Abstract
The cyclin-dependent kinase inhibitor p21Cip1 plays an important role in the cellular response to DNA damage. In normal cells, genotoxic stress activates the ATM-p53 pathway that up-regulates the expression of p21Cip1 leading to cell cycle arrest. However, we have found that in several neoplastic cell lines, ionizing radiation (IR) induces ubiquitin-dependent degradation of p21Cip1. This process is independent of the ATM pathway as it occurs in immortalized A-T fibroblasts. Knockdown of Skp2, an F-box protein capable of regulating the normal turnover of p21Cip1, does not prevent the IR-induced degradation. Instead, this process requires the Cul4-DDB1Cdt2 E3 ligase as knockdown of either DDB1 or Cdt2 rescues p21Cip1 degradation after IR. Mutating the proliferating cell nuclear antigen-binding site of p21Cip1 also prevents its IR-induced degradation suggesting that the p21Cip1-proliferating cell nuclear antigen interaction is critical for this event. Although ectopic expression of a nondegradable p21Cip1 did not by itself affect the clonogenic survival of HEK293 cells after IR, the degradation of p21Cip1 and other targets of the Cul4-DDB1Cdt2 E3 ligase may collectively contribute to the survival of neoplastic cells after ionizing radiation.
It is important that eukaryotic cells respond appropriately to DNA damage to ensure that the integrity of the genome is maintained. One family of proteins that plays an important role in the coordination of this response is the PI3K3-like family of protein kinases (PIKK), which includes ATM, ATR, and DNA-PK (1). Of these family members, ATM seems to be most important for the cellular response to ionizing radiation (IR) and other agents that generate DNA double strand breaks. Following exposure to IR, ATM kinase activity increases within minutes and leads to the phosphorylation of a number of target proteins that regulate an array of cellular processes, including the activation of cell cycle checkpoints and the initiation of DNA repair (2). One of the most well characterized targets of ATM is the p53 tumor suppressor protein, which is essential for the G1 checkpoint after IR (3, 4). ATM activation leads to phosphorylation of p53 at multiple sites resulting in both the increased stability and increased transcriptional activity of the protein (1). This ultimately leads to the increased expression of many p53 target genes, including the cyclin-dependent kinase inhibitor p21Cip1 (5).
p21Cip1 was initially identified in cyclin D1 immunoprecipitates as a component of a quaternary protein complex that included cyclin D1, cyclin-dependent kinase 2 or 4, and PCNA (6). Additional studies identified p21Cip1 as a potent inhibitor of cyclin-dependent kinases, suggesting that it played an important role in cell cycle regulation (7–9). This was confirmed by several independent studies that identified p21Cip1 as an important mediator of the G1 cell cycle arrest that occurs in response to a variety of cellular stresses (9–11). In particular, it is now well established that p21Cip1 is critical for the p53-dependent G1 arrest that occurs following DNA damage (10, 12–14).
In addition to binding cyclin-dependent kinases, p21Cip1 also directly binds the DNA polymerase processivity factor PCNA through its C-terminal region. This association has been shown to lead to inhibition of PCNA-dependent DNA replication in vitro (15–17). Thus, up-regulation of p21Cip1 following DNA damage may potentially target both the cell cycle machinery and the DNA replication machinery. However, high levels of p21Cip1 may also inhibit PCNA-dependent repair following DNA damage. In fact, it has been shown that p21Cip1 is degraded following UV radiation and that this degradation is required to facilitate DNA repair (18). This UV-induced degradation of p21Cip1 was shown to require ATR, a PIKK family member known to be activated in response to UV (1). In addition, the F-box protein Skp2, which functions as an adaptor protein for the SCF E3 ligase, was also shown to be required, although several recent reports have suggested that the Cul4A-DDB1 complex may actually be the E3 ligase responsible for this event (19, 20).
Here we report that in many transformed cell lines, ionizing radiation leads to the degradation of p21Cip1. We show that this IR-inducible degradation is dependent on the Cul4A-DDB1 E3 ligase but is independent of ATM and other PIKK family members.
EXPERIMENTAL PROCEDURES
Reagents—Cycloheximide, epoxomicin, wortmannin, N-ethylmaleimide, and 1,10-phenanthroline were from Sigma. Z-VAD and MG132 were from EMD Biosciences. Complete protease inhibitor tablets were from Roche Applied Science. KU-55933 was a gift from Graeme Smith (Kudos Pharmaceuticals). Phleomycin was a gift from Richard Kolodner (University of California, San Diego).
Plasmids—All HA-p21 plasmids were constructed using PCR to introduce the HA tag and necessary restriction sites into p21, p21ΔPCNA, or p21K6R. All fragments were then cloned into the multiple cloning site of pcDNA3.1+ (Invitrogen). The original p21K6R plasmid was a gift from Jim Roberts (Fred Hutchinson Cancer Research Center).
Cell Culture—All cells were supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin/streptomycin (Mediatech) unless otherwise indicated. HEK293, COS-1, HeLa, and HCT116 cells were cultured in DMEM with high glucose and l-glutamine (Mediatech). A-T fibroblasts (Coriell Repository) were grown in DMEM and supplemented with 100 μg/ml hygromycin B (Sigma). BJ normal human foreskin fibroblasts (between population doublings 30–45) were grown in Eagle's minimal essential media with Earle's balanced salt solution (ATCC). Saos-2 cells were grown in McCoy's 5a medium (Invitrogen) supplemented with 15% fetal bovine serum and 1% penicillin/streptomycin. Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunoblotting—Cells were lysed in RIPA buffer, and protein concentration was determined using the DC protein assay (Bio-Rad). Equal amounts of protein were run on 5, 12, or 4–20% Tris-glycine gels (Invitrogen). Proteins were transferred to PVDF membranes (Millipore) for 1 h at 100 V or overnight at 40 V using a cooling coil. Membranes were incubated in primary antibody for 2 h at room temperature or overnight at 4 °C and in secondary antibody for 1 h at room temperature. Where LICOR analysis was used, PVDF-FL membranes (Millipore) were used.
Immunoprecipitation—Primary antibodies were conjugated to A/G UltraLink resin (Pierce) for at least 4 h at 4 °C, and 1.0 μg of conjugated antibody was incubated with 1.0 mg of whole cell lysate overnight at 4 °C. The resin was washed three times in RIPA buffer and resuspended in 25 μl of 3× sample buffer.
