projection matching was evaluated and compared with theoretical values calculated for actin filaments with 13/6 symmetry. The average departure from 13/6 symmetry of adjacent segments was 18.9° while 55% of segments were assigned angles within ±20° of the expected value and 78% within ±40° (supplemental Fig. S2). Cumulative angular disorder is known to perturb the total angle traversed by the actin one-start helix Φj (after N subunits) (32) according to Equation 1.
 |
(Eq. 1) |
The average angle between adjacent subunits is
 (167°). Reported values of
drms the root mean square angular deviation per subunit
for actin are 5°-6°
(33). For adjacent segments
centered on troponin complexes (N = 14) the cumulative disorder is
thus 19°-22°, which compares closely with the value of 18.9°
observed in our studies. The regulatory proteins are believed to increase
rigidity of the thin filament, but even in this case substantial angular
disorder has been observed
(32).
(167°). Reported values of
drms the root mean square angular deviation per subunit
for actin are 5°-6°
(33). For adjacent segments
centered on troponin complexes (N = 14) the cumulative disorder is
thus 19°-22°, which compares closely with the value of 18.9°
observed in our studies. The regulatory proteins are believed to increase
rigidity of the thin filament, but even in this case substantial angular
disorder has been observed
(32).
Location of Troponin—Extra density in the central region of the segments on opposing sides of the filament was apparent in most of the initial class averages: this density can be attributed to troponin. After subsequent rounds of refinement, as the data became better aligned, the density attributed to troponin became easier to visualize. The identification of densities attributed to troponin depends primarily on the orientation of the filament around its long axis. Although particularly clear in a number of classes (Fig. 1C, γ =-115°) density caused by troponin is less readily visualized in others (Fig. 1C, γ = 162°). This can be explained by the orientation of the filament: it was observed that troponin was less readily identified in orientations where it superimposes directly on top of the actin subunits. It was possible to check the location of troponin in a class average from the one-dimensional density profile (Fig. 1B). The presence of troponin was indicated by a significant peak close to the midpoint. This approach was used to assess class averages for inclusion in the three-dimensional analysis. A second peak was also often observed at a distance from the first peak close to the 385-Å troponin repeat.
Comparison of Two-dimensional Class Averages with Reprojections of the Three-dimensional Map—The level of detail that can be seen in an individual class average was used as the selection criterion for inclusion in the three-dimensional analysis. Another measure of the quality of a class average and the accuracy of its angular assignment was by comparison with the reprojection of the resulting three-dimensional reconstruction at the same viewing angle. Visual similarity was used to assess the accuracy of angular assignment and the effectiveness of classification. Five typical class averages with corresponding reprojections and surface views of the three-dimensional reconstruction are displayed in Fig. 1C. The central portion of each class average is asymmetric. The class averages (Fig. 1, Ci) show good visual agreement with reprojections of the three-dimensional reconstruction (Fig. 1, Cii) as well high levels of correlation (correlation coefficients ∼0.9).
Refinement of the structure was achieved by iterating over the image processing steps of alignment, classification, angle assignment, three-dimensional reconstruction, and reprojection (see “Experimental Procedures”) until no significant changes in three-dimensional density distribution were observed. The 38 class averages used in the final three-dimensional reconstruction were taken from a set of 225: the average number of segments per class in this set was ∼12. The polarity of the segments was checked for consistency with respect to other segments from the same filament: 13% proved to be inconsistent and were removed from the final refinement. The helical arrangement of the troponin complexes was exploited to improve the signal to noise ratio of our analysis. Addition of the two actin strands by translating and rotating along troponin helical path allowed a 2-fold averaging of the structure in a novel way (see Experimental Procedures). The alignment was refined further using routines in Spider (34) and the two strand averaging repeated. Prior to two-strand averaging, a quantitative assessment of the resolution of the final map was made using the Fourier shell correlation criterion. The 0.5 criteria (35, 36) returned a resolution of 32 Å and the ½ bit criteria (37) returned a resolution of 27 Å (Fig. 1D). These values are close to a qualitative estimate of ∼35 Å obtained by visually comparing our final density map to model data filtered to a known resolution.
