Abstract
The amyloid precursor protein (APP) is implied both in cell growth and differentiation and in neurodegenerative processes in Alzheimer disease. Regulated proteolysis of APP generates biologically active fragments such as the neuroprotective secreted ectodomain sAPPα and the neurotoxic β-amyloid peptide. Furthermore, it has been suggested that the intact transmembrane APP plays a signaling role, which might be important for both normal synaptic plasticity and neuronal dysfunction in dementia. To understand APP signaling, we tracked single molecules of APP using quantum dots and quantitated APP homodimerization using fluorescence lifetime imaging microscopy for the detection of Förster resonance energy transfer in living neuroblastoma cells. Using selective labeling with synthetic fluorophores, we show that the dimerization of APP is considerably higher at the plasma membrane than in intracellular membranes. Heparan sulfate significantly contributes to the almost complete dimerization of APP at the plasma membrane. Importantly, this technique for the first time structurally defines the initiation of APP signaling by binding of a relevant physiological extracellular ligand; our results indicate APP as receptor for neuroprotective sAPPα, as sAPPα binding disrupts APP dimers, and this disruption of APP dimers by sAPPα is necessary for the protection of neuroblastoma cells against starvation-induced cell death. Only cells expressing reversibly dimerized wild-type, but not covalently dimerized mutant APP are protected by sAPPα. These findings suggest a potentially beneficial effect of increasing sAPPα production or disrupting APP dimers for neuronal survival.
The amyloid precursor protein (APP)4 is known both for its important role in the development and plasticity of the nervous system (1–6) and for its involvement in Alzheimer disease (AD) (7, 8). Despite intensive research efforts, the initial events that lead to the prevalent sporadic, i.e. non-familial, forms of AD are still unclear. Furthermore, although a higher gene dose of APP (9) or the presence of pathological APP mutations is sufficient to induce familial AD (for review, see Ref. 10), the exact pathological mechanism that is triggered by APP is still under debate.
Some fragments of APP, such as the β-amyloid peptide (Aβ), are thought to contribute to synaptic dysfunction and neurotoxicity (11, 12). On the other hand, the α-secretase-derived extracellular fragment of APP (sAPPα), which is present at lower levels in AD patients than in controls (13), has been shown to be beneficial for memory function, to possess neuroprotective properties, and to counteract the effects of Aβ (14–18).
Signaling by transmembrane APP may directly contribute to neurodegeneration in AD (19–24); however, the signal transduction pathway for transmembrane APP remains unknown, although several potential regulatory proteins, glycosaminoglycans, and metal ions are known to bind with high affinity to APP and sAPPα (25, 26). The most common form of signal transduction for single-pass transmembrane proteins is the ligand-induced perturbation of a monomer/dimer equilibrium. Indeed, the dimerization of transmembrane APP has been implied several times in the past. Several studies have investigated the effects of presumed dimer-breaking perturbations on biological read-outs, such as the production of Aβ (27, 28), but without directly measuring the APP aggregation state, or have investigated the aggregation state of APP subdomains, often reconstituted in cell-free systems (27–32). Dimerization interfaces in both the extracellular and the transmembrane domain have been suggested.
In the studies investigating the aggregation state of full-length APP, most of the employed methods, such as chemical cross-linking and co-immunoprecipitation, do not lend themselves readily to a rigorous quantitative analysis of the abundance of potentially instable dimers (31, 33), whereas in other cases the use of chimeras may have influenced the dimerization potential or precluded the search for a natural stimulus (23, 34). The only previously reported direct observation of APP dimerization by Förster resonance energy transfer (FRET) microscopy uses an assay in which the FRET efficiency varies with the level of overexpression (35). Therefore, a concentration-dependent FRET component due to nonspecific stochastic encounters cannot be excluded in this study.
Most importantly, as none of the published procedures permitted the selective detection of APP dimers on the surface of live cells, where they would encounter ligands, they could not differentiate between subpopulations of APP. This may be one reason why no natural ligand of APP has ever been shown to signal via modulation of its monomer/dimer equilibrium.
Another elusive goal is the identity of the receptor for neuroprotective sAPPα (36–39). The ligand-dependent dimerization of sAPPα in solution (40) and its origination from transmembrane APP suggest that APP might serve as receptor for sAPPα, but this binding has never been experimentally shown.
EXPERIMENTAL PROCEDURES
Plasmids—pcDNA3-mGFP was obtained by cloning monomerized enhanced GFP (L222K mutation (41) produced from pEGFP-N1 (Clontech, Mountain View, CA) using QuikChange (Stratagene, La Jolla, CA)), flanked by NotI and XhoI primers, into the NotI and SalI sites of pcDNA3 (all restriction enzymes were from New England Biolabs, Ipswich, MA). APP-mGFP was derived by inserting APP695 into the EcoRV and NotI sites of pcDNA3-mGFP; an AAAG linker connects the C terminus of APP to the N terminus of mGFP. For substitution of the fluorophore, mGFP was excised using NotI and ApaI; mCherry ((42), a kind gift of S. Eimer, European Neuroscience Institute Göttingen) or REACh2 (43) was amplified using NotI and ApaI primers and inserted in its place. The K624C mutants were produced by using QuikChange on the XhoI-NotI fragment of APP-mGFP and inserting the mutant fragment back into a wild-type vector (K624C-APP-mGFP and K624C-APP-mCherry). APP-ACP-mGFP and K624C-APP-ACP-mGFP were obtained by silent mutation of APP695 codons 294–295 in the EcoRV-XhoI fragment of APP-mGFP to introduce an NheI site (GCCAGT to GCTAGC) and insertion of acyl carrier protein (ACP; Covalys Biosciences AG, Witterswil, Switzerland), and amplified using NheI-encoding forward and backward primers into the corresponding site thus created. ACP was, therefore, inserted in a similar position as the Kunitz protease inhibitor domain, which occurs in non-neuronal isoforms of APP. The EcoRV-XhoI fragment was then inserted into either wild-type APP-mGFP or K624C-APP-mGFP. To eliminate mGFP from these vectors, wild-type APP-ACP and K624C-APP-ACP were obtained by digestion with NotI and PspOMI followed by religation. The entire open reading frames of all constructs were confirmed by sequencing (SeqLab, Göttingen, Germany).
