Abstract
Liver glucokinase (LGK) plays an essential role in controlling blood glucose levels and maintaining cellular metabolic functions. Expression of LGK is induced mainly regulated by insulin through sterol regulatory element-binding protein-1c (SREBP-1c) as a mediator. Since LGK expression is known to be decreased in the liver of liver X receptor (LXR) knockout mice, we have investigated whether LGK might be directly activated by LXRα. Furthermore, we have studied interrelationship between transcription factors that control gene expression of LGK. In the current studies, we demonstrated that LXRα increased LGK expression in primary hepatocytes and that there is a functional LXR response element in the LGK gene promoter as shown by electrophoretic mobility shift and chromatin precipitation assay. In addition, our studies demonstrate that LXRα and insulin activation of the LGK gene promoter occurs through a multifaceted indirect mechanism. LXRα increases SREBP-1c expression and then insulin stimulates the processing of the membrane-bound precursor SREBP-1c protein, and it activates LGK expression through SREBP sites in its promoter. LXRα also activates the LGK promoter by increasing the transcriptional activity and induction of peroxisome proliferator-activated receptor (PPAR)-γ, which also stimulates LGK expression through a peroxisome proliferator-responsive element. This activation is tempered through a negative mechanism, where a small heterodimer partner (SHP) decreases LGK gene expression by inhibiting the transcriptional activity of LXRα and PPARγ by directly interacting with their common heterodimer partner RXRα. From these data, we propose a mechanism for LXRα in controlling the gene expression of LGK that involves activation through SREBP-1c and PPARγ and inhibition through SHP.
Several tissues are involved in maintaining optimal blood glucose levels. Among these, the liver plays a major role by maintaining the balance between the storage and release of glucose (1). Conversion of glucose to glucose 6-phosphate is a rate-controlling reaction in the hepatic glucose metabolism. If the hepatic glycogen stores are replete, glucose 6-phosphate enters the glycolytic pathway to produce pyruvate for de novo lipogenesis.
Glucose entering the liver is phosphorylated by hepatic glucokinase (liver glucokinase; LGK)4 (ATP:d-glucose 6-phosphotransferase; EC 2.7.1.1). Since the Km value of LGK is considerably higher than normal blood glucose concentrations and LGK is not subjected to allosteric regulation by the end product, the rate of glucose phosphorylation is directly proportional to the blood glucose level. Thus, LGK is considered to play an essential role for sensing and maintaining proper blood glucose levels (2, 3). GK is mainly expressed in the liver, pancreatic β cells, and neuroendocrine cells of the brain. Two alternate promoters regulate the tissue-specific expression of the GK gene in the liver and pancreatic β cells (4). An upstream promoter regulates β cell-specific GK expression, whereas the promoter that regulates LGK gene expression is controlled by a downstream promoter. In the liver, LGK gene expression is regulated in response to fasting and refeeding (5), with insulin and glucagon serving as the mediators of this response. Insulin stimulates LGK gene expression in primary cultured hepatocytes regardless of glucose concentration, and glucagon inhibits LGK gene expression (6, 7). The action of insulin on the up-regulation of LGK transcription is mediated by the SREBP-1c (8, 9).
LXRs are nuclear receptors that sense oxysterols and regulate cholesterol and lipid metabolism (10, 11). The LXR family consists of the LXRα and LXRβ isoforms. LXRα is expressed primarily in liver, adipose tissue, kidney, macrophage and intestine, whereas LXRβ is present ubiquitously (12). LXRs form heterodimers with RXRα and bind to target DNA sequences known as the LXR response element (LXRE) (13). In response to oxysterols, LXRs activate genes involved in reverse cholesterol transport and hepatic cholesterol metabolism (14). LXRα also increases the synthesis of fatty acids by either up-regulating SREBP-1c or binding to the promoters of some lipogenic genes directly (15–17). Other diverse roles of LXRα include the inhibition of the expression of gluconeogenic genes, such as phosphoenolpyruvate carboxykinase and glucose-6-phosphatase (18). In addition, LXRα stimulates adipocyte differentiation through induction of PPARγ expression (19).
SHP is an atypical nuclear receptor that lacks a conventional DNA binding domain (20, 21). It is known to interact with other transcription factors and represses transcriptional activity by either competing with other coactivators for binding to an affected transcription factor or by recruiting corepressors directly to the transcriptional repression domain of SHP (22). SHP is expressed in liver and plays an important role in the regulation of cholesterol homeostasis (23, 24). Also, it is known that SHP regulates LXR transcriptional activity and augments the transcriptional activity of PPARγ (25, 26).
In this study, we identified an interactive mechanism for regulation of LGK expression through LXRα with critical roles for SREBP-1c, PPARγ, and SHP. LXRα directly activated LGK expression by binding to the LXRE in the LGK promoter, and LXRα-mediated LGK gene expression was also indirectly up-regulated by increasing SREBP-1c gene expression and increasing transcriptional activity of PPARγ. We also show that SHP inhibited LXRα-dependent transcriptional activation of LGK by interacting with RXRα. These results suggest a multicomponent mechanism for regulation of LGK expression by diverse nuclear receptors.