Antibodies—Antibodies from Santa Cruz Biotechnology were as follows: p21Cip1 (C-19 and H164), Chk1 (G4), Chk2 (A12), SV40 LT (polyclonal antibody 101), and rabbit IgG. Antibodies from Cell Signaling were as follows: p21 (DCS60 and 12D1), ATR (2790), DNA-PK (4602), and Skp2 (4358). DDB1 was from Pharmingen (612488). GFP was from Covance (B34). PCNA was from EMD Biosciences (PC10). ATM pS1981 was from Rockland Immunochemicals. ATM 5C2 was a gift from Dr. Eva Lee (University of California, Irvine). Secondary antibodies were from Pierce and LICOR.
IR Experiments—Cells were seeded at 75–90% confluence in 60-mm dishes, and media were changed at least every 48 h. Cells were irradiated 2.5 or 4.5 days after seeding with a JL Shepherd Mark 1 137Cs irradiator using a dose rate of 4 Gy/min.
Ubiquitination Assay—To preserve endogenously ubiquitinated p21Cip1, we adopted a method developed for the detection of endogenously ubiquitinated IκB.4 Cells were immediately lysed in 500 μl of 95 °C ubiquitin lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% SDS, 45 μm N-ethylmaleimide, 60 μm 1,10-phenanthroline, 2× protease inhibitor) and boiled for 10 min. Lysates were sonicated continuously for 30 s and spun at 14,000 rpm for 30 min. Clarified lysates were diluted in dilution buffer (lysis buffer without SDS) to a final volume of 5 ml. Lysates were immunoprecipitated with 1.0 μg of p21Cip1 polyclonal antibody (Santa Cruz Biotechnology H-164) conjugated to 10 μl of protein A/G UltraLink resin (Pierce) overnight at 4 °C. The resin was then washed in 20 mm Tris, pH 7.5, 10% glycerol containing 150, 300, and 500 mm NaCl (washed two times with each buffer). The resin was washed once more in wash buffer with 150 mm salt prior to being run on a 4–20% Tris-glycine gel and transferred to a PVDF membrane and incubated with a monoclonal primary antibody (Cell Signaling DCS60).
Lentiviral shRNA—All shRNAs were from Sigma. The shRNA sequences for DDB1 experiments were as follows: CCGGCGACCGTAAGAAGGTGACTTTCTCGAGAAAGTCACCTTCTTACGGTCGTTTTTG, CCGGCGTGTACTCTATGGTGGAATTCTCGAGAATTCCACCATAGAGTACACGTTTTTG, and CCGGCCTATCACAATGGTGACAAATCTCGAGATTTGTCACCATTGTGATAGGTTTTTG. The skp2 shRNA sequence was CCGGGCCTAAGCTAAATCGAGAGAACTCGAGTTCTCTCGATTTAGCTTAGGCTTTTT. For virus production, the shRNA plasmids were cotransfected along with the pMDL, pRev, and pVSVG packaging plasmids into 293FT cells (Invitrogen) using the calcium phosphate method. Media were changed 12 h after transfection, and viral supernatants were collected 36 h later. Viral supernatants were filtered, and Polybrene (Sigma) was added to 8 μg/ml prior to infection of target cells. Target cells were infected for 48 h and selected with 2.0 μg/ml puromycin until control cells were dead and completely detached from the dish.
siRNA—siRNA sequences were as follows: Skp2, CCUAUCGAACUCAGUUAUAdTdT and CCUUAGACCUCACAGGUAAdTdT (Ambion); ATR, CGAGACUUCUGCGGAUUGCdTdT (Dharmacon); DNA-PK, CAAGCGACUUUAUAGCCUUdTdT (Ambion); and DDB1, ACUAGAUCGCGAUAAUAAAdTdT (Qiagen). The siRNAs were transfected at 50 nm with 10 μl of Lipofectamine 2000 (Invitrogen) 60 h prior to irradiation.
Quantitative PCR—Total RNA was extracted from HEK293 cells with the RNeasy kit (Qiagen) and reverse-transcribed into cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems). Real time PCR was run on the 7900 HT Fast real time PCR system (Applied Biosystems) using the Power SYBR Green PCR master mix (Applied Biosystems). Primers were as follows: actin, CGAGAAGATGACCCAGATCATGTT (forward) and CCTCGTAGATGGGCACAGTGT (reverse); p21Cip1, GGCGGGCTGCATCCA (forward) and AGTGGTGTCTCGGTGACAAAGTC (reverse).
Clonogenic Survival—5 × 106 HEK293 cells were cotransfected with 50 ng of GFP and 100 ng of the indicated p21Cip1 constructs. Cells were irradiated 60 h later. Cells were trypsinized 6 h post-IR, and the GFP-positive cells were sorted directly into 6-well plates containing 2 ml of conditioned media using a FACSAria cell sorter. Colonies of >50 cells were counted 10–14 days later.
RESULTS
p21Cip1 Is Degraded following IR—We have observed that irradiation of confluent HEK293 cells leads to a transient reduction in p21Cip1 protein levels between 30 and 90 min after IR (Fig. 1A). Although reduction in p21Cip1 levels is observed at lower densities, confluent HEK293 cells were used because these cells, which normally express p21Cip1 at low levels, up-regulate p21Cip1 as cell density increases (supplemental Fig. S1) making it easier to evaluate the extent of p21Cip1 degradation. As all other cell lines used in this study (many of which express p21Cip1 at low levels presumably because of impaired p53 function) were also found to up-regulate p21Cip1 with increasing cell density (data not shown), we conducted all subsequent experiments with confluent cultures of cells.
FIGURE 1.
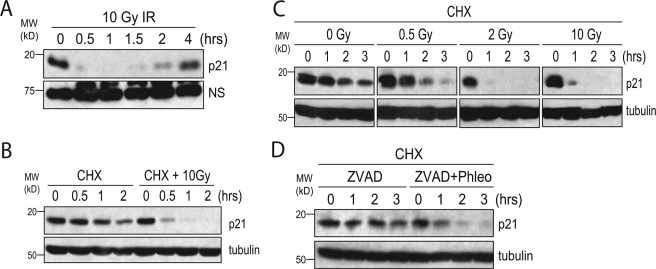
Ionizing radiation induces degradation of p21Cip1. A, HEK293 cells were irradiated with 10 Gy IR and collected at the indicated times. A Western blot was performed on equal amounts of whole cell lysate. NS is a nonspecific band of the p21Cip1 antibody (C-19) used as a loading control. B, Western blot of HEK293 cells pretreated with 25 μg/ml CHX prior to treatment with no IR or 10 Gy IR. C, Western blot of HEK293 cells pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 0, 0.5, 2, or 10 Gy IR. D, Western blot of HEK293 cells pretreated with 50 μm Z-VAD-fmk, 25 μg/ml CHX, and 100 μg/ml phleomycin (or vehicle) for 1 h.