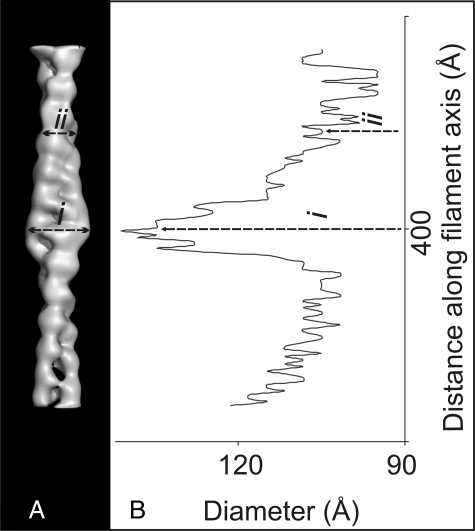
Three-dimensional Reconstruction of the Thin Filament—The three-dimensional reconstruction of the thin filament is displayed in Fig. 2 as a surface-rendered map (2A) and a wire mesh (2C). Using the known stoichiometry of binding (actin/troponin/tropomyosin, 7:1:1), we calculated the expected particle molecular mass (∼1630 kDa). Maps are contoured to include 125% of this weight assuming a protein density of 0.844 dalton/Å3. It has been reported that three-dimensional reconstructions of intermediate resolution frequently require thresholding at a higher molecular weight primarily due to the degree to which the low-spatial frequencies are represented (38, 39). A range of molecular weights were trialled up to 140% and the contours of the three-dimensional map inspected carefully. An increased molecular mass improved the connectivity of the density attributed to tropomyosin up to a value of 125%. Increasing the volume further produced a noisier map. The filament has an average width of 110 ± 7 Å and at its widest point measures 141 ± 7 Å (Fig. 3B). Two strands of actin subunits clearly twist around each other in the well recognized right-handed, long pitch actin helix. The interstrand cleft is particularly clear and the subunits appear bilobed. There is an area of localized mass on each actin strand around the midpoint of the reconstruction which can be attributed to the globular part of the troponin complex. The exact axial stagger of the density peaks attributed to the globular head of troponin was found to be 27.5 Å through the alignment procedure prior to averaging the two strands.
FIGURE 2.
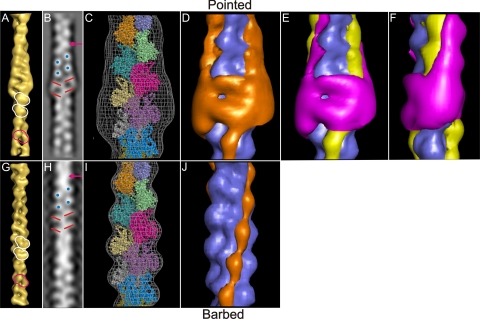
A comparison of the single particle based reconstruction and the helical reconstruction calculated from the same data. A–F, single particle reconstruction. G–J, helical reconstruction. A and G, are shown as surface-rendered maps. B and H, two-dimensional projections of the two reconstructions. C and I, are shown as wire meshes with atomic F-actin models docked. D, E, F, and J, surface-rendered difference maps. D and J, are obtained by subtracting F-actin models from the single particle and helical reconstructions, respectively. E and F, are composite difference maps which display the difference density calculated by subtracting an F-actin model with docked tropomyosin from the single particle reconstruction and the difference density displayed in D. Extra mass in the central region of the single particle reconstruction (A and C) surrounds the central actin subunits whereas the helical reconstruction (G and I) gives a much tighter fit to the crystal structure. Two actin subunits are encircled in white in each surface-rendered representation (A and G). The incorrect fit of the inverted F-actin model would give rise to the assignment of actin subunits illustrated in the pink outline (A and G). The crossover point of the two long pitch actin strands (arrowed), a slight gap surrounded by four high density bright regions (blue) and bands of density running diagonally out from the center of the filament (red) are visible in the two-dimensional projections of both the single particle and helical reconstructions (B and H). These features confirm the polarity inferred from fitting of the F-actin model. The difference density maps D and J calculated from single particle and helical reconstructions respectfully highlight the non-actin density (orange). This density attributable to tropomyosin and troponin is highly asymmetric in the single particle reconstruction. The composite difference density maps E and F highlight the contribution of troponin (magenta) and tropomyosin (yellow) related by a 90° rotation about the filament axis.
FIGURE 3.
Filament width. A, surface-rendered map of the single particle-based reconstruction. B, plot displaying the diameter of the filament against distance along the filament axis. The widest point of the filament is in the center of the segment (i =∼141 Å) the density begins to taper as toward the pointed end of the filament (ii =∼115 Å).