Cell Culture—B103 neuroblastoma cells were a kind gift from Dr. David Schubert (Salk Institute). APP or APP-like protein 2 cannot be detected in these cells (44). Cells were plated at 105 cells/well in Dulbecco's modified Eagle's medium + 10% fetal calf serum on poly-l-ornithine-coated glass coverslips in 24-well plates (Corning Life Sciences, Lowell, MA) and transfected using 0.2–0.8 μg of plasmid DNA per well and magnetic nanoparticle MaTra beads on a 24-magnet plate (beads and magnet from IBA GmbH, Göttingen, Germany). The medium was changed 1–2 h after transfection, and expression was allowed to proceed for a further 16–24 h. MaTra transfection was superior to several liposomal transfection methods in replicating the expression pattern of endogenous APP and in maintaining cell morphology (data not shown). For mGFP imaging, coverslips were washed by a short immersion in phosphate-buffered saline and mounted on a drop of imaging buffer (135 mm NaCl, 10 mm KCl, 0.4 mm MgCl2, 2 mm CaCl2, 10 mm HEPES, 0.1% bovine serum albumin, 20% glucose) using a homemade silicone rubber gasket on a glass slide. For ACP imaging, coverslips were incubated for 20 h at 16 °C in a humidified atmosphere in imaging buffer containing 10 mm MgCl2, 1 μm wild-type ACP synthase (purified according to instructions from Covalys Biosciences AG, Witterswil, Switzerland), and CoA dyes (Covalys) at a concentration of 1 μm CoA-488 and 3 μm CoA-547, where indicated. They were then thoroughly washed twice by immersion in phosphate-buffered saline and draining off the excess liquid and mounted in imaging buffer as for mGFP imaging. Where indicated, 1 μm recombinant human sAPPα695 or sAPPa770 (45) was added to the imaging buffer after removal of the dyes. The measurements started ∼10 h after the addition of sAPPα to the cells. To remove cell surface heparin, cells were incubated for 2 h with 2 units/ml heparinase I (Sigma-Aldrich) in complete medium before labeling; heparinase was also present during labeling with CoA dyes.
For quantum dot experiments, 1 μm CoA coupled to biotin was used. After washing, the cells were incubated with the indicated concentration of streptavidin-coated 585-nm and/or 655-nm emitting quantum dots (Invitrogen) (46).
In survival experiments, B103 cells were transfected with 0.2 μg of a yellow fluorescent protein construct (TN-XXL, a kind gift of Oliver Griesbeck, MPI for Biochemistry, Munich) and 0.2 μg of either pcDNA3, pcDNA3-APP, or pcDNA3-K624C-APP per well using MaTra. 6 h after transfection wells were washed once with phosphate-buffered saline and filled either with imaging buffer or with imaging buffer containing 16 nm sAPPα770. In some experiments 24 h after transfection another 16 nm were added to the same wells.
Microscopy—Fluorescence lifetime imaging measurements were performed on a frequency domain setup (47) using an argon laser (Innova 304C, Coherent, Dieburg, Germany) at 488 nm (mGFP) or 496 nm (CoA-488). The presence of other fluorophores was confirmed using mercury lamp excitation. All filter cubes were from AHF (Tübingen, Germany) with the following components: mGFP dichroic LP495, emission BP 515/30; CoA-488 dichroic LP515, emission BP 535/30; CoA-547 excitation BP 545/30, dichroic LP565, emission BP615/75; mCherry excitation BP530–585, dichroic LP600, emission BP615. mGFP imaging was performed at 22 °C; ACP imaging was performed at 16 °C to slow down internalization of the labeled APP, necessary to image APP at the cell surface. Eight-phase step images were acquired in random order. Confocal microscopy was performed at room temperature on a Zeiss LSM-510 Meta instrument using laser excitation at 488 or 633 nm and a 63× water immersion objective or on a programmable array microscope fitted on an Olympus IX71 microscope using laser excitation at 488 nm and a 150× oil immersion objective (48). Quantum dot tracking was performed using the View5D plugin (Rainer Heintzmann, King's College, London) for ImageJ (National Institutes of Health), and diffusion coefficients were calculated from the exported coordinates as described before (49).
Statistical Analysis—In mGFP images, the optimal threshold was set automatically (opthr module for MatLab by F. T. Marti). All further operations were performed with custom-written routines (Alessandro Esposito and M. Gralle) in MatLab (Math-Works, Natick, MT). ACP images were manually threshold to the smallest mask that includes the entire cell body; a contour mask was then applied that extended 10 pixels inward from the boundaries of the threshold mask to isolate the plasma membrane. The lifetime and total intensity values of each pixel in the threshold mask (mGFP) or contour mask (ACP) were stored together with an estimate of the background fluorescence (median intensity of the pixels outside the threshold mask). For each pixel, the signal to noise ratio (SNR) was calculated as the quotient of its total intensity over the background fluorescence in the image. SNRs and apparent lifetimes for all pixels in the appropriate masks of all cells were joined for each experimental condition and imported into IgorPro (WaveMetrics, Lake Oswego, Oregon) for fitting.
ACP Regression Analysis—As the ACP labeling creates a small but non-negligible nonspecific background fluorescence, the lifetime of specifically labeled APP (τ) and the nonspecific background (τb) for each condition were estimated by fitting SNR and apparent lifetimes (τapp) in all pixels to
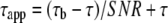 |
(Eq. 1) |
The parameter τb was linked in a global fit across all experimental conditions, returning a value of 3.55 ns. Histograms show the corrected lifetimes, obtained by inversion of (1),
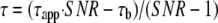 |
(Eq. 2) |
As the mean of the distribution of corrected lifetimes is independent of SNR but its width increases with lower SNR (data not shown), only pixels with a signal-to-noise >4 were included in the histograms for display purposes. These histograms were fitted to a Gaussian distribution using the IgorPro inbuilt function.
RESULTS
We investigated the oligomerization state of APP in living B103 neuroblastoma cells by measuring FRET (50) using fluorescence lifetime imaging microscopy (51). This technique is able to detect the association of proteins in their natural, unperturbed environment. The association of the donor with the acceptor fluorophore is reflected in a characteristic reduction in the excited state lifetime of the donor. In our initial experiments, we used APP-mGFP as donor and APP-mCherry as acceptor. APP-mGFP displays a largely vesicular staining localized predominantly to perinuclear membranes, most likely the secretory compartments (endoplasmic reticulum and Golgi apparatus), which is identical to the distribution of APP in cells that express it endogenously (52) (Fig. 1, A, B, and E). APP-mCherry localizes to the same cell compartments (Fig. 1F). The excited state lifetime of the mGFP fluorophore in APP-mGFP in the absence of acceptor is homogeneous, with a mean lifetime of 2.37 ± 0.01 ns (Fig. 1D). In the presence of APP-mCherry, however, the lifetime of APP-mGFP decreases to a mean value of 2.17 ± 0.01 ns, corresponding to a FRET efficiency of 8.3 ± 0.6% (Fig. 1G). This indicates that a portion of the co-localizing APP-mGFP and APP-mCherry molecules is closer together than 10 nm, i.e. physically interacts (50). Coexpression of APP-mGFP with APP-REACh2 (43) resulted in similar results (data not shown). It is important to note that much less FRET was observed in formaldehyde-fixed cells. This suggests that the association of APP molecules is labile and can easily be disturbed and disrupted by changes in the membrane environment. This apparent instability could explain why only very low amounts of dimerized APP are observed in cell lysates (31).