To determine whether the IR-induced decrease in p21Cip1 protein level was due to protein degradation, we pretreated HEK293 cells with cycloheximide (CHX) and looked at the half-life of p21Cip1 with or without IR (Fig. 1B). We noticed that following a dose of 10 Gy IR, the half-life of p21Cip1 decreased from more than an hour to less than 30 min. Importantly, we observed no decrease in the rate of p21Cip1 transcription in these cells during this same time period, suggesting that the decrease in p21Cip1 protein level is entirely due to a decrease in protein stability (supplemental Fig. S1).
To establish whether degradation of p21Cip1 occurred over a broad range of IR doses, we irradiated HEK293 cells with doses ranging from 0.5 to 20 Gy. Although a dose of 0.5 Gy had only a very modest effect, doses between 2 and 20 Gy all led to a significant reduction in p21Cip1 half-life (Fig. 1C) (data not shown). In addition, we were also able to detect a decrease in p21Cip1 half-life in HEK293 cells following treatment with the DNA-damaging agent phleomycin, suggesting that degradation of p21Cip1 may occur in response to other agents that induce DNA double strand breaks (Fig. 1D). Although p21Cip1 has been shown to be cleaved by caspase-3 in some cell lines following IR (21), both the IR-induced and phleomycin-induced degradation of p21Cip1 occurred in the presence of the broad specificity caspase inhibitor Z-VAD-fmk (Fig. 1D), demonstrating that the degradation we observe is not because of caspase cleavage of p21Cip1.
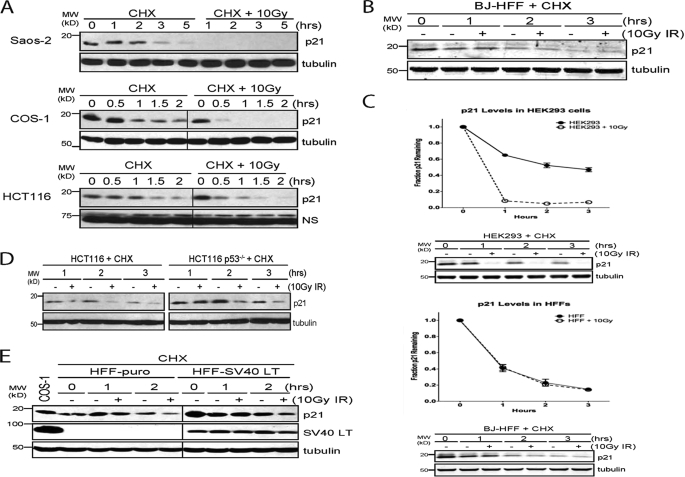
p21Cip1 Is Degraded in Transformed Cell Lines after IR—We examined p21Cip1 levels in a number of cell lines following IR and observed that COS-1, Saos-2, and HeLa cells all rapidly degraded p21Cip1 after IR (Fig. 2A) (data not shown). In contrast, HCT116 cells exhibited only a modest reduction in p21Cip1 after IR (Fig. 2A). Furthermore, irradiation of BJ human fibroblasts had no effect on p21Cip1 levels (Fig. 2B). Quantification of p21 levels showed that the half-life of p21 in HEK293 cells went from 2 h to less than 30 min after IR (Fig. 2C). Similar experiments with BJ fibroblasts confirmed that there was no change in the half-life of p21 in these cells after IR (Fig. 2C).
FIGURE 2.
IR-induced degradation of p21Cip1 occurs in transformed cells. A and B, indicated cells were pretreated with 25 μg/ml CHX and left untreated or irradiated with 10 Gy IR. All immunoblots were run using equal amounts of whole cell lysate and are representative of multiple experiments. C, same as in A and B, except LICOR secondary antibodies were used, and LICOR imaging software was used to quantify p21 levels. p21 levels were normalized to tubulin. Graphs are from three independent experiments with representative experiments shown below. D, Western blot of HCT116 or HCT116 p53-/- cells pretreated with 25 μg/ml CHX for 1 h prior to 10 Gy IR. E, Western blot of BJ human foreskin fibroblasts infected with a control retrovirus or a retrovirus expressing SV40 T-antigen. Infected cells were selected with 2.0 μg/ml puromycin until control cells were dead and detached from the dish. Selected cells were cultured to confluence and pretreated with 25 μg/ml CHX for 1 h prior to 10 Gy IR. Vertical lines indicate gel lanes that were spliced together.
We noticed that the cell lines that degrade p21Cip1 most efficiently after IR all lack functional p53 and Rb proteins, either because of impairment by viral proteins (HEK293, COS-1, HeLa) or by way of genetic losses (Saos-2). To test whether the lack of p53 function was responsible for the different rates of p21Cip1 degradation, we compared the half-life of p21Cip1 in wild-type and p53-deficient HCT116 cells. Following irradiation, p53-deficient HCT116 cells exhibited only a modest decrease in p21Cip1 stability, and this decrease was similar to that seen in the parental HCT116 cells (Fig. 2D). Thus, p53 loss alone does not permit p21Cip1 degradation after IR. To determine whether the combined loss of p53 and Rb function would cause a more robust degradation of p21Cip1, we infected BJ fibroblasts with a control retrovirus or a retrovirus expressing the SV40 large T-antigen, which impairs both p53 and Rb (22). Cells expressing T-antigen exhibited no increase in the rate of p21Cip1 degradation after IR relative to control cells (Fig. 2E), demonstrating that although the IR-induced degradation of p21Cip1 appears to be most robust in cells that lack functional p53 and Rb, other unidentified factors must also influence whether p21Cip1 is degraded after IR.