Agreement with Actin—To interpret the structure at a molecular level an F-actin atomic model was computationally docked into the three-dimensional density (Fig. 2C). The actin model was a 13/6 helical filament as described in Ref. 40. The docking programs Situs (30) and URO (31) were both used to evaluate different orientations of this high resolution filamentous model into the low resolution EM map. The best fit recovered in each case was virtually identical, indicating a single solution. The only alternative fits were discrete subunit translations along the short pitch filament helix. In no case was there a reversal of polarity. As a test, docking was repeated after rotating the F-actin model by 180° perpendicular to the filament axis. Although neither software package managed to invert the F-actin model, the calculated fit for the inverted model is inconsistent with the three-dimensional reconstruction: empty regions in the three-dimensional reconstruction are filled by density from the model and vice versa. Moreover the actin interstrand gap of the model structure was placed inside the actin subunit density of the three-dimensional reconstruction, and the actin subunit density of the model bridged the interstrand gap of the three-dimensional reconstruction (Figs. 2A and 3G). These conclusions are strongly supported by comparing the correlation coefficients of the masked fits for the best orientation (0.92) with that for the inverted orientation (0.68). Thus we can be confident in rejecting this alternative orientation of the actin backbone. By viewing the computed fit in stereo the agreement between the atomic model and our structure is clear as anticipated from the strong correlation between the fitted model and the data. Additionally the bi-lobed appearance of the subunits corresponds directly to the cleft between subdomains 3&4 and 1&2 of the actin subunit (Fig. 2C).
F-actin Fitting in a Helical Reconstruction—Further evidence for the polarity assignment in our single particle reconstruction was obtained from a helical reconstruction of the thin filament data. Fourier Bessel helical reconstruction (41) using 13/6 helical symmetry was applied to twelve selected straightened thin filament images. The relative polarity of each straightened filament was identified. Within the helical reconstruction the characteristic asymmetric actin monomer shape, subunit cleft and interstrand gap are easily identified in each subunit. The difference between the helical and single particle reconstruction is readily apparent, namely, the substantial extra density halfway along the single particle reconstruction (Fig. 2, A and C), which we attribute to troponin. The same atomic coordinate F-actin structure was docked into both the helical (Fig. 2I) and the single particle-based reconstruction (Fig. 2C). The F-actin fit to the single particle and helical reconstructions are virtually identical with the asymmetric shape of the actin subunit clearly identified in both maps (Fig. 2, C and I).
The orientation of the thin filament in the sarcomere is known. It is necessary to orient the three-dimensional reconstruction in this frame of reference to fully evaluate the biological significance of our results. The polarity of actin and the thin filament is often described by the directionality and arrow shape that the myosin heads provide with the barbed end of the filament situated at the Z-band of the sarcomere. This convention has been used in Figs. 2, 3, 4. The characteristic pattern of two-dimensional projections also indicates the filament polarity. The order of three distinct features of the filament detailed below characterizes the polarity of the underlying actin, as outlined in previous studies (42). Viewing an actin model filtered to 35 Å in a similar manner highlights the agreement between the atomic coordinates and the two-dimensional projections calculated from our helical reconstruction and the single particle full and half-data set reconstructions (supplemental Fig. S1, and Fig. 2, B and H). Subdomains 1 and 3 form a band of density running diagonally out from the center of the filament. If lines are drawn highlighting these subdomains, a mini arrow-like pattern is seen. It is noted that this is in the opposite direction to the arrow-like pattern seen when myosin subfragment-1 heads label actin. Two other strong features are visible in projection, the crossover point of the two long pitch strands and a slight gap between the strands. The crossover is the narrowest point of the projected image and is a high density bright area. The gap is surrounded by four subunits viewed from the side which results in four small high density bright regions. These features in the two-dimensional projection images (Fig. 2, B and H) support the orientation of the docked F-actin model in both of the three-dimensional reconstructions, allowing us to unambiguously determine the polarity of the data, in each case.
FIGURE 4.
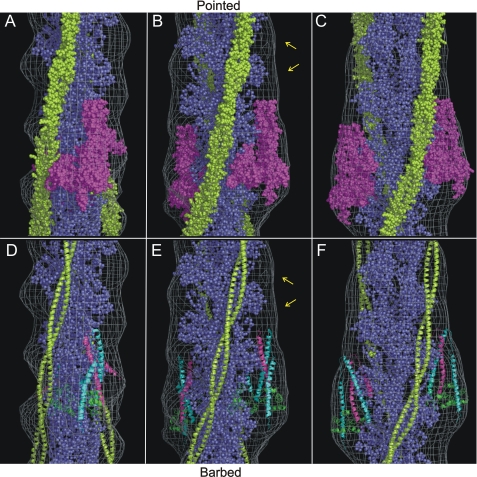
Docking of troponin and tropomyosin structures. The single particle three-dimensional map is shown as a wire mesh. The actin atomic model (38) has been docked into the three-dimensional map (Cα atoms in blue). The orientation of the map in C and F is consistent with Fig. 2, A–F. The map has been rotated by -80° about the filament axis in A and D and by -40° in B and E with respect to original position shown in C and F. A–C, space-filling model of the partial troponin crystal structure (4) (displayed in magenta, PDB code 1yv0), and the tropomyosin crystal structure (2) (displayed in yellow, PDB code 1c1g) are docked into the electron density envelope. D–F, troponin and tropomyosin displayed as a ribbon diagram: the three troponin components are displayed in different colors TnC, green; TnT2, pink; and TnI, cyan. Unassigned density (yellow arrows) running alongside tropomyosin can be seen in B and E.