EXPERIMENTAL PROCEDURES
Plasmids and Materials—Construction of the luciferase reporter of the rat LGK promoter, pRGKL-1448, and its mutants m2 and mab were described earlier (9, 27). Mutant clones m4, m5, m6, and m7 were constructed by introducing substitution mutations into pRGKL-1448. Expression plasmids for mPPARγ, mRXRα, and β-galactosidase and their control vectors were previously described (9, 28). Human SHP expression vector was a kind gift from Dr. Heung-Sik Choi, and mouse LXRα expression vector was a kind gift from Dr. David J. Mangelsdorf (29, 30). The expression plasmid for myc-His-tagged human LXRα was received from Dr. J. B. Kim (Seoul National University, Korea). LGK-PPRE×3tkLuc and LGK-LXRE×3tkLuc were prepared by inserting three copies of -119/-98 and -56/-32 regions of the LGK promoter, respectively. APRE or GPRE was prepared by inserting three copies of PPRE from the acyl-CoA oxidase promoter (APRE) or β cell-specific GK promoter (GPRE) in front of the thymidine kinase minimal promoter in the luciferase reporter (ptkLuc). To generate Gal4DBD-PPARγ, Gal4DBD-RXRα, Gal4DBD-SHP, VP16-PPARγ, VP16-RXRα, and VP16-SHP constructs used in the mammalian two-hybrid assay, full-length cDNAs of PPARγ, RXRα, and SHP were prepared by PCR amplification, and then these fragments were subcloned into pM and pVP16 vectors, respectively (Clontech). All of the plasmid constructs were confirmed by DNA sequencing. Rosiglitazone was a kind gift from GlaxoSmithKline Korea (Seoul, Korea), and 9-cis-retinoic acid (9-CR), 22-hydroxycholesterol, and insulin were purchased from Sigma. TO-901317 (T1317) was purchased from Cayman (Ann Arbor, MI).
Cell Culture and Transient Transfection Assay—Alexander cells (human hepatoma cell lines; ATCC number CRL8024) and CV-1 cells (Monkey kidney cell lines; ATCC number CCL70) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The culture medium for Alexander cells was supplemented with 50 μg/ml tylosine for an anti-pleuropneumonia-like organisms (PPLO) agent. Primary cultured hepatocytes were isolated from Sprague-Dawley rats (∼200 g) and cultured as described previously (9). Transient transfection and luciferase assays were performed using Lipofectamine PLUS reagent (Invitrogen) and a luciferase assay kit (Promega, Madison, WI) (27). Luciferase activities were normalized with β-galactosidase activities to adjust transfection efficiency. Normalized luciferase activities are shown as the mean ± S.D. of three independent experiments performed in triplicate and are expressed as -fold increases relative to the basal activity of the reporters in the absence of overexpression vectors.
Preparation of Recombinant LXRα Protein and Anti-LXRα Antibody—To prepare bacterial recombinant fusion proteins, we cloned cDNA of mouse LXRα into pET21 bacterial expression plasmid and transformed into Escherichia coli (BL21-DE3). The recombinant LXRα expression was induced for 4 h with 0.5 mm isopropyl-β-d-thiogalactopyranoside. The recombinant protein containing polyhistidine (His) tag was purified by Ni2+-nitrilotriacetic acid-agarose resin chromatography (Peptron, Taejeon, Korea). One milligram of recombinant LXRα protein was suspended in 1 ml of phosphate-buffered saline and emulsified with 1 mm Freund's complete adjuvant (Sigma), which was injected subcutaneously into the dorsal side of a New Zealand White rabbit. The second and third immunizations were performed in the same way to boost production of anti-LXRα antibody. One month after the third immunization, we collected blood from immunized rabbit and prepared serum. Specificity of immunized rabbit serum (IgG) was confirmed by Western blot using myc-His-LXRα-overexpressed HEK293 cell extract and used for the experiment.
Isolation of Total RNA, Reverse Transcription, and Quantitative PCR—Total RNA was extracted from primary cultured hepatocytes using the Illustra RNAspin Mini RNA isolation kit (GE Healthcare) according to the manufacturer's protocol. First strand cDNA was synthesized from 2 μg of total RNA in a 20-μl volume using random hexamers and ImProm II Reverse transcriptase (Promega, Madison, WI). Real time quantitative PCR was performed using the ABI PRISM 7500 sequence detection system instrumentation and software (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol with minor modifications. Briefly, the appropriate amount of the reverse transcription reaction mixture was amplified with specific primers using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). Expression levels of target genes were determined by generating a five-point serial standard curve. This curve was used to calculate the amount of target gene mRNA in vehicle- and ligand-treated samples based on real time PCR. RNA samples were normalized by determining β-actin mRNA level. Primers used in PCR were as follows: LGK-f, 5′-CTCAGGAGTCAGGAACATCT-3′; LGK-r, 5′-TGACCAGCATCACTCTGAAG-3′; SREBP-1c-f, 5′-GGAGCCATGGATTGCACATT-3′; SREBP-1c-r, 5′-AGGAAGGCTTCCAGAGAGGA-3′; LXRα-f, 5′-GAGAAGCTGGTGGCTGCCCA-3′; LXRα-r, 5′-AGCTGTAGGAAGCCAGGGAG-3′; PPARγ-f, 5′-TCCGTGATGGAAGACCACTC-3′; PPARγ-r, 5′-CCCTTGCATCCTTCACAAGC-3′; FAS-f, 5′-AGCCTAACACCTCTGTGCAGT-3′; FAS-r, 5′-TCCTTGCAGCCATCTGTGTTC-3′; SHP-f, 5′-CCTCTTCAACCCAGATGTGCC-3′; SHP-r, 5′-GTTCAGTGATGTCAACATCTCC-3′; β-actin-f, 5′-TTGTAACCAACTGGGACGATATGG-3′; β-actin-r, 5′-CGACCAGAGGCATACAGGGACAAC-3′.