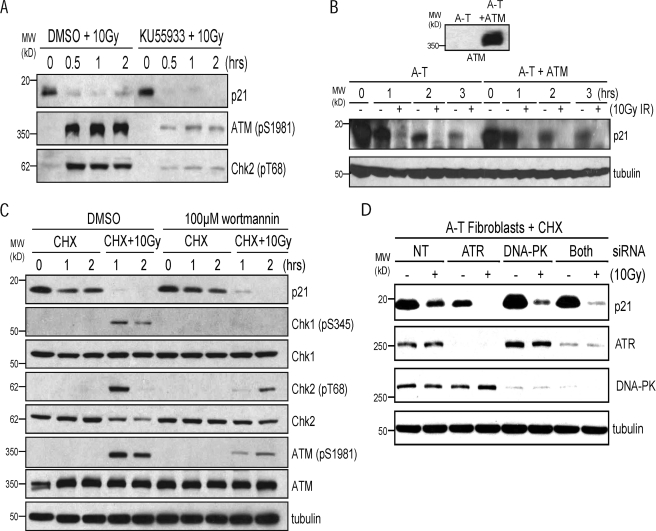
IR-induced Degradation of p21Cip1 Is Independent of ATM—It is well established that the ATM protein plays an important role in orchestrating the cellular response to ionizing radiation (1). To determine whether ATM is required for the IR-induced degradation of p21Cip1, we treated HEK293 cells with KU55933, a small molecule inhibitor of the ATM kinase that does not target other PIKK family members (23). Pretreatment of cells with this inhibitor led to a significant reduction in the phosphorylation of multiple ATM target sites after IR but failed to prevent the IR-induced degradation of p21Cip1 (Fig. 3A), suggesting this event may occur independently of ATM. To further rule out a role for ATM in this response, we compared the IR-induced degradation of p21Cip1 in a pair of SV40 immortalized A-T fibroblasts, one of which has been reconstituted with human ATM. Following irradiation, the degradation of p21Cip1 occurred with nearly identical kinetics in both cell lines, showing that ATM is not required for the IR-induced degradation of p21Cip1 (Fig. 3B).
FIGURE 3.
IR-induced degradation of p21Cip1 is independent of ATM. A, Western blot of whole cell lysates from HEK293 cells pretreated for 1 h with DMSO or 10 μm KU55933 prior to 10 Gy IR. Phospho-specific antibodies to ATM or the ATM target Chk2 were used to verify the efficacy of the inhibitor. B, Western blot of whole cell lysates from immortalized A-T fibroblasts or reconstituted A-T fibroblasts pretreated with 25 μg/ml cycloheximide for 1 h prior to 10 Gy IR. C, Western blot of whole cell lysate from HEK293 cells pretreated with 25 μg/ml CHX and DMSO or 100 μm wortmannin for 1 h prior to 10 Gy IR. Phospho-specific antibodies to ATM or ATR target sites were used to verify the efficacy of the inhibitor. D, A-T cells were transfected with siRNAs to ATR or DNA-PK, either individually or in combination. 60 h post-transfection, cells were pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy IR. Cells were collected 1 h after IR, and equal amounts of whole cell lysate were analyzed by Western blot.
The possibility exists that in the absence of ATM, other PIKK family members, such as ATR, may orchestrate the IR-induced degradation of p21Cip1. To address this possibility, we treated HEK293 cells with high concentrations (100 μm) of the fungal metabolite wortmannin, a general inhibitor of the PI3K family of kinases. At these concentrations, wortmannin has been shown to inhibit ATM, ATR, and DNA-PK (24). As shown in Fig. 3C, the IR-induced phosphorylation of ATM target sites was significantly reduced in the presence of 100 μm wortmannin, whereas phosphorylation of an ATR target site on Chk1 was undetectable. Despite the apparent inhibition of both ATM and ATR under these conditions, p21Cip1 was still degraded in these cells after IR (Fig. 3C). To further rule out compensation from other PIKK family members, we also transfected A-T cells with siRNAs against both ATR and DNA-PK. Cells receiving both siRNAs, which exhibit about 90% knockdown of both proteins, still degrade p21Cip1 after IR (Fig. 3D). This suggests that the IR-induced degradation of p21Cip1 is independent of each of the PI3K-related kinases.
Degradation of p21Cip1 Is Dependent on the Ubiquitin-Proteasome System—To assess the proteasome dependence of p21Cip1 degradation after IR, we treated HEK293 cells with the proteasome inhibitor MG132 for 1 h prior to IR. We found that pretreatment with this inhibitor completely prevented the IR-induced degradation of p21Cip1 (Fig. 4A). As MG132 inhibits proteases other than the proteasome (25), we also treated cells with epoxomicin, a much more specific inhibitor of the proteasome (26). Pretreatment with this inhibitor also prevented degradation of p21Cip1 (Fig. 4B), demonstrating that the IR-induced degradation of p21Cip1 is proteasome-dependent.
FIGURE 4.
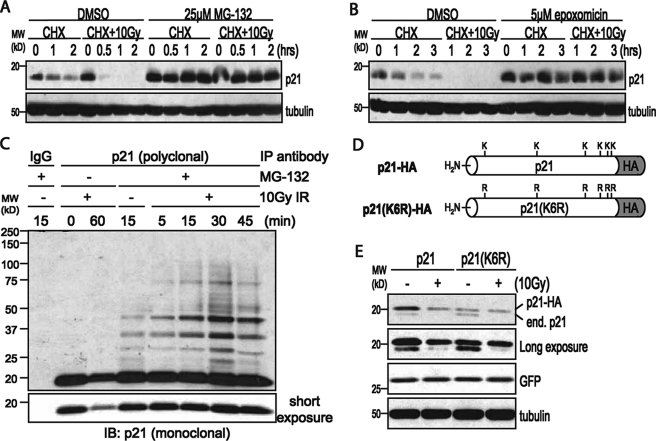
p21Cip1 is degraded by the ubiquitin-proteasome system following IR. A, Western blot of whole cell lysates from HEK293 cells pretreated with 25 μg/ml CHX and DMSO or 25 μm MG132 for 1 h prior to 10 Gy IR. B, Western blot of whole cell lysates from HEK293 cells pretreated with 25 μg/ml CHX and DMSO or 5 μm epoxomicin for 1 h prior to 10 Gy IR. C, HEK293 cells were pretreated with DMSO or 25 μm MG132 for 30 min prior to IR. Cells were collected at the indicated time points as described under “Experimental Procedures.” p21Cip1 was immunoprecipitated from 1.0 mg of whole cell lysate using a polyclonal antibody to p21Cip1, and a Western blot was performed using a p21Cip1 monoclonal antibody. D, schematic of the p21-HA and p21(K6R)-HA constructs. E, HEK293 cells were transfected with 5.0 μg of total DNA, including 50 ng of GFP and 100 ng of p21-HA or p21(K6R)-HA. Cells were pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy IR. Cells were collected 1 h after IR, and equal amounts of whole cell lysate were analyzed by Western blot.