Difference Density Analysis: Assignment of Troponin—The docked atomic co-ordinates of F-actin and F-actin with tropomyosin were used to create an electron density models. The F-actin model was then subtracted from the three-dimensional reconstruction to create a difference map which highlights all non-actin density (Fig. 2D). For comparison the equivalent difference density map was calculated between the helical reconstruction and the docked actin model (Fig. 2J). A second difference map was calculated using the F-actin tropomyosin density model subtracted from the single particle reconstruction. The density in the reconstruction with no counterpart in actin or tropomyosin is highlighted: this density can be assigned to troponin. A composite difference density map is shown in Fig. 2, E and F.
In the single particle reconstruction two bulbous areas of density label each strand of actin with a 27.5 Å axial stagger. The exact stagger was determined as part of the averaging procedure. Troponin has been described as a “comma-shaped” molecule (7, 43). This appearance is similar to the shape of the two regions of density we see in the reconstruction. The observed density is located halfway along the filament and tapers away toward the pointed end. Measurements of the filament width agree with this observation (Fig. 3B). We interpret the shape of the complex to mean that the globular part of troponin (TnI, TnC, and TnT2) is the extra mass in the central region of the filament and the tapering density running up the filament axis is the tail of troponin (TnT1). TnT1 is the Ca2+-invariant anchor point of troponin onto the thin filament through its binding of tropomyosin (44). In our interpretation, the tail of troponin runs toward the pointed end of the filament and is very closely associated with tropomyosin. In the currently accepted model of the thin filament the globular head is thought to bind closer to the pointed end of the actin filament with the tail pointing toward the barbed/Z-band end (45). Our present structural analysis of the thin filament has resulted in a three-dimensional reconstruction in which the natural interpretation of the observed troponin density is that the polarity of the troponin complex is reversed with respect to this accepted model.
Difference Density Analysis: Assignment of Tropomyosin—In addition to regions of bulbous density assigned to troponin, continuous strands of density running diagonally up the filament in the cleft of the actin subunit are visible in both the single particle and helical reconstructions (Fig. 2, E, F, and J). This density can be interpreted as tropomyosin and in the helical reconstruction tropomyosin and a smeared out troponin. Tropomyosin in both the helical and single particle reconstructions appears to have slight swellings that repeat with the same repeat as the actin subunits. We believe that the swellings and the general appearance of tropomyosin is due to the resolution of the maps. By docking the 7-Å crystal structure of tropomyosin (PDB code 1c1g) (2) into these regions of density in the single particle map we have traced the likely path of tropomyosin, which is seen to lie over subdomain 1 of actin. The average position of the docked tropomyosin in both the single particle and helical maps agrees with the blocked or B-state described by Refs. 9, 10.
Troponin Crystal Structure Orientation—A space-filling model of the skeletal troponin crystal structure in a Ca2+-free state (4) was manually positioned in the EM density envelope. Fig. 4 displays our favored orientation. The dimensions of the crystal structure match the size of the extra density region that we have ascribed to the globular head of the complex. The α-helical coiled-coil formed between TnI and TnT (the IT arm) runs approximately parallel to the filament axis. The N-terminal lobe of TnC is located over subdomain 1 of actin, resulting in TnI 96 and TnI 117 being almost equidistant from actin 374 at 44 Å and 47 Å, respectively. These are close to the distances (based on FRET measurements) used to create the published atomic model of the thin filament (5). The linker between the two domains of TnC is disordered in this structure. The inherent flexibility of this disordered region allowed us to incorporate a rotation of ∼30° of the N-terminal lobe relative to the rest of the crystal structure.
The “core” structure (4) contains ∼60% of the mass of the troponin complex. The missing regions include the C terminus of TnI (residues 144–181) and the whole of TnT1 (residues 1–159 of TnT). Docking the core structure and the present difference density analysis highlights areas that are not occupied. An area of density directly above the troponin core (Fig. 4, B and E), lying alongside tropomyosin, is an obvious location for TnT1 due to the Ca2+ invariant interactions known to occur between these two proteins (44). Additional unoccupied density appears to “arc” around the face of the actin subunit, which we propose could be the location of the C terminus of TnI also missing from the crystallized core structure. A connection between the two strands of actin via the C terminus of TnI has recently been reported (46).
DISCUSSION
We have used electron microscopy and single particle analysis to study the structure of the native muscle thin filament. The quality of imaging and filament preservation means that regions of density corresponding to the troponin complex were evident from the beginning of our analysis and used for the accurate identification of filament segments centered on the troponin density. An adapted single particle method (25) has been applied successfully to reveal the location and shape of troponin in relaxed thin filaments. This work represents the first reference-free structural analysis of the thin filament (i.e. with no prior assumptions with respect to the shape or location of troponin).