FIGURE 1.
Investigation of APP dimerization using APP-mGFP. A, confocal image of a B103 cell expressing APP-mGFP. B–G, wide-field images of B103 cells expressing APP-mGFP alone (B–D) or in combination with APP-mCherry (E–G). B and E, mGFP channel. C and F, mCherry channel. D and G, mGFP lifetime. Scale bars: 10 μm. H, histograms of FRET efficiencies in different experimental conditions. Histograms (thin lines) were fitted to Gaussian curves (thick lines). APP donor only, 36 cells; APP FRET, 48 cells; K624C-APP donor only, 27 cells; K624C-APP FRET, 21 cells. PDF, probability density function.
To allow an interpretation of FRET efficiency in terms of degree of association, we employed a standard that is quantitatively dimerized. The K624C mutant of APP has been shown to form disulfide-bridged dimers and to dimerize quantitatively (31). Furthermore, as the B103 cell line does not contain detectable levels of endogenous APP or related APP-like proteins (44), it can be assumed that APP-mCherry only competes with other APP-mGFP molecules for binding to APP-mGFP. Mutant APP-mGFP displayed a mean lifetime of 2.06 ± 0.02 ns in the presence of mutant acceptor APP-mCherry, corresponding to a FRET efficiency of 13.1 ± 1.0% (Fig. 1H). The FRET efficiency of wild-type APP corresponds to ∼65% of the quantitatively dimerized mutant. Because the fluorophores are expected to undergo little orientation restriction at the ends of the unstructured cytoplasmic domain of APP (53), it is reasonable to assume that the presence of the mutation in the transmembrane domain of APP does not noticeably affect the FRET coupling of the fluorophores. Moreover, as the proportion of APP-mGFP to APP-mCherry is the same for both mutant and wild-type forms, the degree of association is proportional to the FRET efficiency. It can, therefore, be concluded that wild-type cellular APP is 35% monomeric (Figs. 1H and 3), with the rest being dimers, as suggested from x-ray diffraction and scattering studies of APP fragments (32, 40) or higher oligomers.
FIGURE 3.
Proportion of dimerized APP in different experimental conditions. Left, total cellular APP (cf. Fig. 1). Right, cell surface APP (cf. Fig. 2). Black, quantitatively dimerized K624C mutant of APP, normalized to 100%. Dark gray, K624C-APP in the presence of sAPPα. Light gray, wild-type APP in the absence of biological ligands. White, wild-type APP in the presence of sAPPα. Error bars indicate the S.E. (left) or the error of fit (right). *, p < 0.05; **, p < 0.001.
Although intracellular APP may already be cleaved to biologically active fragments (54, 55) and may serve additional roles (56), only plasmalemmal APP can act as receptor for extracellular molecules or metal ions. The strong signal of intracellular APP-mGFP prevented us from detecting an effect for several extracellular ligands on the aggregation state of APP (Fig. 3 and data not shown). The quantity of cell surface APP is tightly regulated by its very rapid turnover (57) and varies considerably between different cell types and in different activation states, independently of the total quantity of cellular APP (58, 59). The reason for this tight regulation might be that the activation of cell surface APP is toxic under certain circumstances (19, 20, 22–24). It is, therefore, essential to address the aggregation state of cell surface APP separate from total cellular APP.
We did not succeed in isolating the very faint fluorescence lifetime imaging microscopy signal of plasma membrane APP-mGFP. The use of antibodies on living cells may have a clustering effect on the antigen or influence their internalization; furthermore, the FRET efficiency of dyes coupled to anti-APP antibodies was too low for the reliable quantification of its oligomerization behavior, probably because of the large inter-antibody distances involved (60). Therefore, we decided to use a recently described site-directed biolabeling approach that selectively and covalently labels cell-surface proteins carrying an ACP domain (61, 62). Using this system, it was indeed possible to selectively measure the signal of cell-surface APP-ACP even in the presence of much larger quantities of unlabeled intracellular APP-ACP (Fig. 2, A–D). The lifetime of specifically labeled APP-ACP was derived from the apparent lifetime by regression analysis of the fluorescence intensity (Fig. 2K; see the “Experimental Procedures” for details on the regression). This procedure eliminated the very low, but a non-negligible contribution of unbound or non-specifically bound CoA-488 and permitted the use of all plasma membrane pixels, even those at extremely low APP concentrations. Co-labeling of membrane-exposed APP-ACP with a mixture of CoA-488 donor and CoA-547 acceptor dyes establishes the FRET couple for the detection of APP dimers. An advantage of this technique is that the dye-labeled FRET partners can be “clamped” to a known and invariant stoichiometry, improving the comparability between different conditions.
FIGURE 2.
Investigation of cell surface APP dimers using APP-ACP. A–D, confocal image of a B103 cell expressing APP-ACP-mGFP and labeled with CoA-biotin and 1 nm quantum dots (QD655). A, mGFP fluorescence; B, QD655-labeled APP fluorescence. C, phase contrast image; untransfected cells are devoid of quantum dot labeling. D, overlay of images A and B. Note the selective plasma membrane staining by CoA-647-labeled APP-ACP (red) compared with APP-mGFP (green). E–J, wide-field images of APP-ACP-expressing B103 cells labeled with CoA-488 alone (E–G) or in combination with CoA-547 (H–J). The focal plane was chosen to give the strongest signal from the plasma membrane, and therefore, does not include neurites. E and H, CoA-488 channel. F and I, CoA-547 channel. G and J, lifetime of CoA-488-labeled APP-ACP. Scale bars:10 μm. K, regression analysis of the fluorescence lifetime of CoA-488-labeled APP-ACP-expressing B103 cells. The lifetime of each pixel was multiplied with its SNR and plotted against SNR. The slope of the linear fit gives the background-corrected lifetime (see “Experimental Procedures”). Different experimental conditions were vertically displaced for easier display, and parallels (broken lines) to the APP donor-only fit (red) were drawn to facilitate comparison between the fits. Black, APP donor-only; orange, APP FRET exposed to sAPPα; green, APP FRET; red, K624C-APP FRET. L, histograms of FRET efficiencies in different experimental conditions. Histograms (thin lines) were fitted to Gaussian curves (thick lines). APP donor-only, 55 cell; APP FRET, 98 cells; APP FRET + sAPPα, 35 cells; K624C-APP donor-only, 29 cells; K624C-APP FRET, 50 cells. PDF, probability density function.