To determine whether p21Cip1 is ubiquitinated after IR, we immunoprecipitated endogenous p21Cip1 from MG132-treated HEK293 cells in the presence of N-ethylmaleimide and 1,10-phenanthroline, two inhibitors of deubiquitinating enzymes (27). As shown in Fig. 4C, in the absence of MG132, significantly less p21Cip1 was immunoprecipitated from irradiated cells, again showing that p21Cip1 is degraded after IR (Fig. 4C, lower panel). In cells pretreated with MG132, we observed several higher molecular weight forms of p21Cip1, consistent with reports that the normal turnover of p21Cip1 can involve ubiquitination (28, 29). Following irradiation of MG132-treated cells, we observed a time-dependent increase in the higher molecular weight forms of p21Cip1 (Fig. 4C), suggesting that p21Cip1 is ubiquitinated after IR. Importantly, the increase in these higher molecular weight forms of p21Cip1 peaks around 30 min, which correlates well with the kinetics of p21Cip1 degradation after IR (Fig. 1A).
To show that ubiquitination is required for p21Cip1 degradation following IR, we transfected HEK293 cells with wild-type p21Cip1 or a p21(K6R) mutant in which the six lysines of p21Cip1 have all been mutated to arginines (28). These lysine mutations eliminate all potential ubiquitination sites within the protein (with the possible exception of the amine group at the N terminus) and therefore should prevent any process that requires p21Cip1 to be ubiquitinated. Because it has been reported that N-terminal tags may affect the stability of p21Cip1 (29, 30), both the wild-type p21Cip1 and p21(K6R) constructs were designed with HA tags on the C terminus (Fig. 4D). Following irradiation of cells expressing wild-type p21-HA, there was a significant reduction in both the endogenous and HA-tagged forms of p21Cip1. In contrast, irradiation of cells expressing p21(K6R)-HA led only to a reduction in endogenous p21Cip1 levels (Fig. 4E), indicating that that the p21(K6R)-HA mutant is protected from degradation, and suggesting that ubiquitination of p21Cip1 is indeed necessary for its degradation after IR.
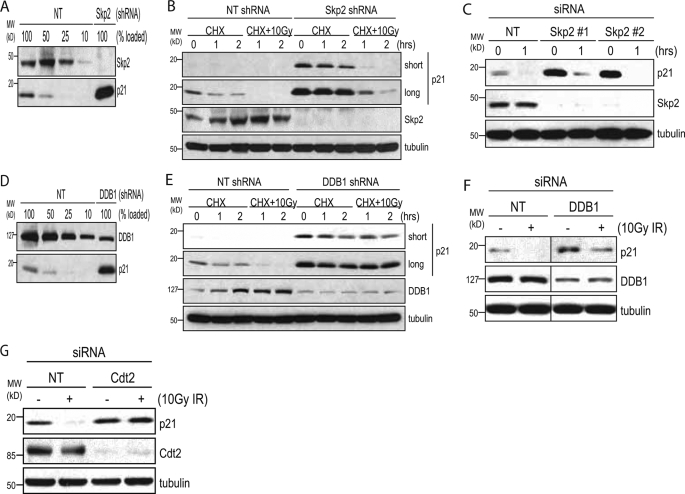
Skp2 Is Dispensable for p21Cip1 Degradation after IR—Skp2 is an F-box protein, which functions as an adaptor for the Cul1-Skp1 E3 ligase and has previously been reported to be involved in both the normal and inducible turnover of p21Cip1 (18, 31–33). Because the IR-induced degradation of p21Cip1 appeared to be ubiquitin-dependent, we asked if it was also Skp2-dependent. To investigate the role of Skp2 in this process, we infected HEK293 cells with a lentiviral shRNA against Skp2, or a nontarget shRNA that targets no known or predicted human gene. Immunoblotting showed that Skp2 protein levels were depleted by more than 90% in cells infected with the Skp2 shRNA. In addition, these cells also had significantly elevated levels of p21Cip1 suggesting that Skp2 function was in fact compromised by the knockdown (Fig. 5A). Despite this, irradiation of these cells still resulted in the degradation of p21Cip1 (Fig. 5B). We obtained similar results using Skp2 siRNAs (Fig. 5C), suggesting that although Skp2 is involved in the normal turnover of p21Cip1, it is dispensable for the IR-induced degradation of the protein.
FIGURE 5.
DDB1, but not Skp2, is required for the IR-induced degradation of p21Cip1. A, HEK293 cells were infected with a nontarget (NT) lentiviral shRNA or an shRNA against Skp2. Infected cells were selected with 2.0 μg/ml puromycin. Knockdown of Skp2 was assessed by Western blot using dilutions of the nontarget sample. B, nontarget or Skp2 knockdown cells were pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy IR. Cells were collected at the indicated time points and analyzed by Western blot. C, HEK293 cells were transfected with a nontarget siRNA or two separate siRNAs against Skp2. 60 h post-transfection, cells were irradiated with 10 Gy. Cells were collected 1 h after IR, and whole cell lysates were analyzed by Western blot. D, HEK293 cells were infected with a nontarget lentiviral shRNA or an shRNA against DDB1 and assessed for DDB1 knockdown and p21Cip1 up-regulation as in A. E, nontarget or DDB1 knockdown cells were pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy IR. Cells were collected at the indicated time points and analyzed by Western blot. F, HEK293 cells were transfected with a nontarget siRNA or an siRNAs against DDB1. 60 h post-transfection, cells were pretreated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy. Cells were collected 1 h after IR and whole cell lysates were analyzed by Western blot. Vertical lines indicate gel lanes that were spliced together. G, HEK293 cells were transfected with a nontarget siRNA or an siRNAs against Cdt2. 60 h post-transfection, cells were irradiated with 10 Gy. Cells were collected 1 h after IR, and whole cell lysates were analyzed by Western blot.