The actin backbone of the filament is well resolved in the single particle three-dimensional reconstruction: docking the atomic model of F-actin into the three-dimensional map emphasizes the agreement between the reconstruction and the model. The docked F-actin model and the docked tropomyosin crystal structure were used to calculate a difference structure in which mass present in the reconstruction with no counterpart in the F-actin or tropomyosin density is evident (Fig. 2, E and F). The three-dimensional map and difference structure reveal a significant accumulation of density halfway along the filament attributable to the troponin complex. The density assigned to individual troponin complexes appears bulbous at the barbed end of the filament and tapers along the filament toward the pointed end.
The exact stagger between pairs of troponin complexes was established to be 27.5 ± 0.5 Å confirming the 1972 model proposed by Ebashi (1). This arrangement allows the troponin and tropomyosin molecules to make equivalent interactions with actin on both strands. Why this stagger is 27.5 Å as opposed to alternatives such as a stagger of 3 × 27.5 Å is not yet clear. One possibility is that as the thin filament assembles the positions of the first two troponin molecules on opposite strands may be related to pseudo 2-fold interactions with Z-band components. Alternatively, binding of the first troponin could make a local conformational change in the actin subunits, which promotes the binding of the next troponin complex 27.5 Å away on the opposite strand. Subsequent troponin complexes would label the filament at this regular spacing and if binding occurred in the wrong place it might be weak or may result in an unstable filament structure consequently unraveling the filament.
The troponin complex is highly asymmetric and has a specific polarity when bound to the thin filament. The widely accepted orientation of troponin is such that the rod-like tail of the complex is closer to the Z-band than the globular head. Our single particle based reconstruction exhibits an asymmetric arrangement of troponin density. Docking the F-actin model allows us to establish the orientation of our reconstruction. However, this orientation places the widest region of troponin closest to the Z-band and troponin tapers off in the direction of the M-band. This suggests that the rod-like tail of troponin, which we now know to consist mainly of TnT1, is positioned closer to the M-band than the Z-band. A similar interpretation of troponin polarity can also be inferred from the shape of the thin filament in an electron tomogram of fast frozen stretched rigor insect flight muscle fibers (47). We consider below whether this reverse polarity, as observed in our reconstruction, can be reconciled with the earlier observations.
Strong evidence exists for the polarity of tropomyosin on the actin filament. Tropomodulin binds tropomyosin and caps the actin filament at the M-band end (48). This information combined with the identification of tropomodulin binding site at the N-terminal region of tropomyosin provides a polarity for tropomyosin in the thin filament (49). Evidence for two well-separated binding sites of troponin on tropomyosin is also well documented. The globular head region of troponin has been localized to a region spanning residues 150–180 of tropomyosin (50) and in the vicinity of Cys-190 (51, 52). The tail region of troponin is known to bind to the C-terminal region of tropomyosin (50–52). However, there is no direct evidence from these studies showing whether the tail of troponin binds in an anti-parallel or parallel manner to tropomyosin, although the anti-parallel arrangement has generally been assumed (7). Individual troponin subfragments were labeled (45) with antibodies to TnT1, T2, C, and I in electron microscope images of isolated assemblies containing I-bands and Z-bands. This gave rise to the idea that TnT1 is closer to the Z-band than TnT2. Antibody striations are evident in the micrographs. However, subsequent work with Fab fragments disagreed with the initial findings (43). Here it was reported that the staining of TnT1 and TnT2 is in the same position and that possible discrepancies may be due to the size of the antibody. The estimated length of the troponin complex also increased considerably from 10 nm to 26.5 nm over the period 1968–1988. The initial length measurements were taken from paracrystal data (53, 54) and later direct measurements were made on rotary shadowed samples (7, 43). This 2-fold increase in length could result in a different interpretation of the early immunoelectron microscopy data, and possibly a reversal in the orientation of troponin on the actin filament. The geometry of binding of TnT to troponin has been investigated in double diamond co-crystals of TnT and tropomyosin (55, 56). Despite the relatively similar appearance of the crystals, different interpretations have been drawn over the exact arrangement of the component proteins. Because the distance between identical nodes of the crystal is ∼400 Å (the length of tropomyosin) Carr et al. (56) concluded that tropomyosin runs between these identical nodes: as previously observed in the pure tropomyosin Bailey crystals. The N-terminal region of TnT was interpreted as being located at the acute vertex of the double diamond which corresponded with the C-terminal of tropomyosin. The mutant TnT used by Cabral-Lily (55) lacked the N-terminal region. As a result it was suggested that the C-terminal of TnT must bind at the acute vertex of the diamond, and the N-terminal region of TnT extended into the long-arm of the large diamond to the C-terminal of tropomyosin. This arrangement is possible if the head to tail overlap of two tropomyosin molecules is placed in the middle of the long arm and not at the nodes of the crystal as favored by Carr et al. (56). Nevertheless the polarity of tropomyosin in the double diamond co-crystals has not been established directly, so the relative orientation of TnT and tropomyosin cannot be ascertained from these studies.