Electrophoretic Mobility Shift Assay—In vitro translated proteins of LXRα-myc-His and RXRα-FLAG were prepared by using a coupled transcription/translation kit (Promega, Madison, WI). Synthesis of full-length LXRα-myc-His and RXRα-FLAG proteins was confirmed by Western blotting. An oligonucleotide covering the -59/-29 region of the rat LGK promoter was used as a wild-type probe. The probes were labeled as previously described (31). Briefly, 10 pmol of single-stranded sense oligonucleotide was labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs Inc., Ipswich, MA) and annealed with a 5-fold molar excess of antisense oligonucleotide. 32P-Labeled double-stranded oligonucleotides were purified with Sephadex G25 spin columns (Amersham Biosciences). Ten thousand cpm (∼0.08 pmol) of probe was incubated with the in vitro translated protein for 30 min in 10 mm Hepes (pH 7.9), 1 mm MgCl2, 30 mm KCl, 1 mm dithiothreitol, 5% glycerol. One microgram of poly(dI-dC) was added to each reaction to suppress nonspecific binding. For competition assays, excessive unlabeled GLUT4 LXRE oligonucleotides were added to the reaction mixture (32). Two μl of anti-myc antibody were added to the reaction for the supershift assay (Cell signaling, Denver, MA). The oligonucleotides used in the electrophoretic mobility shift assay were as follows: wild type, 5′-CTGGCCCTGACCTTGTGACACTAGGCAGGG-3′; GLUT4 LXRE, 5′-CAGCCCCGGGTTACTTTGGGGCATTgCTCC-3′.
Chromatin Immunoprecipitation Assay—The chromatin immunoprecipitation assay was performed based on a previously described method with minor modification (33). Proteins were cross-linked to DNA by adding formaldehyde directly to the culture medium to a final concentration of 1% and incubating for 10 min at 37 °C. The cells were harvested after washing and lysed with SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1). The livers of adult male rats (ad libitum) were perfused with serum-free Dulbecco's modified Eagle's medium for 5 min through the portal vein and then cross-linked with 3% formaldehyde in serum-free Dulbecco's modified Eagle's medium. Chromatin complexes in the supernatants were immunoprecipitated overnight with 2 μg of preimmune serum or antibody specific for LXRα and RXRα (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4 °C with rotation. The GLUT2 promoter was used as a negative control (34). LGK promoter-specific primers were as follows: sense, 5′-ACCAGTGTTCTGTCATC-3′; antisense, 5′-GGTCTGTCTGGCTGAGT-3′. A known LXR binding site from the ABCA1 promoter was used as a positive control (35).
Small Interfering RNA Experiments—The siRNAs sequences for rat LXRα and scramble are AUUAGCAUCCGUGGGAACAUCAGUC and CCUACGCCACCAAUUUCGU-dTdT, respectively. For siRNA transfection, rat primary cultured hepatocytes were incubated with Dulbecco's modified Eagle's medium containing 10% fetal bovine serum in the absence of antibiotics for 12 h and transfected with 200 nm siRNA-scramble or siRNA-LXRα using Lipofectamine RNAiMAX following the manufacturer's protocol (Invitrogen). After 5 h, the media containing the respective ligands were changed and incubated for 24 h.
Adenoviral Transduction—The cDNA encoding full-length human SHP into pAdTrack-CMV shuttle vector was received from Dr. I. K. Lee (36). The cDNA encoding SREBP-1c-DN was described (9). The recombinant adenoviruses were amplified in 293A cells and purified using CsCl gradient centrifugation. Isolated primary hepatocytes were infected with 10 plaque-forming units/cell of Ad-SHP, Ad-DN-SREBP-1c, or Ad-null virus and incubated for 3 h.
Western Blot—Cell lysates were prepared using passive lysis buffer (Promega, Madison, WI) containing protease inhibitors. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare). The membranes were blocked and incubated with anti-HA (Covance Inc., CA), anti-α-tubulin (Calbiochem, Darmstadt, Germany) in fresh phosphate-buffered saline containing 1% nonfat dry milk overnight at 4 °C with agitation. Incubated membranes were washed and treated with horseradish peroxidase-conjugated secondary antibody. Then horseradish peroxidase signal was detected by using ECL substrate (GE Healthcare).
GST Pull-down Assay—GST and GST-RXRα proteins were expressed in E. coli (BL21-DE3) and conjugated with glutathione-agarose 4B beads (Peptron, Taejeon, Korea). In vitro translated SHP-HA was incubated with GST or GST-RXRα in HEMG buffer (40 mm Hepes (pH 7.9), 100 mm KCl, 0.2 mm EDTA, 5 mm MgCl2, 1.5 mm dithiothreitol, 0.1% Nonidet P-40, 10% glycerol) containing protease inhibitors. Beads were washed six times in HEMG buffer, boiled in SDS-PAGE sample buffer, and subjected to Western blotting.