The lifetime distribution, thus, derived from CoA-488-labeled APP-ACP in the absence of acceptor was homogeneous, with a mean value of 3.31 ± 0.02 ns (Fig. 2, E–G), but it was reduced considerably in the presence of the FRET acceptor CoA-547-labeled APP-ACP (Fig. 2, H–J); the mean value of 2.97 ± 0.02 ns corresponds to a FRET efficiency of 10.2 ± 0.6%. The covalent dimer K624C mutant of APP-ACP labeled with CoA-488 and CoA-547 exhibited a mean FRET efficiency of 11.2 ± 0.5%. Therefore, even wild-type APP is almost completely dimerized in the plasma membrane (Figs. 2L and 3).
The insertion by the ACP labeling method of conformationally unconstrained fluorophores between the two described dimerization interfaces in the extracellular domain of APP (32, 63, 64) makes it very unlikely that the FRET coupling in the APP dimer is sensitive to the K624C mutation in the transmembrane domain of APP. Furthermore, the good fit of the data to our two-species model (specifically CoA-488-labeled APP-ACP undergoing FRET and nonspecific CoA-488 background) over a scale of more than an order of magnitude of concentration (Fig. 2K) showed that there was no appreciable relationship between the concentration of labeled APP and FRET efficiency. If APP monomers frequently underwent stochastic encounters in the plasma membrane or if they formed higher order aggregates in addition to dimers, lifetime should reduce more at higher APP concentrations because of the increased formation of FRET-generating higher order clusters. The observed stringent linearity over a scale of more than an order of magnitude of fluorescence signal, thus, strongly supports the interpretation that the associated APP molecules detected by fluorescence lifetime imaging microscopy at the plasma membrane are dimers.
To directly investigate the presence of specific and stable APP dimers in the plasma membrane, single molecule experiments were undertaken. After coupling quantum dots to APP-ACP, APP molecules could be tracked over several minutes (Fig. 4 and supplemental Movie 1) because of the high photostability of the quantum dots. Single molecules were observed because the concentration of quantum dots was low enough for sparse coupling to APP, and unspecific staining of untransfected cells was not observed. Fig. 4B shows that at room temperature some quantum dots undergo endocytosis, as judged by the increase in velocity and directionality because of active endosomal transport, and others that had been endocytosed before the beginning of measurement remained in the interior of the cell (cf. Fig. 4B, inset) for prolonged periods. However, for further analysis only quantum dots on the cell surface were examined. Because of the sparse coupling, it is not to be expected that in all APP dimers both molecules are coupled to quantum dots, but in several cases joint movement of quantum dots of different colors does show a stable association between APP molecules (Fig. 4, B and C). Such association persisted for up to 110 s. Two experimental observations suggest that the jointly moving cell surface APP molecules are dimers. First, in several tracks (e.g. Fig. 4C), each of the two color channels displays the blinking behavior typical of single quantum dots (65). The quantum dots were tracked in all three spatial dimensions, and the images show maximum projections of all z planes, excluding loss of focus as explanation for the blinking behavior. Second, the diffusion coefficient calculated for cell surface quantum dot-marked APP molecules is 3.8 ± 0.5 · 10-11 cm2/s (n = 17 quantum dots in 3 cells), which is within the range observed for other plasma membrane resident protein dimers (66). These results thus provide independent support for dimers as the prevailing interaction state of plasma membrane APP.
FIGURE 4.
Tracking of APP using quantum dots. A, confocal image of a B103 cell expressing APP-ACP labeled with CoA-biotin and a 100 pm concentration of a mixture of quantum dots emitting at 585 nm (green) and 655 nm (red). Note the colocalization of both quantum dots at several spots. The z plane is through the middle of the cell. B, time tracks of the quantum dots shown in panel A, projected into the xy plane (duration ≤190 s). Inset, APP marked with quantum dots in a lower focal plane (z = 0 μm, t = 0 s) delineates the contour of the cell. Cell surface APP monomers and dimers display random walks. *, APP dimer undergoing endocytosis after 28 s; note the transition from random movement to fast directional transport. Ellipse, internalized quantum dots. Scale bar, 1 μm. C, APP dimer shown in the white square in panels A and B at higher magnification (1.2-μm side length) at different time points. Images are maximum projections into the xy plane. Blinking of 655-nm quantum dot (t = 2.2 s and 30.4 s) and 585-nm quantum dot (t = 17.4s) indicates that these are single quantum dots.
Having isolated the signal of cell-surface APP from the much higher amounts of intracellular APP, it was now possible to measure the reasons for the higher degree of dimerization at the cell surface and to search for modulators of the aggregation state of cell surface APP. The presence of high concentrations of heparan sulfate side chains in the cell glycocalyx might be responsible for facilitating APP dimerization in the plasma membrane, as heparin has been shown to quantitatively dimerize sAPPα (40). Indeed, heparinase treatment of live B103 cells reduces the proportion of wild-type APP dimers in the plasma membrane by 44 ± 6% (Fig. 5). Heparinase treatment had no significant effect on the aggregation state of plasma membrane K624C-APP dimers (Fig. 5).
FIGURE 5.
Removal of heparin reduces APP dimerization. A–H, wide-field images of APP-ACP-expressing B103 cells labeled with CoA-488 and CoA-547. The focal plane was chosen to give the strongest signal from the plasma membrane and, therefore, does not include neurites. A, C, E, and G, CoA-488 intensity channel. B, D, F, and H, lifetime of CoA-488-labeled APP-ACP. A and B, wild-type APP-ACP in control condition. C and D, wild-type APP-ACP after heparinase treatment. E and F, K624C-APP-ACP in control condition. G and H, K624C-APP after heparinase treatment. Scale bars: 10 μm. I, proportion of dimerized APP. Left, K624C mutant of APP-ACP; right, wild-type APP-ACP. Black, control condition; white, after incubation with heparinase. Error bars indicate the S.E. (*, p = 0.011, n = 10–20 cells per condition).