These apparent inconsistencies bring us to question the evidence behind the previously accepted model of the thin filament leading us to propose a reversed polarity of the troponin complex. The reversed polarity would imply a parallel arrangement for the interaction between TnT1 and tropomyosin in which troponin would retain its established interaction with the C terminus of tropomyosin while spanning the region between the N terminus and residue 190 of the adjacent tropomyosin molecule. The importance of a reversal in polarity would be paramount when considering the mechanism of regulation. The conformational changes that occur within troponin and the thin filament when Ca2+ binds to TnC hold the key to understanding regulation of muscle contraction. Without locating directly the troponin subunits on actin in both the high and low Ca2+ states we cannot begin to reveal the mechanism.
Our novel analysis methods have resulted in two-dimensional class averages and a three-dimensional reconstruction with clear contributions from troponin and tropomyosin. The region of density that we attribute to troponin in our reconstruction is significantly larger and more continuous than the volume indicated in the 2001 Narita map (22). Their map could be enclosed in a cylinder with a diameter of 120 Å, whereas the diameter of the reconstruction presented here at its widest point is ∼141 Å. A model-based single particle approach was taken by Pirani et al. in 2005 (23), and the final three-dimensional map strongly resembles the initial model with an additional area of density which was interpreted as the C terminus of TnI. Although these observations provide some support for their conclusion, it would be more evident that model bias had been avoided if in each case (many orientations of the troponin complex were tested) the same end result was obtained: this apparently was not observed. The differences between the image processing methods used in our studies and previous analyses of the structure (22, 23) offer some explanation for a number of the differences in our conclusions. In particular the objective reference-free alignment procedure used to obtain our first reconstruction.
Many issues remain unaddressed concerning the structure of the thin filament. High resolution EM structures of the filament at high and low Ca2+ are necessary to allow accurate molecular models to be built. The existing crystal structures of troponin are incomplete and previous EM reconstructions have not identified the missing regions. The important role of the 40% of troponin that remains un-crystallized is becoming apparent. As well as the widely reported anchoring function of TnT1, the activation of actomyosin ATPase activity is reported to be mediated by a direct interaction between TnT1, tropomyosin, and actin (57). The number of mutations in ∼TnT1 identified as causing heart disease is also increasing (58). We need to locate the exact position of these mutations to begin to understand their pathophysiology. We have identified the likely path of the troponin tail in our reconstruction. In future work we aim to identify unambiguously the position of TnT1 and all the components in the thin filament at the molecular level.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Minorities Basic Research Support Grant S06 GM08224 (to R. W. K.) and a Research Centers in Minority Institutions Award G12RR-03051 from the NCRR. This work was also supported by a BHF studentship (13962, to D. P.), a BHF Project Grant 23480 (to E. P. M. and J. M. S.), a Wellcome Trust Grant 066418 (to E. P. M.), and the European MYORES Network on Muscle Development (to J. M. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: EM, electron microscopy; TnC, Ca2+-binding subunit of troponin; TnI, inhibitory subunit of troponin; TnT, tropomyosin-binding subunit of troponin; PDB, Protein Data Bank.