IR-induced Degradation of p21Cip1 Requires DDB1Cdt2 and PCNA Binding—One complex with an established role in the IR-induced degradation of proteins is the Cul4-DDB1Cdt2 E3 ligase, which has been shown to degrade the replication licensing factor Cdt1 after both UV and IR (34–37). Interestingly, DDB1Cdt2 was also recently implicated in both the normal and UV-inducible turnover of p21Cip1 (19, 20, 38). To address the role of DDB1Cdt2 in the IR-induced degradation of p21Cip1, we infected HEK293 cells with a nontarget shRNA or three separate shRNAs against DDB1. Similar to what was seen with knockdown of Skp2, knockdown of DDB1 led to an up-regulation of p21Cip1 in untreated cells (Fig. 5D), suggesting that DDB1 contributes to the normal turnover of p21Cip1. However, in contrast to what was seen with Skp2, knockdown of DDB1 also prevented the IR-induced degradation of p21Cip1 (Fig. 5E and supplemental Fig. S2). Similar results were obtained with siRNA (Fig. 5F). To examine the role of the Cdt2 adaptor protein, we transfected cells with an siRNA against Cdt2. Knockdown of this adaptor protein also prevented the IR-induced degradation of p21Cip1 (Fig. 5G). Taken together, these results demonstrate that although both Skp2 and DDB1Cdt2 are capable of regulating the normal turnover of p21Cip1, only DDB1Cdt2 is required for its IR-induced degradation.
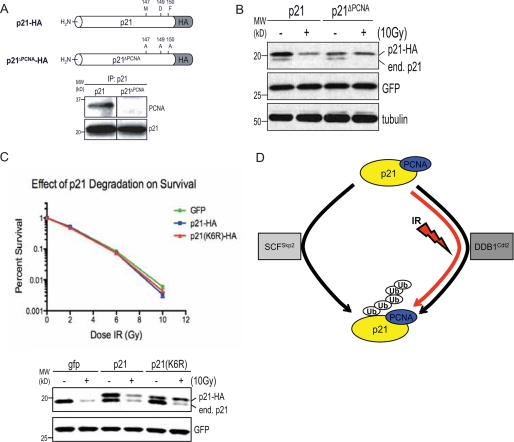
IR-induced p21Cip1 Degradation Is PCNA-dependent—The Cul4-DDB1Cdt2 E3 ligase is known to be required for the DNA damage-induced degradation of Cdt1 (36). Like p21Cip1, Cdt1 binds PCNA, and it has been shown that this interaction is required for degradation of Cdt1 following UV and IR (34, 39). To determine whether PCNA binding is required for the DDB1-dependent degradation of p21Cip1 following IR, we transfected cells with p21-HA or p21ΔPCNA-HA, which harbors three point mutations in the C terminus and has been shown to be defective in PCNA binding (Fig. 6A). Irradiation of cells transfected with wild-type p21-HA led to degradation of both the endogenous and HA-tagged p21Cip1. In contrast, irradiation of cells transfected with p21ΔPCNA-HA led only to degradation of endogenous p21Cip1 (Fig. 6B), suggesting that PCNA binding is critical for the IR-induced degradation of p21Cip1.
FIGURE 6.
p21Cip1 lacking the ability to bind PCNA is not degraded following IR. A, HEK293 cells were transfected with 5.0 μg of DNA, including 500 ng of p21-HA or p21ΔPCNA-HA and 100 ng of GFP. 60 h post-transfection, p21Cip1 was immunoprecipitated from 1.0 mg of whole cell lysate, and immunoblots were performed with antibodies to p21Cip1 or PCNA. B, HEK293 cells were transfected with 5.0 μg of DNA, including 100 ng of p21-HA or p21ΔPCNA-HA and 50 ng of GFP. 60 h after transfection, cells were treated with 25 μg/ml CHX for 1 h prior to irradiation with 10 Gy of IR. Samples were collected 1 h after IR. Equal amounts of whole cell lysate were analyzed by Western blot for expression of the indicated proteins. C, upper panel, HEK293 cells were transfected with 5.0 μg of DNA, including 100 ng of p21-HA or p21(K6R)-HA and 50 ng of GFP. 60 h later cells were irradiated with the indicated doses of IR. 6 h post-IR, cells were trypsinized and GFP+ cells were sorted (in triplicate) directly into 6-well dishes containing conditioned DMEM. 10–14 days later, surviving colonies were counted as those with >50 cells. Shown are the results of an experiment representative of multiple independent repeats. Lower panel, Western blot of 10 Gy samples taken at 1 h post-IR. In this experiment, 30% of cells were GFP+, so exogenous p21 levels are about 3-fold higher than shown. D, model depicting degradation of p21Cip1. Both Skp2 and DDB1Cdt2 can regulate the normal turnover of p21Cip1, but only DDB1Cdt2 is responsible for the IR-inducible turnover.
Recent reports showing that Cul4-DDB1Cdt2 is responsible for the UV-induced degradation of p21Cip1 have also demonstrated that this process is dependent on the p21Cip1-PCNA interaction (19, 20, 40) and postulated that this functions to facilitate PCNA-dependent DNA repair (18). This led us to ask whether transformed cells that degrade p21Cip1 do so to increase DNA repair and promote survival. To assess the biological significance of p21Cip1 degradation after IR, we performed a clonogenic survival experiment with HEK293 cells expressing wild-type p21Cip1 or the nondegradable p21(K6R) mutant. We observed no reduction in survival for cells expressing nondegradable p21Cip1 suggesting that although p21Cip1 degradation may contribute to increased PCNA-dependent repair, the ectopic expression of p21(K6R) is not sufficient to block PCNA-dependent repair processes (Fig. 6C). This may indicate that other PCNA-bound proteins may also need to be degraded to permit PCNA-dependent repair following DNA damage.
DISCUSSION
We have observed that in many transformed cell lines, ionizing radiation leads to the degradation of the cyclin-dependent kinase inhibitor p21Cip1. In addition, phleomycin, a DNA-damaging agent that produces both single and double strand breaks (41), also leads to p21Cip1 degradation. Based on these results, it seems probable that many agents that produce DNA strand breaks may trigger p21Cip1 degradation in these cells. However, as we have been unable to detect degradation of p21Cip1 following treatment with either doxorubicin or etoposide (data not shown), two other DNA-damaging agents capable of generating strand breaks, it seems that the type of damage and/or the kinetics with which the damage occurs likely influence whether p21Cip1 is degraded following damage.