RESULTS
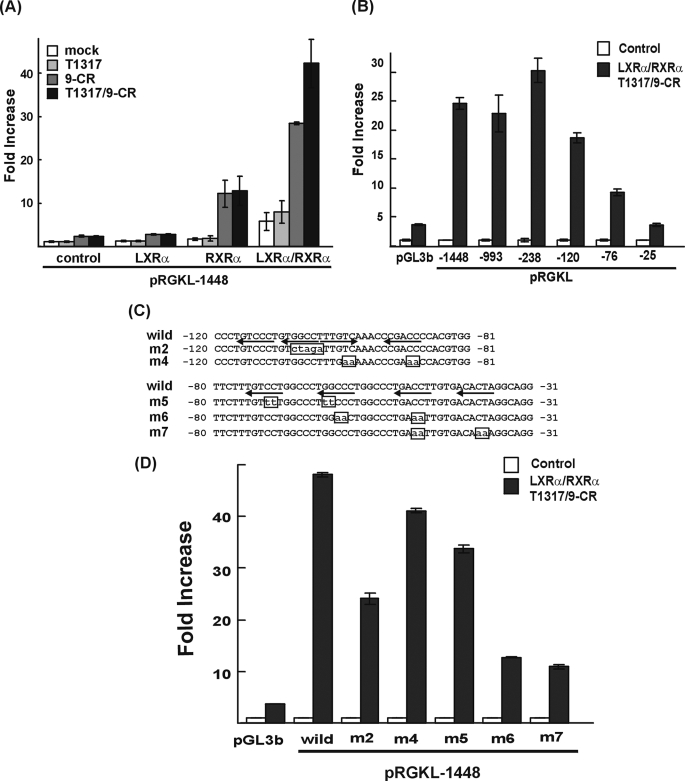
LXRα Activates LGK Expression Directly—SREBP-1c is known to mediate the regulation of LGK gene expression by insulin. Both LGK and SREBP-1c gene expressions are known to be increased in the livers of mice after administration of LXRα ligand for 3 days, and the effect was lost in LXR null mice (37, 38). Oral administration of LXRα ligand for 12 h did not increase LGK gene expression in the liver of SREBP-1c null mice, which suggested that SREBP-1c is required for LGK gene expression by LXRα ligand (39). However, we observed that LXRα activated the rat LGK promoter in Alexander cells, where SREBP-1a is predominantly expressed instead of SREBP-1c. As shown in Fig. 1, LGK promoter activity was not activated by T1317 and/or 9-CR when LXRα and/or RXRα expression vector were not transfected. Treatment of 9-CR resulted in an increase in the LGK promoter activity in the Alexander cells with the expression of RXRα. The coexpression of LXRα and RXRα activated the LGK promoter in the absence of ligands, probably due to a low concentration of endogenous oxysterol LXR ligands. The promoter activity was further increased by T1317/9-CR with the expression of LXRα and RXRα, suggesting that the LGK promoter was directly activated by the LXRα/RXRα heterodimer.
FIGURE 1.
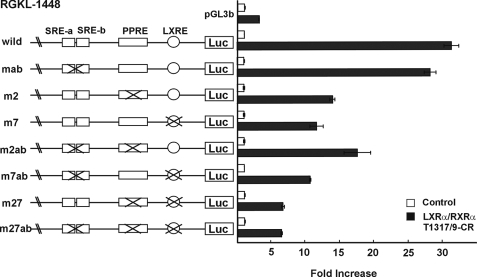
Identification of LXRE in the LGK gene promoter. A, Alexander cells were cotransfected with 100 ng of expression vectors for LXRα and/or RXRα and luciferase reporters under the control of rat LGK promoters (pRGKL-1448). After 24 h, the cells were treated with T1317 (1 μm) and/or 9-CR (1 μm) for 24 h. White bar, negative control; gray bar, T1317; dark gray bar, 9-CR; black bar, T1317- and 9-CR-treated group. B, 5′ serial deletion constructs of the LGK gene promoter reporter were transfected into Alexander cells. The positions of mutations are shown in Fig. 2C. D, luciferase reporter constructs containing mutants in the LGK gene promoter were transfected into Alexander cells. The cells were cotransfected with the expression vectors for LXRα and RXRα in the presence or absence of T1317 and 9-CR. White bar, no transfection of LXRα and RXRα and no treatment with 9-CR and T1317; black bar, transfection of LXRα and RXRα and treatment with T1317 and 9-CR (B and D). Normalized luciferase activities are shown as the mean ± S.D. of three independent experiments performed in triplicate and are expressed as -fold increases relative to the basal activity.