When searching for the effects of extracellular ligands on the aggregation state of APP, sAPPα is of particular interest because it has a well known neuroprotective effect and has been suggested to counteract the detrimental neurotoxic effects of Aβ in Alzheimer disease (14–18). The addition of 1 μm sAPPα to wtAPP-ACP-expressing cells decreased the FRET efficiency to 7.0 ± 0.5%. If this diminished FRET efficiency derives from a physical separation of a subpopulation of APP with an accompanying complete loss of FRET, it corresponds to a ∼30% loss of dimeric APP by reference to the quantitatively dimerized K624C-APP-ACP measurements (Figs. 2L and 3). This reduction is comparable with the reduction achieved by heparinase treatment (Fig. 5). We cannot exclude the possibility that heparinase and/or sAPPα treatment leads to a conformational change in the APP dimers (e.g. by disrupting only the extracellular dimer interfaces). In this case the fraction of participating dimers would be even higher. However, the addition of 1 μm sAPPα had no significant effect on FRET efficiency in covalent K624C-APP dimers (Fig. 3), again in agreement with their resistance against heparinase treatment, and this together with functional data (see below) suggests that sAPPα indeed completely disrupts wtAPP dimers and not only changes their conformation.
The effect of exogenous sAPPα on APP dimers raises the question of whether an endogenous sAPP-ACP fragment generated from APP-ACP by the action of secretases might have contributed to our measurements. We, therefore, quantified sAPP in the medium conditioned by APP-ACP-transfected B103 cell directly before the addition of the dyes and found the concentration to be in the subnanomolar range (supplemental Fig. 1). Because endogenous sAPP-ACP was further reduced by the washing steps before a measurement (see “Experimental Procedures”), it is very unlikely that it might have influenced the results.
We note that in vivo concentrations of sAPPα in cerebrospinal fluid are also in the low nanomolar range (67). The sAPPα-induced modulation of APP dimerization and signaling, requiring higher concentrations, is thus expected to remain restricted to spatially confined compartments surrounding axons and synapses, where sAPPα is generated (68).
A possible functional role for APP dissociation in response to sAPPα binding was tested by measuring the survival of neuron-derived cells expressing APP under metabolic stress, both in the absence and in the presence of sAPPα. Changes in the metabolic state of the brain, especially a lower availability of intracellular glucose, accompany aging and have been suggested to contribute to the genesis of sporadic Alzheimer disease (69). When the B103 neuroblastoma cell line, which contains no endogenous APP or APP-like proteins (44), was grown for 2 days in the absence of nutrients and serum, cell survival was low; the number of mock-transfected cells surviving after 2 days under metabolic stress was ∼20% that of the number of mock-transfected cells surviving after 2 days in complete medium. The addition of sAPPα did not significantly change the number of surviving mock-transfected cells (Fig. 6). However, when wild-type APP-transfected cells were exposed to sAPPα, cell survival was more than twice as high as in mock-transfected cells (Fig. 6). Importantly, this effect was specific for wild-type APP, as the presence of sAPPα did not have a significant effect on the survival of cells transfected with the covalent dimer mutant of APP (Fig. 6). Together, these results show that for sAPPα to have a neuroprotective function, it must be able to dissociate transmembrane APP dimers in the plasma membrane. This reinforces the hypothesis that the reduction in FRET efficiencies indeed corresponds to a complete disruption of wtAPP dimers. Furthermore, the negligible difference in survival rates between mock-transfected and wtAPP-transfected B103 cells in the absence of exogenous sAPPα corroborates that only very low concentrations of neuroprotective endogenous sAPP fragment are generated in our model system.
FIGURE 6.
Effect of sAPPα on survival of neuroblastoma cells. Cells transfected with control plasmid (left), wild-type APP (middle), or K624C-APP (right) and a fluorescent marker plasmid were starved of nutrients and serum for 48 h in the absence (open bars) or the presence of sAPPα (solid bars). Cell numbers are normalized to control cells in the absence of sAPPα. S.E. are indicated. **, p < 0.05; ***, p < 0.001 (n = 20 fields of view, 9 wells).
DISCUSSION
Since its discovery over 20 years ago (7), APP has posed many challenges for its investigation both with regard to its role in Alzheimer disease and during normal development and neuronal plasticity. One of the more difficult questions has been the role of intact, full-length APP in these processes. Although a large proportion of APP is cleaved in various cell compartments (55) and the fragments are secreted or degraded, a small proportion of transmembrane APP remains stably localized to the plasma membrane (70). Different lines of evidence have suggested both growth-promoting and toxic roles for transmembrane APP (19, 20, 22–24, 71), whereas there is unequivocal evidence for a neuroprotective role of sAPPα (14–18). Both inappropriate signaling by transmembrane APP and a lowered sAPPα concentration may, therefore, contribute to neurodegeneration.
Our detection of APP dimerization by FRET/fluorescence lifetime imaging microscopy and single-molecule tracking using site-directed bioconjugation of surface-exposed APP, the invariant concentration ratio of donor and acceptor labels inherent to the labeling method, the independence of FRET in our assay on expression level and label density more than one order of magnitude, and the comparison with the constitutively dimerized K624C-APP mutant control condition permitted for the first time the rigorous statistical analysis of the monomer/dimer relationship of plasma membrane APP. The most important novelty in our approach is the successful separation of cell surface APP from the much greater pool of intracellular APP. We establish an almost complete dimerization of cell surface APP, apparently involving the presence of heparan sulfate on the cell surface, and its partial monomerization upon binding of sAPPα. We further show that the sAPPα-mediated disruption of transmembrane APP dimers is required for the neuroprotective effects of sAPPα to occur. The inability of sAPPα to exert the same protective effect in cells expressing the irreversibly dimerized K624C-APP mutant points to an underlying mechanism that depends on the monomerization of APP in the membrane.
Because sAPPα can dimerize in solution (32, 40) and is almost completely equivalent to the extracellular domain of APP, the dependence of sAPPα effects on the dissociation of transmembrane APP dimers is most easily explained by a direct interaction model (Fig. 7). In this model the dimerization interface of the heterodimer sAPPα/transmembrane APP would replace the dimerization interface between transmembrane APP molecules. Whether interactions of Nogo-66, TAG1, and integrin β1 with APP and/or sAPPα also affect APP dimerization (26, 72, 73) remains to be investigated.
FIGURE 7.
Hypothetical model of sAPPα action. Left, in the basal state APP mainly exists as dimers in the plasma membrane, which may activate cell death pathways. Part of transmembrane APP is cleaved by α-secretase, liberating sAPPα into the extracellular space. Right, when sAPPα binds to transmembrane APP, the dimers are disrupted, leading to increased cell survival.