References
- 1.Ebashi, S. (1972) Nature 240 217-218 [DOI] [PubMed] [Google Scholar]
- 2.Whitby, F. G., and Phillips, G. N., Jr. (2000) Proteins 38 49-59 [PubMed] [Google Scholar]
- 3.Holmes, K. C., Popp, D., Gebhard, W., and Kabsch, W. (1990) Nature 347 44-49 [DOI] [PubMed] [Google Scholar]
- 4.Vinogradova, M. V., Stone, D. B., Malanina, G. G., Karatzaferi, C., Cooke, R., Mendelson, R. A., and Fletterick, R. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5038-5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirani, A., Vinogradova, M. V., Curmi, P. M., King, W. A., Fletterick, R. J., Craig, R., Tobacman, L. S., Xu, C., Hatch, V., and Lehman, W. (2006) J. Mol. Biol. 357 707-717 [DOI] [PubMed] [Google Scholar]
- 6.Takeda, S., Yamashita, A., Maeda, K., and Maeda, Y. (2003) Nature 424 35-41 [DOI] [PubMed] [Google Scholar]
- 7.Flicker, P. F., Phillips, G. N., Jr., and Cohen, C. (1982) J. Mol. Biol. 162(2), 495-501 [DOI] [PubMed] [Google Scholar]
- 8.Brown, J. H., and Cohen, C. (2005) Adv. Protein Chem. 71 121-159 [DOI] [PubMed] [Google Scholar]
- 9.Lehman, W., Craig, R., and Vibert, P. (1994) Nature 368 65-67 [DOI] [PubMed] [Google Scholar]
- 10.Lehman, W., Vibert, P., Uman, P., and Craig, R. (1995) J. Mol. Biol. 251 191-196 [DOI] [PubMed] [Google Scholar]
- 11.Squire, J. M., and Morris, E. P. (1998) Faseb. J. 12 761-771 [DOI] [PubMed] [Google Scholar]
- 12.van Heel, M. (1984) Ultramicroscopy 13 165-183 [DOI] [PubMed] [Google Scholar]
- 13.van Heel, M. (1989) Optik 82 114-126 [Google Scholar]
- 14.van Heel, M. (1987) Ultramicroscopy 21 111-123 [DOI] [PubMed] [Google Scholar]
- 15.Goncharov, A. B., and Gefland, M. S. (1988) Ultramicroscopy 25 317-328 [Google Scholar]
- 16.Radermacher, M. (1994) Ultramicroscopy 53 121-136 [DOI] [PubMed] [Google Scholar]
- 17.Radermacher, M. (1988) J. Electron. Microsc. Tech. 9 359-394 [DOI] [PubMed] [Google Scholar]
- 18.Harauz, G., and van Heel, M. (1986) Optik 73 146-156 [Google Scholar]
- 19.Egelman, E. H. (2000) Ultramicroscopy 85 225-234 [DOI] [PubMed] [Google Scholar]
- 20.Woodhead, J. L., Zhao, F. Q., Craig, R., Egelman, E. H., Alamo, L., and Padron, R. (2005) Nature 436 1195-1199 [DOI] [PubMed] [Google Scholar]
- 21.Al-Khayat, H. A., Morris, E. P., Kensler, R. W., and Squire, J. M. (2006) J. Struct. Biol. 155 202-217 [DOI] [PubMed] [Google Scholar]
- 22.Narita, A., Yasunaga, T., Ishikawa, T., Mayanagi, K., and Wakabayashi, T. (2001) J. Mol. Biol. 308 241-261 [DOI] [PubMed] [Google Scholar]
- 23.Pirani, A., Xu, C., Hatch, V., Craig, R., Tobacman, L. S., and Lehman, W. (2005) J. Mol. Biol. 346 761-772 [DOI] [PubMed] [Google Scholar]
- 24.Burgess, S., Walker, M., Knight, P. J., Sparrow, J., Schmitz, S., Offer, G., Bullard, B., Leonard, K., Holt, J., and Trinick, J. (2004) J. Mol. Biol. 341 1161-1173 [DOI] [PubMed] [Google Scholar]
- 25.Paul, D., Patwardhan, A., Squire, J. M., and Morris, E. P. (2004) J. Struct. Biol. 148 236-250 [DOI] [PubMed] [Google Scholar]
- 26.Kensler, R. W., and Stewart, M. (1989) J. Cell Sci. 94 391-401 [DOI] [PubMed] [Google Scholar]
- 27.Kocsis, E., Trus, B. L., Steer, C. J., Bisher, M. E., and Steven, A. C. (1991) J. Struct. Biol. 107 6-14 [DOI] [PubMed] [Google Scholar]
- 28.van Heel, M., Gowen, B., Matadeen, R., Orlova, E. V., Finn, R., Pape, T., Cohen, D., Stark, H., Schmidt, R., Schatz, M., and Patwardhan, A. (2000) Q Rev. Biophys. 33 307-369 [DOI] [PubMed] [Google Scholar]
- 29.Patwardhan, A., Paul, D., Al-Khayat, H. A., and Morris, E. P. (2004) J. Mol. Biol. 344 707-724 [DOI] [PubMed] [Google Scholar]
- 30.Wriggers, W., Milligan, R. A., and McCammon, J. A. (1999) J. Struct. Biol. 125 185-195 [DOI] [PubMed] [Google Scholar]
- 31.Navaza, J., Lepault, J., Rey, F. A., Alvarez-Rua, C., and Borge, J. (2002) Acta Crystallogr D Biol. Crystallogr 58 1820-1825 [DOI] [PubMed] [Google Scholar]