To identify the region responsible for the activation by LXRα, serial deletion constructs of LGK promoter-reporter were transfected into Alexander cells with the LXRα and RXRα expression vectors in the presence or absence of added T1317/9-CR (Fig. 1B). Deletion of the promoter down to -120 resulted in slight loss of LXRα-dependent LGK promoter activity; however, the LGK promoter was still substantially activated by LXRα. Further deletion down to -76 and -25 resulted in almost complete loss of LXRα responsiveness. If LXRα-dependent activation of LGK promoter is entirely mediated by SREBP-1c, deletion down to -120 is likely to result in complete loss of LXRα-dependent activation, because two SREs are present between bp -238 and -128 of the rat LGK promoter. But this deletion study suggested that the region between -120 and -25 was more responsible for the LXRα-dependent activation of the LGK promoter. Considering that a PPRE is present in the -120/-76 bp region of the LGK gene promoter (27) and that PPARγ is also categorized as a permissive binding partner of RXRα, the region between bp -76 and -25 is more likely to contain an LXRE. Unfortunately, this region did not contain an easily identifiable DR4, which is the high affinity LXRE (16). In order to find putative LXRE, five scanning mutants were prepared by introducing substitution mutations into the putative nuclear receptor half-site elements contained within the -120/-25 bp region (Fig. 1C), and the mutants were tested for responsiveness to LXRα (Fig. 1D). The PPRE mutation (m2) showed a 50% decrease in LXRα-dependent activation of LGK promoter, but m4 and m5 mutations retained LXRα responsiveness. Mutations at the m6 and m7 sites resulted in a 75% decrease in LXRα-dependent activation, suggesting the presence of a functional LXRE in this region. From these data, it is assumed that the -52/-37 bp region may be an LXRE, and LGK could be activated by LXRα through this site and the PPRE.
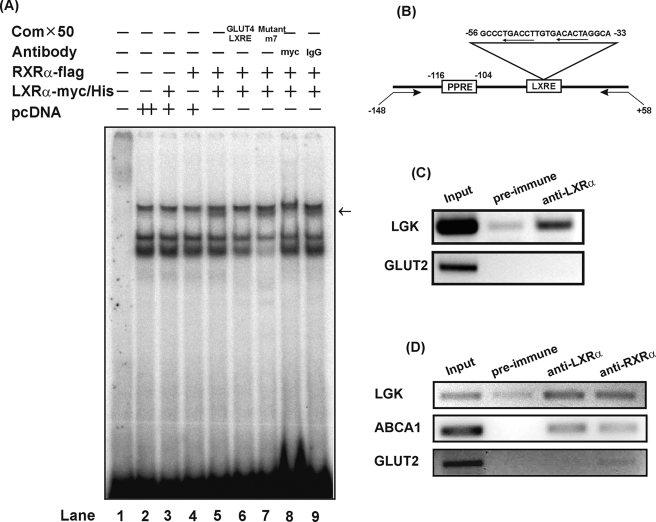
To know whether LXRα could bind to the putative LXRE in the -52/-37 bp region of the LGK promoter, we performed electrophoretic mobility shift assays using in vitro translated LXRα-myc-His and RXRα-FLAG. As shown in Fig. 2A, neither LXRα nor RXRα formed specific protein-DNA complex alone (lanes 3 and 4). When the probes were incubated with both LXRα and RXRα, a specific shifted band was observed, indicating the binding of an LXRα/RXRα heterodimer to the LGK-LXRE (lane 5). The addition of an excessive amount of unlabeled competitor LXRE (50-fold) from the GLUT4 promoter completely blocked LXRα/RXRα binding to the -59/-29 bp region of LGK promoter (lane 6), but unlabeled m7 mutant did not compete the binding of LXRα/RXRα to LXRE (lane 7). The specificity of the shifted complex was confirmed once again by anti-myc antibody (lane 8). The direct binding of LXRα to LGK-LXRE was further confirmed by a chromatin immunoprecipitation assay in both primary cultured hepatocytes (Fig. 2C) and liver (Fig. 2D). In addition to identifying the LXRE in LGK promoter through both in vitro and in vivo approaches, we needed to confirm whether the LGK-LXRE itself could respond to LXRα/RXRα, because its sequence was different from the conventional LXRE consensus sequence. To this end, three copies of LGK-LXRE were subcloned in front of the thymidine kinase minimal promoter (tk-Luc) and used to test its responsiveness to LXRα/RXRα. As shown in Fig. 7, LGK-LXRE was well activated by LXRα/RXRα, indicating that LGK-LXRE is functional both in the LGK promoter and the artificial promoter context. These results suggested that LXRα/RXRα could bind to the LGK promoter and directly activate LGK transcription in the liver.
FIGURE 2.
Binding of LXRα/RXRα to the LXRE in the LGK gene promoter. A, an electrophoretic mobility shift assay using in vitro translated protein from pcLXRα-myc-His and pcRXRα-FLAG expression vectors. The oligonucleotides covering the -59/-29 bp region (wild) was used as probes. 32P-labeled probe was incubated with 4 μl of the in vitro translated protein. Unlabeled GLUT4 LXRE and mutant LXRE(m7) with a 50-fold excess were used as a competitor (Com). A chromatin immunoprecipitation assay was adopted to confirm the binding of LXRα/RXRα to the LGK promoter in primary cultured hepatocytes (C) and liver (D). Primary cultured hepatocytes isolated from rats were treated with T1317 and 9-CR for 24 h and then were cross-linked using formaldehyde. The liver was fixed with 3% formaldehyde by perfusing the portal vein. Chromatins were incubated with anti-LXRα antiserum and anti-RXRα antibody. DNA in the presence or in the absence of antibody was immunoprecipitated, and PCR amplification of the DNA fragments was performed using primer pairs specific to the -148/+50 bp region of the rat LGK gene (B and D).
FIGURE 7.