In our studies we did not differentiate between the APP modulatory effect of sAPPα and sAPPβ mainly because of a lack of means of producing sAPPβ in a eukaryotic system. Given the importance of secretase switching under stress and pathological conditions, this represents an interesting aspect. However, the action of sAPP to dimerize with transmembrane APP most likely does not depend on the C-terminal 16 amino acids that are exclusively present in sAPPα and absent in sAPPβ, as both the dimerization interfaces known from atomic resolution studies (32, 63, 64) are present in both secreted APP forms. Furthermore, the proportion of APP cleaved by α-secretase is much higher than that cleaved by β-secretase in brain and can further increase severalfold (17) to the detriment of the major fraction of APP degraded intracellularly (74), Therefore, under conditions favorable for α-secretase cleavage (75), not only the ratio sAPPα/sAPPβ but also the total amount of sAPP is increased and may influence APP dimerization in the plasma membrane.
As it had not been shown before that cell surface APP is already almost entirely predimerized, the known neurotoxic effect of certain antibodies (19–22, 24) could have been assumed to be exerted through the dimerization of cell surface APP. Rather, our novel data suggest that these antibodies act by retaining increased amounts of APP dimers at the cell surface and/or by inducing higher order APP aggregates. This hypothesis is supported by our observation that the fluorescence intensity of cell surface APP is consistently higher in cells treated with the antibody 22C11 (data not shown). It is possible that sAPPα, on the other hand, accelerates APP internalization by disrupting dimers. Another artificial peptide ligand of APP has been shown to monomerize smaller APP domains and to modulate Aβ production but without explicitly demonstrating an effect on the dimerization status of transmembrane APP (27). It has been speculated that other natural ligands of APP, e.g. Cu2+, may also induce the monomerization of plasmalemmal APP (31). Unfortunately, Cu2+ quenches the fluorescence of CoA-488-labeled APP (data not shown), which precluded the investigation of its effects by our fluorescence-based methods. Such dimer-disrupting agents might be beneficial in that they could decrease both transmembrane APP signaling and the production of Aβ (27, 31), and our experimental model is compatible with a systematic screen for such agents.
In conclusion, our results help to clarify the discussion on toxic versus protective roles of APP by the recognition of different pools of transmembrane APP in living cells and indicate a critical role of the concentration of cell surface APP dimers for cell behavior. Furthermore, they predict a beneficial effect of treatments that would decrease cell surface APP dimer concentrations and reinforce the importance of increasing sAPPα production as a treatment option for Alzheimer disease symptoms.
Supplementary Material
Acknowledgments
We thank Joachim Dichter and Drs. Wouter Caarls, Thomas Jovin, and Donna Arndt-Jovin (Laboratory for Cellular Dynamics, Max Planck Institute for Biophysical Chemistry, Göttingen) for kind help with ACP staining and the programmable array microscope. We also thank Dr. David Schubert (Salk Institute) for a gift of the B103 cells and generous advice.
This work was supported by the German Federal Ministry for Education and Research (Bundesministerium für Bildung und Forschung) for the project “FLI-Cam” in the Biophotonik III program and from the European Neuroscience Institute Göttinge.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie 1 and Fig. 1.
Footnotes
The abbreviations used are: APP, amyloid precursor protein; AD, Alzheimer disease; Aβ, β-amyloid peptide; sAPPα, α-secretase-derived extracellular fragment of APP; ACP, acyl carrier protein; FRET, Förster resonance energy transfer; SNR, signal to noise ratio; wt wild type; mGFP, monomeric green fluorescent protein.
References
- 1.Heber, S., Herms, J., Gajic, V., Hainfellner, J., Aguzzi, A., Rulicke, T., Kretzschmar, H., von Koch, C., Sisodia, S., Tremml, P., Lipp, H. P., Wolfer, D. P., and Müller, U. (2000) J. Neurosci. 20 7951-7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herms, J., Anliker, B., Heber, S., Ring, S., Fuhrmann, M., Kretzschmar, H., Sisodia, S., and Müller, U. (2004) EMBO J. 23 4106-4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leyssen, M., Ayaz, D., Hebert, S. S., Reeve, S., De Strooper, B., and Hassan, B. A. (2005) EMBO J. 24 2944-2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang, P., Yang, G., Mosier, D. R., Chang, P., Zaidi, T., Gong, Y. D., Zhao, N. M., Dominguez, B., Lee, K. F., Gan, W. B., and Zheng, H. (2005) J. Neurosci. 25 1219-1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priller, C., Bauer, T., Mitteregger, G., Krebs, B., Kretzschmar, H. A., and Herms, J. (2006) J. Neurosci. 26 7212-7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young-Pearse, T. L., Bai, J., Chang, R., Zheng, J. B., LoTurco, J. J., and Selkoe, D. J. (2007) J. Neurosci. 27 14459-14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum, J. M., Masters, C. L., Grzeschik, K. H., Multhaup, G., Beyreuther, K., and Müller-Hill, B. (1987) Nature 325 733-736 [DOI] [PubMed] [Google Scholar]
- 8.Goate, A., Chartier-Harlin, M. C., Mullan, M., Brown, J., Crawford, F., Fidani, L., Giuffra, L., Haynes, A., Irving, N., James, L., et al. (1991) Nature 349 704-706 [DOI] [PubMed] [Google Scholar]