- 32.Stokes, D. L., and DeRosier, D. J. (1987) J. Cell Biol. 104 1005-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egelman, E. H., and DeRosier, D. J. (1992) Biophys. J. 63 1299-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M., and Leith, A. (1996) J. Struct. Biol. 116 190-199 [DOI] [PubMed] [Google Scholar]
- 35.Beckmann, R., Bubeck, D., Grassucci, R., Penczek, P., Verschoor, A., Blobel, G., and Frank, J. (1997) Science 278 2123-2126 [DOI] [PubMed] [Google Scholar]
- 36.Bottcher, B., Wynne, S. A., and Crowther, R. A. (1997) Nature 386 88-91 [DOI] [PubMed] [Google Scholar]
- 37.van Heel, M., and Schatz, M. (2005) J. Struct. Biol. 151 250-262 [DOI] [PubMed] [Google Scholar]
- 38.Frank, J., and Agrawal, R. K. (1998) Biophys. J. 74 589-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouche, F., Zhu, Y., Pulokas, J., Potter, C. S., and Carragher, B. (2003) J. Struct. Biol. 144 301-312 [DOI] [PubMed] [Google Scholar]
- 40.Mendelson, R. A., and Morris, E. (1994) J. Mol. Biol. 240 138-154 [DOI] [PubMed] [Google Scholar]
- 41.Moore, P. B., Huxley, H. E., and DeRosier, D. J. (1970) J. Mol. Biol. 50 279-295 [DOI] [PubMed] [Google Scholar]
- 42.Bremer, A., Henn, C., Goldie, K. N., Engel, A., Smith, P. R., and Aebi, U. (1994) J. Mol. Biol. 242 683-700 [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuki, I., Onoyama, Y., and Shiraishi, F. (1988) J. Biochem. (Tokyo) 103 913-919 [DOI] [PubMed] [Google Scholar]
- 44.Greaser, M. L., and Gergely, J. (1973) J. Biol. Chem. 248 2125-2133 [PubMed] [Google Scholar]
- 45.Ohtsuki, I. (1979) J. Biochem. (Tokyo) 86 491-497 [DOI] [PubMed] [Google Scholar]
- 46.Galinska-Rakoczy, A., Engel, P., Xu, C., Jung, H., Craig, R., Tobacman, L. S., and Lehman, W. (2008) J. Mol. Biol. 379 929-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, J., Reedy, M. C., Goldman, Y. E., Franzini-Armstrong, C., Sasaki, H., Tregear, R. T., Lucaveche, C., Winkler, H., Baumann, B. A., Squire, J. M., Irving, T. C., Reedy, M. K., and Taylor, K. A. (2004) J. Struct. Biol. 147 268-282 [DOI] [PubMed] [Google Scholar]
- 48.Gregorio, C. C., Weber, A., Bondad, M., Pennise, C. R., and Fowler, V. M. (1995) Nature 377 83-86 [DOI] [PubMed] [Google Scholar]
- 49.Greenfield, N. J., and Fowler, V. M. (2002) Biophys. J. 82 2580-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, S. P., Cohen, C., and Phillips, G. N., Jr. (1987) Nature 325 826-828 [DOI] [PubMed] [Google Scholar]
- 51.Mak, A. S., and Smillie, L. B. (1981) J. Mol. Biol. 149 541-550 [DOI] [PubMed] [Google Scholar]
- 52.Pearlstone, J. R., and Smillie, L. B. (1982) J. Biol. Chem. 257 10587-10592 [PubMed] [Google Scholar]
- 53.Ohtsuki, I. (1974) J Biochem. (Tokyo) 75 753-765 [DOI] [PubMed] [Google Scholar]
- 54.Ebashi, S., Ohtsuki, I., and Mihashi, K. (1972) Cold Spring Harb. Symp. Quant Biol. 37 215-223 [Google Scholar]
- 55.Cabral-Lilly, D., Tobacman, L. S., Mehegan, J. P., and Cohen, C. (1997) Biophys. J. 73 1763-1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carr, H. J., O'Brien, E. J., and Morris, E. P. (1988) J. Muscle Res. Cell Motil. 9 384-392 [DOI] [PubMed] [Google Scholar]
- 57.Oliveira, D. M., Nakaie, C. R., Sousa, A. D., Farah, C. S., and Reinach, F. C. (2000) J. Biol. Chem. 275 27513-27519 [DOI] [PubMed] [Google Scholar]
- 58.Gomes, A. V., Barnes, J. A., Harada, K., and Potter, J. D. (2004) Mol Cell Biochem. 263 115-129 [DOI] [PubMed] [Google Scholar]
- 59.Crowther, R. A., De Rosier, D. J., and Klug, A. (1970) Proc. Roy. Soc. London A. 317 319-340 [Google Scholar]
- 60.Crowther, R. A., Henderson, R., and Smith, J. M. (1996) J. Struct. Biol. 116 9-16 [DOI] [PubMed] [Google Scholar]
- 61.DeLano, W. L. (2002) The PyMol Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.