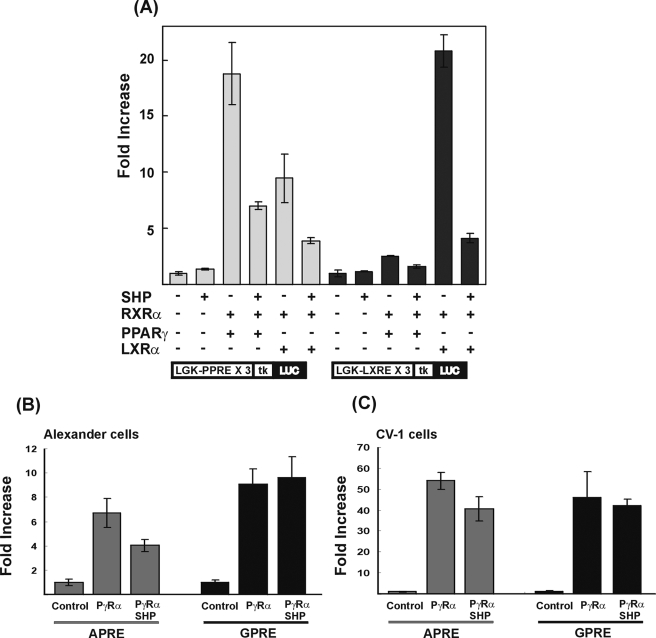
LXRα activates PPRE by increasing the transcriptional activity of the PPARγ. A, Alexander cells were cotransfected with 100 ng of expression vectors for LXRα, PPARγ, RXRα, and SHP and the luciferase reporter containing the three copies of LGK-PPRE or LGK-LXRE in front of a thymidine kinase minimal promoter, as indicated. The LGK-PPRE construct (which includes the -119/-98 region of the LGK promoter) and LGK-LXRE construct, which includes the -56/-32 region, were transfected into Alexander cells. For SHP, a plus or minus sign indicates the presence or absence of SHP overexpression. For LXRα, PPARγ, and RXRα, a plus sign indicates both overexpression of receptors and treatment of appropriate ligands, and a minus sign indicates without overexpression and no treatment of ligands. Alexander cells (B) or CV-1 cells (C) were cotransfected with 100 ng of expression vector for PPARγ, RXRα, and SHP and luciferase reporter containing the three copies of APRE or GPRE in front of the thymidine kinase minimal promoter. The cells were treated with their respective ligands as indicated. Normalized luciferase activities are shown as the mean ± S.D. of three independent experiments performed in triplicate.
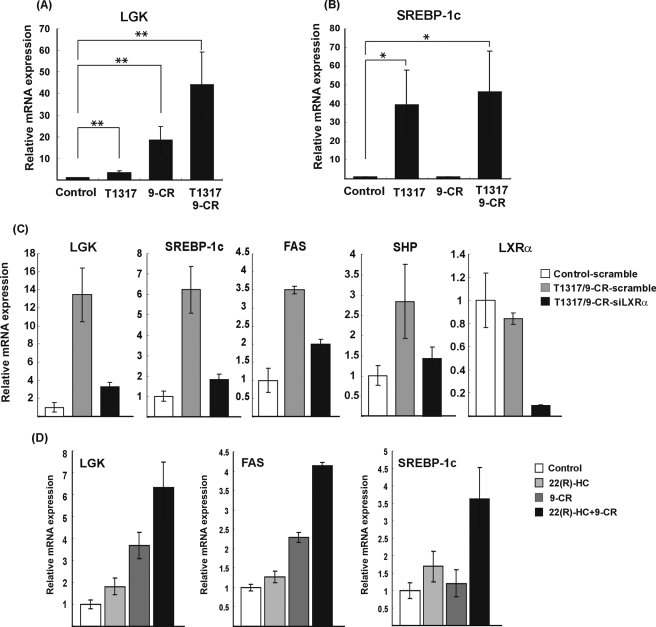
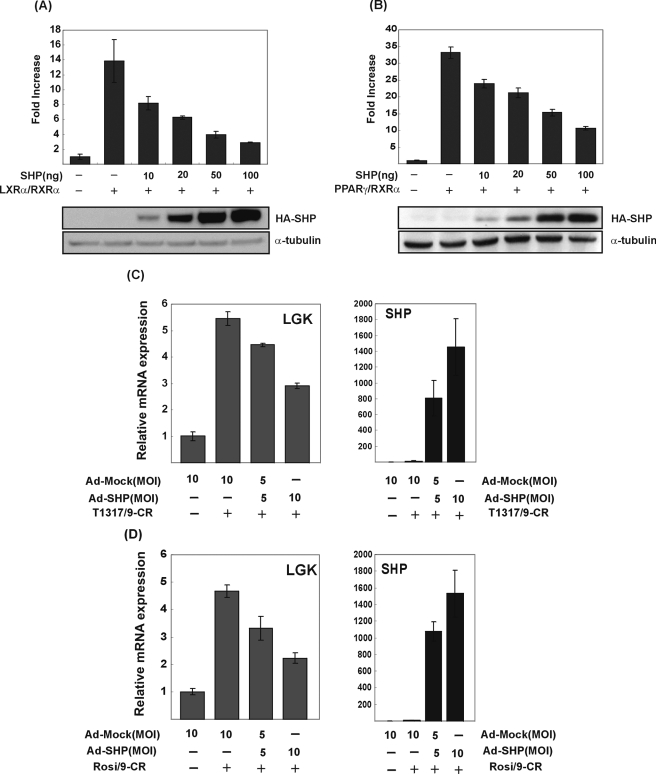
The promoter study led us to explore the precise molecular interplay between LXRα and SREBP-1c in the regulation of LGK gene expression. To delineate the relationship between LXRα and SREBP-1c upon LGK expression, primary hepatocytes were isolated from rat liver and treated with the LXRα ligand (T1317, 1 μm) and/or the RXRα ligand (9-CR, 1 μm) for 24 h. The mRNA level of LGK was increased either by T1317 (3-fold) or 9-CR (18-fold), respectively (Fig. 3A). Furthermore, combined treatment of T1317 and 9-CR resulted in a synergistic increase in LGK mRNA level (43-fold). In contrast, the pattern of SREBP-1c gene expression in response to the LXRα ligand did not match with that of LGK gene expression. SREBP-1c mRNA was increased by T1317 (39-fold), but 9-CR did not increase SREBP-1c gene expression. In addition, the combined treatment of T1317 and 9-CR did not result in the synergistic increase in the SREBP-1c mRNA level (Fig. 3B). If LXRα-dependent transcriptional induction of LGK gene is entirely mediated by SREBP-1c, the transcriptional induction pattern of LGK by the ligands of LXRα and/or RXRα could be similar to that of SREBP-1c. The different pattern of transcriptional induction by the ligands of LXRα and/or RXRα for LGK versus SREBP-1c suggested again that LXRα could activate LGK expression independently of SREBP-1c. We also confirmed the effect of LXRα on LGK expression by knocking down LXRα in primary hepatocytes. As shown in Fig. 3C, transfection of siRNA for LXRα, siLXRα, resulted in a 90% decrease in the LXRα expression. Knockdown of LXRα could block the increased gene expression of LGK as well as SREBP-1c by T1317/9-CR, and it also blocked the LXRα-induced gene expression of FAS and SHP, which are well known targets of LXRα (16, 40). In addition, we tested endogenous ligands of LXRα on LGK expression in primary hepatocytes. 22-Hydroxycholesterol increased the LGK mRNA level by 2-fold, and combined treatment of 22-hydroxycholesterol and 9-CR increased LGK mRNA level synergistically (Fig. 3D). Taken together, we concluded that T1317/9-CR could stimulate LGK expression through direct activation of LXRα in hepatocytes.