- 9.Rovelet-Lecrux, A., Hannequin, D., Raux, G., Le Meur, N., Laquerrière, A., Vital, A., Dumanchin, C., Feuillette, S., Brice, A., Vercelletto, M., Dubas, F., Frebourg, T., and Campion, D. (2006) Nat. Genet. 38 24-26 [DOI] [PubMed] [Google Scholar]
- 10.Selkoe, D. J. (2001) Physiol. Rev. 81 741-766 [DOI] [PubMed] [Google Scholar]
- 11.Gong, Y. S., Chang, L., Viola, K. L., Lacor, P. N., Lambert, M. P., Finch, C. E., Krafft, G. A., and Klein, W. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10417-10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacor, P. N., Buniel, M. C., Chang, L., Fernandez, S. J., Gong, Y. S., Viola, K. L., Lambert, M. P., Velasco, P. T., Bigio, E. H., Finch, C. E., Krafft, G. A., and Klein, W. L. (2004) J. Neurosci. 24 10191-10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sennvik, K., Fastbom, J., Blomberg, M., Wahlund, L. O., Winblad, B., and Benedikz, E. (2000) Neurosci. Lett. 278 169-172 [DOI] [PubMed] [Google Scholar]
- 14.Mattson, M. P., Cheng, B., Culwell, A. R., Esch, F. S., Lieberburg, I., and Rydel, R. E. (1993) Neuron 10 243-254 [DOI] [PubMed] [Google Scholar]
- 15.Meziane, H., Dodart, J. C., Mathis, C., Little, S., Clemens, J., Paul, S. M., and Ungerer, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12683-12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein, T. D., Anders, N. J., DeCarli, C., Chan, S. L., Mattson, M. P., and Johnson, J. A. (2004) J. Neurosci. 24 7707-7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postina, R., Schroeder, A., Dewachter, I., Bohl, J., Schmitt, U., Kojro, E., Prinzen, C., Endres, K., Hiemke, C., Blessing, M., Flamez, P., Dequenne, A., Godaux, E., van Leuven, F., and Fahrenholz, F. (2004) J. Clin. Investig. 113 1456-1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell, K. F. S., Zheng, L., Fahrenholz, F., and Cuello, A. C. (2008) Neurobiol. Aging 29 554-565 [DOI] [PubMed] [Google Scholar]
- 19.Rohn, T. T., Ivins, K. J., Bahr, B. A., Cotman, C. W., and Cribbs, D. H. (2000) J. Neurochem. 74 2331-2342 [DOI] [PubMed] [Google Scholar]
- 20.Sudo, H., Jiang, H., Yasukawa, T., Hashimoto, Y., Niikura, T., Kawasumi, M., Matsuda, S., Takeuchi, Y., Aiso, S., Matsuoka, M., Murayama, Y., and Nishimoto, I. (2000) Mol. Cell. Neurosci. 16 708-723 [DOI] [PubMed] [Google Scholar]
- 21.Sudo, H., Hashimoto, Y., Niikura, T., Shao, Z. J., Yasukawa, T., Ito, Y., Yamada, M., Hata, M., Hiraki, T., Kawasumi, M., Kouyama, K., and Nishimoto, I. (2001) Biochem. Biophys. Res. Commun. 282 548-556 [DOI] [PubMed] [Google Scholar]
- 22.Mbebi, C., Sée, V., Mercken, L., Pradier, L., Müller, U., and Loeffler, J. P. (2002) J. Biol. Chem. 277 20979-20990 [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto, Y., Niikura, T., Chiba, T., Tsukamoto, E., Kadowaki, H., Nishitoh, H., Yamagishi, Y., Ishizaka, M., Yamada, M., Nawa, M., Terashita, K., Aiso, S., Ichijo, H., and Nishimoto, I. (2003) J. Pharmacol. Exp. Ther. 306 889-902 [DOI] [PubMed] [Google Scholar]
- 24.Bouron, A., Mbebi, C., Loeffler, J. P., and De Waard, M. (2004) Eur. J. Neurosci. 20 2071-2078 [DOI] [PubMed] [Google Scholar]
- 25.Gralle, M., and Ferreira, S. T. (2007) Prog. Neurobiol. 82 11-32 [DOI] [PubMed] [Google Scholar]
- 26.Ma, Q. H., Futagawa, T., Yang, W. L., Jiang, X. D., Zeng, L., Takeda, Y., Xu, R. X., Bagnard, D., Schachner, M., Furley, A. J., D., K., Watanabe, K., Dawe, G. S., and Xiao, Z. C. (2008) Nat. Cell Biol. 10 283-294 [DOI] [PubMed] [Google Scholar]
- 27.Kaden, D., Münter, L. M., Joshi, M., Treiber, C., Weise, C., Bethge, T., Voigt, P., Schaefer, M., Beyermann, M., Reif, B., and Multhaup, G. (2008) J. Biol. Chem. 283 7271-7279 [DOI] [PubMed] [Google Scholar]
- 28.Kienlen-Campard, P., Tasiaux, B., Van Hees, J., Li, M., Huysseune, S., Sato, T., Fei, J. Z., Aimoto, S., Courtoy, P. J., Smith, S. O., Constantinescu, S. N., and Octave, J. N. (2008) J. Biol. Chem. 283 7733-7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman, P. M., Kim, S., Guo, M., Melnyk, R. A., McLaurin, J., Frase, P. E., Bowie, J. U., and Chakrabartty, A. (2008) BMC Neurosci. 9 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, T., Tang, T. C., Reubins, G., Fei, J. Z., Fujimoto, T., Kienlen-Campard, P., Constantinescu, S. N., Octave, J. N., Aimoto, S., and Smith, S. O. (2009) Proc. Natl. Acad. Sci. U. S. A. 106 1421-1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheuermann, S., Hambsch, B., Hesse, L., Stumm, J., Schmidt, C., Beher, D., Bayer, T. A., Beyreuther, K., and Multhaup, G. (2001) J. Biol. Chem. 276 33923-33929 [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., and Ha, Y. (2004) Mol. Cell 15 343-353 [DOI] [PubMed] [Google Scholar]
- 33.Soba, P., Eggert, S., Wagner, K., Zentgraf, H., Siehl, K., Kreger, S., Lower, A., Langer, A., Merdes, G., Paro, R., Masters, C. L., Muller, U., Kins, S., and Beyreuther, K. (2005) EMBO J. 24 3624-3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, C. D., Oh, S. Y., Hinman, J. D., and Abraham, C. R. (2006) J. Neurochem. 97 30-43 [DOI] [PubMed] [Google Scholar]
- 35.Münter, L. M., Voigt, P., Harmeier, A., Kaden, D., Gottschalk, K. E., Weise, C., Pipkorn, R., Schaefer, M., Langosch, D., and Multhaup, G. (2007) EMBO J. 26 1702-1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ninomiya, H., Roch, J. M., Jin, L. W., and Saitoh, T. (1994) J. Neurochem. 63 495-500 [DOI] [PubMed] [Google Scholar]
- 37.Barger, S. W., and Mattson, M. P. (1995) Biochem. J. 311 45-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann, J., Pietrzik, C. U., Kummer, M. P., Twiesselmann, C., Bauer, C., and Herzog, V. (1999) J. Histochem. Cytochem. 47 373-382 [DOI] [PubMed] [Google Scholar]