Degradation of p21Cip1 following ionizing radiation occurs in both a proteasome- and ubiquitin-dependent manner but is surprisingly independent of ATM as it occurs in immortalized A-T fibroblasts. It is unlikely that this ATM independence is due to compensation by other ATM family members as degradation of p21Cip1 also occurs in the presence of the general PIKK inhibitor wortmannin and in A-T cells depleted of ATR and DNA-PK by siRNA. In an attempt to determine whether other serine/threonine kinases play a role in the IR-induced degradation of p21Cip1, we have treated cells with the general kinase inhibitor staurosporine, but have observed no effect on p21Cip1 degradation (data not shown). In addition, we have used two-dimensional gel electrophoresis to search for IR-induced modifications of p21Cip1, but have been unable to detect any changes in the isoelectric point or mobility shift of p21Cip1 (data not shown). Taken together, these results suggest that p21Cip1 may not need to be covalently modified prior to its ubiquitination and degradation after IR.
We show here that the IR-induced degradation of p21Cip1 requires DDB1Cdt2 and is dependent on the p21Cip1-PCNA interaction. The best characterized target of the Cul4-DDB1Cdt2 E3 ligase is the replication licensing factor Cdt1, which is degraded by Cul4-DDB1Cdt2 both during S-phase and following DNA damage induced by UV or IR (34–37). Similar to p21Cip1, Cdt1 binds PCNA, and this interaction is necessary for both its S-phase and DNA damage-induced degradation (34, 39, 42). Interestingly, several studies published during preparation of this manuscript have also shown that Cul4-DDB1 is responsible for the degradation of p21Cip1 both during S-phase and following UV damage (19, 20, 40). In each of these reports, the degradation of p21Cip1 was also shown to be dependent on its interaction with PCNA. Thus, PCNA seems to serve as a critical intermediary for the two most well characterized DDB1 substrates.
Interestingly, PCNA has been shown to be recruited to chromatin within minutes of IR, and this recruitment has been shown to be both transient and ATM-independent (43, 44). As we have shown that the IR-induced degradation of p21Cip1 is also transient and ATM-independent, it is possible that PCNA-mediated recruitment of p21Cip1 to chromatin is the rate-limiting step in p21Cip1 degradation after IR. Because DDB1Cdt2 has been shown to degrade a number of chromatin-bound proteins (45), this model would also help explain why the IR-induced degradation of p21Cip1 is dependent on Cul4-DDB1Cdt2, although the normal turnover of the protein can be regulated by both Cul4-DDB1Cdt2 and SCFSkp2.
A unique feature of the IR-induced degradation of p21Cip1 is that although it occurs in most of the cell lines we have tested, it is significantly more robust in a subset of transformed cell lines that lack functional p53 and Rb proteins. We have observed that loss of p53 alone does not increase the rate at which p21Cip1 is degraded after IR. Introduction of the SV40 large T-antigen, which impairs both the p53 and Rb proteins, also failed to accelerate p21Cip1 degradation after IR. Thus, the increased rate of degradation we observe in cells lacking functional p53 and Rb may be a unique feature of these cells or it may be a result of a more complex transformation process.
Expression of a nondegradable version of p21Cip1 did not lead to reduced survival in irradiated HEK293 cells. This would seem to conflict with a previously proposed model in which damage-induced degradation of p21Cip1 promotes PCNA-dependent DNA repair (18). However, based on the abundance of PCNA in most cells, it seems unlikely that endogenous or near endogenous (in the case of the exogenous p21(K6R)) levels of p21Cip1 would alone be capable of inhibiting PCNA-dependent processes (46). Thus, there may be a number of PCNA-bound proteins that need to be degraded to mobilize PCNA to participate in DNA repair. Such proteins might also be targets of Cul4-DDB1Cdt2.
Previous results have demonstrated that activation of the Cul4-DDB1Cdt2 E3 ligase is important in the cellular response to damage (35). The fact that the Cul4-DDB1Cdt2-dependent degradation of p21Cip1 is specific for transformed cells raises the intriguing possibility that Cul4-DDB1Cdt2 may be regulated differently in cancer cells. As it would be therapeutically desirable to modulate the response to DNA damage specifically in tumors following IR, this possibility warrants future studies.
Supplementary Material
Acknowledgments
We thank Joe Aguilera at the University of California, San Diego, Radiation Medicine Facility, Dr. Gabriel Pineda for help with the detection of endogenous ubiquitination, and Dr. Xiaodong Huang and Dr. Sam Zeitlin for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA43054 (to J. Y. J. W.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PI3K, phosphatidylinositol 3-kinase; ATM, ataxia telangiectasia mutated; ATR, ATM and Rad3-related protein; IR, ionizing radiation; PCNA, proliferating cell nuclear antigen; Gy, gray; DMEM, Dulbecco's modified Eagle's medium; PVDF, polyvinylidene difluoride; GFP, green fluorescent protein; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; shRNA, short hairpin RNA; siRNA, small interfering RNA; CHX, cycloheximide; Rb, retinoblastoma; E3, ubiquitin-protein isopeptide ligase; PIKK, PI3K-related protein kinase; DNA-PK, DNA-dependent protein kinase.