FIGURE 3.
LXRα increases LGK mRNA expression in primary cultured hepatocytes. Primary cultured hepatocytes isolated from rats were treated with T1317 (1 μm) and/or 9-CR (1 μm) for 24 h in the presence of 10% fetal bovine serum. The mRNA levels of LGK (A) and SREBP-1c (B) were quantitated by real time PCR. C, primary cultured hepatocytes were transfected with 200 nm siRNA-scramble (scramble) or siRNA-LXRα (siLXRα) using Lipofectamine RNAiMAX. After 5 h, the media containing 1 μm T1317, 1 μm 9-CR ligands were changed. After 24 h, mRNA levels of LGK, SREBP-1c, FAS, SHP, and LXRα were quantitated by real time PCR. D, primary hepatocytes were isolated from rats. After attachment of the cells, medium was changed with medium containing (22R)-hydroxycholesterol (10 μm) and/or 9-CR (1 μm) and incubated for 24 h. The quantity of mRNAs was normalized with respect to β-actin mRNA. Data were processed by the comparative CT method and expressed as -fold increase relative to the basal transcription level in the absence of ligands. Statistical significance of differences between untreated and ligand-treated hepatocytes was determined by the Mann-Whitney U test. *, p < 0.05; **, p ≤ 0.005.
LXRα Activates LGK Expression Indirectly—As shown in Fig. 1C, activation of the LGK promoter by LXRα was compromised by a mutation in the previously characterized PPRE (m2). Because LXRα is known to increase PPARγ expression and increase adiposity in white adipose tissue (19), it is likely that some effects of LXRα could be exerted through increasing PPARγ expression. In addition, LXRα is known to increase the expression of SREBP-1c, which in turn binds to two SREs in the LGK promoter and increases LGK gene expression in response to insulin (41). Together, PPARγ and SREBP-1c are likely to be involved in the regulation of the LGK promoter by LXRα. To explore these interrelationships further, we prepared LGK promoter reporter constructs with various combinations of mutations in the SRE, PPRE, and LXRE (Fig. 4). The mutated LGK promoter constructs were cotransfected into Alexander cells with expression vectors for LXRα/RXRα and treated with their respective ligands. Mutation of the PPRE (m2) or the LXRE (m7) resulted in an approximately 60% decrease in LXRα-dependent activation, whereas the SRE mutations (mab) did not cause any decrease in the LXRα-dependent activation. Introduction of SRE mutations into the PPRE mutant (m2ab) or LXRE mutant (m7ab) did not result in a further decrease in LXRα-dependent activation of the LGK promoter when compared with the PPRE mutant (m2) or LXRE mutant (m7). Double mutations in LXRE and PPRE (m27) and triple mutations (m27ab) caused the most significant decrease in the LXRα-dependent activation of LGK promoter. These results indicate that LXRα-dependent activation of the LGK promoter in Alexander cells is partly through the direct activation of the LXRE and partly through indirect activation of PPRE.