- 39.Tikkanen, R., Icking, A., Beicht, P., Waneck, G. L., and Herzog, V. (2002) Biol. Chem. 383 1855-1864 [DOI] [PubMed] [Google Scholar]
- 40.Gralle, M., Oliveira, C. L. P., Guerreiro, L. H., McKinstry, W. J., Galatis, D., Masters, C. L., Cappai, R., Parker, M. W., Ramos, H. I., Torriani, I., and Ferreira, S. T. (2006) J. Mol. Biol. 357 493-508 [DOI] [PubMed] [Google Scholar]
- 41.Zacharias, D. A., Violin, J. D., Newton, A. C., and Tsien, R. Y. (2002) Science 296 913-916 [DOI] [PubMed] [Google Scholar]
- 42.Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., and Tsien, R. Y. (2004) Nat. Biotechnol. 22 1567-1572 [DOI] [PubMed] [Google Scholar]
- 43.Ganesan, S., Ameer-beg, S. M., Ng, T. T. C., Vojnovic, B., and Wouters, F. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4089-4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, D., and Behl, C. (1993) Brain Res. 629 275-282 [DOI] [PubMed] [Google Scholar]
- 45.Gralle, M., Botelho, M. M., de Oliveira, C. L. P., Torriani, I., and Ferreira, S. T. (2002) Biophys. J. 83 3513-3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lidke, D. S., Nagy, P., Jovin, T. M., and Arndt-Jovin, D. J. (2007) Methods Mol. Biol. 374 69-79 [DOI] [PubMed] [Google Scholar]
- 47.Esposito, A., Gerritsen, H. C., and Wouters, F. S. (2005) Biophys. J. 89 4286-4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagen, G. M., Caarls, W., Thomas, M., Hill, A., Lidke, K. A., Rieger, B., Fritsch, C., Van Geest, B., Jovin, T. M., and Arndt-Jovin, D. J. (2007) Proc. SPIE 6441 64410S [Google Scholar]
- 49.Lidke, D. S., Nagy, P., Barisas, B. G., Heintzmann, R., Post, J. N., Lidke, K. A., Clayton, A. H. A., Arndt-Jovin, D. J., and Jovin, T. M. (2003) Biochem. Soc. Trans. 31 1020-1027 [DOI] [PubMed] [Google Scholar]
- 50.Jares-Erijman, E. A., and Jovin, T. M. (2006) Curr. Opin. Chem. Biol. 10 409-416 [DOI] [PubMed] [Google Scholar]
- 51.Esposito, A., Gerritsen, H. C., Oggier, T., Lustenberger, F., and Wouters, F. S. (2006) J. Biomed. Opt. 11 034016 [Google Scholar]
- 52.Weidemann, A., König, G., Bunke, D., Fischer, P., Salbaum, J. M., Masters, C. L., and Beyreuther, K. (1989) Cell 57 115-126 [DOI] [PubMed] [Google Scholar]
- 53.Kroenke, C. D., Ziemnicka-Kotula, D., Xu, J., Kotula, L., and Palmer, A. G., 3rd. (1997) Biochemistry 36 8145-8152 [DOI] [PubMed] [Google Scholar]
- 54.Haass, C., Koo, E. H., Capell, A., Teplow, D. B., and Selkoe, D. J. (1995) J. Cell Biol. 128 537-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skovronsky, D. M., Moore, D. B., Milla, M. E., Doms, R. W., and Lee, E. B. (2000) J. Biol. Chem. 275 2568-2575 [DOI] [PubMed] [Google Scholar]
- 56.Devi, L., Prabhu, B. M., Galati, D. F., Avadhani, N. G., and Anandatheerthavarada, H. K. (2006) J. Neurosci. 26 9057-9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koo, E. H., and Squazzo, S. L. (1994) J. Biol. Chem. 269 17386-17389 [PubMed] [Google Scholar]
- 58.Jung, S. S., Nalbantoglu, J., and Cashman, N. R. (1996) J. Neurosci. Res. 46 336-348 [DOI] [PubMed] [Google Scholar]
- 59.Jung, S. S., Gauthier, S., and Cashman, N. R. (1999) Neurobiol. Aging 20 249-257 [DOI] [PubMed] [Google Scholar]
- 60.Schneider, A., Rajendran, L., Honsho, M., Gralle, M., Donnert, G., Wouters, F. S., Hell, S. W., and Simons, M. (2008) J. Neurosci. 28 2874-2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer, B. H., Martinez, K. L., Segura, J. M., Pascoal, P., Hovius, R., George, N., Johnsson, K., and Vogel, H. (2006) FEBS Lett. 580 1654-1658 [DOI] [PubMed] [Google Scholar]
- 62.George, N., Pick, H., Vogel, H., Johnsson, N., and Johnsson, K. (2004) J. Am. Chem. Soc. 126 8896-8897 [DOI] [PubMed] [Google Scholar]
- 63.Dulubova, I., Ho, A., Huryeva, I., Südhof, T. C., and Rizo, J. (2004) Biochemistry 43 9583-9588 [DOI] [PubMed] [Google Scholar]
- 64.Rossjohn, J., Cappai, R., Feil, S. C., Henry, A., McKinstry, W. J., Galatis, D., Hesse, L., Multhaup, G., Beyreuther, K., Masters, C. L., and Parker, M. W. (1999) Nat. Struct. Biol. 6 327-331 [DOI] [PubMed] [Google Scholar]
- 65.Lidke, K. A., Rieger, B., Jovin, T. M., and Heintzmann, R. (2005) Opt. Express 13 7052-7062 [DOI] [PubMed] [Google Scholar]
- 66.Lidke, D. S., Lidke, K. A., Rieger, B., Jovin, T. M., and Arndt-Jovin, D. J. (2005) J. Cell Biol. 170 619-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lannfelt, L., Basun, H., Wahlund, L. O., Rowe, B. A., and Wagner, S. L. (1995) Nat. Med. 1 829-832 [DOI] [PubMed] [Google Scholar]
- 68.Moya, K. L., Benowitz, L. I., Schneider, G. E., and Allinquant, B. (1994) Dev. Biol. 161 597-603 [DOI] [PubMed] [Google Scholar]
- 69.Hoyer, S. (2000) Exp. Gerontol. 35 1363-1372 [DOI] [PubMed] [Google Scholar]
- 70.Storey, E., Katz, M., Brickman, Y., Beyreuther, K., and Masters, C. L. (1999) Eur. J. Neurosci. 11 1779-1788 [DOI] [PubMed] [Google Scholar]
- 71.Qiu, W. Q., Ferreira, A., Miller, C., Koo, E. H., and Selkoe, D. J. (1995) J. Neurosci. 15 2157-2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park, J. H., Gimbel, D. A., GrandPre, T., Lee, J. K., Kim, J. E., Li, W., Lee, D. H., and Strittmatter, S. (2006) J. Neurosci. 26 1386-1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young-Pearse, T. L., Chen, A. C., Chang, R., Marquez, C., and Selkoe, D. J. (2008) Neural Develop. 3 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporaso, G. L., Gandy, S. E., Buxbaum, J. D., Ramabhadran, T. V., and Greengard, P. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 3055-3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postina, R. (2008) Curr. Alzheimer Res. 5 179-186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