G. Pineda, personal communication.
References
- 1.Shiloh, Y. (2003) Nat. Rev. Cancer 3 155-168 [DOI] [PubMed] [Google Scholar]
- 2.Matsuoka, S., Ballif, B. A., Smogorzewska, A., McDonald, E. R., III, Hurov, K. E., Luo, J., Bakalarski, C. E., Zhao, Z., Solimini, N., Lerenthal, Y., Shiloh, Y., Gygi, S. P., and Elledge, S. J. (2007) Science 316 1160-1166 [DOI] [PubMed] [Google Scholar]
- 3.Kastan, M. B., Zhan, Q., el-Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B., and Fornace, A. J., Jr. (1992) Cell 71 587-597 [DOI] [PubMed] [Google Scholar]
- 4.Kastan, M. B., Onyekwere, O., Sidransky, D., Vogelstein, B., and Craig, R. W. (1991) Cancer Res. 51 6304-6311 [PubMed] [Google Scholar]
- 5.Sherr, C. J., and Roberts, J. M. (1995) Genes Dev. 9 1149-1163 [DOI] [PubMed] [Google Scholar]
- 6.Xiong, Y., Zhang, H., and Beach, D. (1992) Cell 71 505-514 [DOI] [PubMed] [Google Scholar]
- 7.Xiong, Y., Hannon, G. J., Zhang, H., Casso, D., Kobayashi, R., and Beach, D. (1993) Nature 366 701-704 [DOI] [PubMed] [Google Scholar]
- 8.Gu, Y., Turck, C. W., and Morgan, D. O. (1993) Nature 366 707-710 [DOI] [PubMed] [Google Scholar]
- 9.Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K., and Elledge, S. J. (1993) Cell 75 805-816 [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W., and Vogelstein, B. (1993) Cell 75 817-825 [DOI] [PubMed] [Google Scholar]
- 11.Noda, A., Ning, Y., Venable, S. F., Pereira-Smith, O. M., and Smith, J. R. (1994) Exp. Cell Res. 211 90-98 [DOI] [PubMed] [Google Scholar]
- 12.Deng, C., Zhang, P., Harper, J. W., Elledge, S. J., and Leder, P. (1995) Cell 82 675-684 [DOI] [PubMed] [Google Scholar]
- 13.Di Leonardo, A., Linke, S. P., Clarkin, K., and Wahl, G. M. (1994) Genes Dev. 8 2540-2551 [DOI] [PubMed] [Google Scholar]
- 14.Dulic, V., Kaufmann, W. K., Wilson, S. J., Tlsty, T. D., Lees, E., Harper, J. W., Elledge, S. J., and Reed, S. I. (1994) Cell 76 1013-1023 [DOI] [PubMed] [Google Scholar]
- 15.Waga, S., Hannon, G. J., Beach, D., and Stillman, B. (1994) Nature 369 574-578 [DOI] [PubMed] [Google Scholar]
- 16.Gulbis, J. M., Kelman, Z., Hurwitz, J., O'Donnell, M., and Kuriyan, J. (1996) Cell 87 297-306 [DOI] [PubMed] [Google Scholar]
- 17.Chen, J., Jackson, P. K., Kirschner, M. W., and Dutta, A. (1995) Nature 374 386-388 [DOI] [PubMed] [Google Scholar]
- 18.Bendjennat, M., Boulaire, J., Jascur, T., Brickner, H., Barbier, V., Sarasin, A., Fotedar, A., and Fotedar, R. (2003) Cell 114 599-610 [DOI] [PubMed] [Google Scholar]
- 19.Abbas, T., Sivaprasad, U., Terai, K., Amador, V., Pagano, M., and Dutta, A. (2008) Genes Dev. 22 2496-2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishitani, H., Shiomi, Y., Iida, H., Michishita, M., Takami, T., and Tsurimoto, T. (2008) J. Biol. Chem. 283 29045-29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gervais, J. L., Seth, P., and Zhang, H. (1998) J. Biol. Chem. 273 19207-19212 [DOI] [PubMed] [Google Scholar]
- 22.Ahuja, D., Saenz-Robles, M. T., and Pipas, J. M. (2005) Oncogene 24 7729-7745 [DOI] [PubMed] [Google Scholar]
- 23.Hickson, I., Zhao, Y., Richardson, C. J., Green, S. J., Martin, N. M., Orr, A. I., Reaper, P. M., Jackson, S. P., Curtin, N. J., and Smith, G. C. (2004) Cancer Res. 64 9152-9159 [DOI] [PubMed] [Google Scholar]
- 24.Sarkaria, J. N., Tibbetts, R. S., Busby, E. C., Kennedy, A. P., Hill, D. E., and Abraham, R. T. (1998) Cancer Res. 58 4375-4382 [PubMed] [Google Scholar]
- 25.Lee, D. H., and Goldberg, A. L. (1998) Trends Cell Biol. 8 397-403 [DOI] [PubMed] [Google Scholar]
- 26.Meng, L., Mohan, R., Kwok, B. H., Elofsson, M., Sin, N., and Crews, C. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10403-10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guterman, A., and Glickman, M. H. (2004) J. Biol. Chem. 279 1729-1738 [DOI] [PubMed] [Google Scholar]
- 28.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5 403-410 [DOI] [PubMed] [Google Scholar]
- 29.Bloom, J., Amador, V., Bartolini, F., DeMartino, G., and Pagano, M. (2003) Cell 115 71-82 [DOI] [PubMed] [Google Scholar]
- 30.Chen, X., Chi, Y., Bloecher, A., Aebersold, R., Clurman, B. E., and Roberts, J. M. (2004) Mol. Cell 16 839-847 [DOI] [PubMed] [Google Scholar]
- 31.Yu, Z. K., Gervais, J. L., and Zhang, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11324-11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, W., Nacusi, L., Sheaff, R. J., and Liu, X. (2005) Biochemistry 44 14553-14564 [DOI] [PubMed] [Google Scholar]
- 33.Petroski, M. D., and Deshaies, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6 9-20 [DOI] [PubMed] [Google Scholar]
- 34.Higa, L. A., Banks, D., Wu, M., Kobayashi, R., Sun, H., and Zhang, H. (2006) Cell Cycle 5 1675-1680 [DOI] [PubMed] [Google Scholar]
- 35.Higa, L. A., Mihaylov, I. S., Banks, D. P., Zheng, J., and Zhang, H. (2003) Nat. Cell Biol. 5 1008-1015 [DOI] [PubMed] [Google Scholar]
- 36.Hu, J., McCall, C. M., Ohta, T., and Xiong, Y. (2004) Nat. Cell Biol. 6 1003-1009 [DOI] [PubMed] [Google Scholar]
- 37.Jin, J., Arias, E. E., Chen, J., Harper, J. W., and Walter, J. C. (2006) Mol. Cell 23 709-721 [DOI] [PubMed] [Google Scholar]
- 38.Cang, Y., Zhang, J., Nicholas, S. A., Kim, A. L., Zhou, P., and Goff, S. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2733-2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu, J., and Xiong, Y. (2006) J. Biol. Chem. 281 3753-3756 [DOI] [PubMed] [Google Scholar]
- 40.Kim, Y., Starostina, N. G., and Kipreos, E. T. (2008) Genes Dev. 22 2507-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kross, J., Henner, W. D., Hecht, S. M., and Haseltine, W. A. (1982) Biochemistry 21 4310-4318 [DOI] [PubMed] [Google Scholar]
- 42.Arias, E. E., and Walter, J. C. (2006) Nat. Cell Biol. 8 84-90 [DOI] [PubMed] [Google Scholar]
- 43.Miura, M., Sasaki, T., and Takasaki, Y. (1996) Radiat. Res. 145 75-80 [PubMed] [Google Scholar]
- 44.Balajee, A. S., and Geard, C. R. (2001) Nucleic Acids Res. 29 1341-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Connell, B. C., and Harper, J. W. (2007) Curr. Opin. Cell Biol. 19 206-214 [DOI] [PubMed] [Google Scholar]
- 46.Prives, C., and Gottifredi, V. (2008) Cell Cycle 7 3840-3846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.