FIGURE 4.
Functional relationships among SREBP-1c, PPARγ, and LXRα in the activation of the LGK gene promoter. Luciferase reporter constructs of the LGK promoter and their mutants were transfected into Alexander cells. The positions of the mutations are shown. The cells were cotransfected with the expression vectors for LXRα and RXRα in the presence or absence of T1317 (1 μm) and/or 9-CR (1 μm). White bar, no transfection of LXRα and RXRα and no treatment with 9-CR and T1317; black bar, transfection of LXRα and RXRα and treatment with T1317 and 9-CR. Normalized luciferase activities are shown as the mean ± S.D. of three independent experiments performed in triplicate and are expressed as -fold increases relative to the basal activity.
To test our hypothesis for indirect activation of the PPRE in the LGK promoter by LXRα, three copies of each of the LGK-PPRE and LGK-LXRE were subcloned in front of a thymidine kinase minimal promoter in the luciferase reporter, and the resulting constructs were tested for responsiveness to LXRα/RXRα or PPARγ/RXRα (Fig. 7). LGK-PPRE was activated both by LXRα/RXRα and PPARγ/RXRα, and the LGK-LXRE was activated only by LXRα/RXRα, suggesting the indirect activation of PPRE by LXRα.
In an attempt to confirm the effects of LXRα in connection with SREBP-1c, primary hepatocytes were treated with T1317/9-CR for 24 h in the presence or absence of insulin and with or without expression of a dominant negative form of SREBP-1c (DN-SREBP-1c). As shown in Fig. 5A, T1317/9-CR increased LGK mRNA (7-fold) without insulin, and insulin increased LGK mRNA level (6-fold). The combination of T1317/9-CR/insulin increased the LGK mRNA level synergistically (22-fold). Although expression of DN-SREBP-1c inhibited the stimulation of LGK expression by T1317/9-CR in the presence of insulin, it did not inhibit its expression in the absence of insulin. We also observed that treatment of T1317/9-CR and/or insulin and the expression of DN-SREBP-1c had similar effects on the gene expression of FAS, which is known to be activated by both SREBP-1c and LXRα (Fig. 5B). These results suggest that LXRα directly activates LGK gene expression. In addition, the activation occurs indirectly by increasing SREBP-1c gene expression.
FIGURE 5.
Effects of LXRα and insulin on the LGK and FAS mRNA levels. Primary cultured hepatocytes were isolated from rat liver and infected with Ad-GFP (multiplicity of infection of 10) or Ad-DN-SREBP-1c (multiplicity of infection of 10) for 3 h. Cells were treated with insulin (100 nm) and/or T1317 (1 μm)/9-CR (1 μm) for 24 h in the absence of fetal bovine serum as indicated. mRNA levels of LGK (A) and FAS (B) were quantitated by real time PCR. The quantity of mRNAs was normalized with respect to β-actin mRNA. Data were processed by the comparative CT method and expressed as -fold increase relative to the basal transcription level in the absence of ligands.
SHP Functions as a Negative Modulator of LGK Gene Transcription—SHP interacts with various transcriptional factors and represses their transcriptional activity either by competing with coactivators or recruiting corepressors (22, 42). SHP is also known to interact with LXRα and RXRα and inhibit the transcriptional activity of these receptors (25, 42). In contrast to LXRα and RXRα, SHP was reported to augment PPARγ transcriptional activity by competing with the binding of a corepressor (26). These opposite effects of SHP on LXRα and PPARγ led us to explore the role of SHP in transcriptional regulation of LGK by PPARγ and LXRα.
To examine the effect of SHP on LGK, the LGK promoter reporter construct (pRGKL-1448) was transfected to Alexander cells, and an expression vector for SHP was cotransfected with vectors for LXRα or PPARγ. As shown in Fig. 6A, LXRα-dependent activation of LGK promoter was decreased by SHP in a dose-dependent manner. When equal amounts of SHP and LXRα were transfected, most of the LXRα-dependent transcriptional activation was abolished. SHP also inhibited PPARγ-dependent activation of the LGK promoter (Fig. 6B). To know whether these repressive effects of SHP on LXRα- and PPARγ-dependent activation of the LGK promoter could inhibit the activation of LGK gene expression by LXRα and PPARγ, we transduced adenovirus expressing SHP into primary hepatocytes and treated cells with the ligands of PPARγ and LXRα. As shown in Fig. 6, C and D, adenoviral expression of SHP inhibited the LXRα- and PPARγ-dependent stimulation of LGK gene expression in primary hepatocytes. The repressive effect of SHP was further confirmed by using LGK-PPRE and LGK-LXRE artificial reporter promoter constructs (Fig. 7A). LGK-PPRE was activated by either PPARγ or LXRα, and both of the activations were inhibited by SHP. In contrast, LGK-LXRE was activated by LXRα, which was also inhibited by SHP. These results suggested that SHP could inhibit the LXRα- and PPARγ-dependent transactivation of LGK promoter.
FIGURE 6.
SHP inhibits LXRα and PPARγ-mediated LGK promoter activity. Alexander cells were cotransfected with expression vectors for LXRα (100 ng) and RXRα (100 ng) (A) or PPARγ (100 ng) and RXRα (100 ng) (B) with SHP expression vector and LGK promoter-luciferase reporter (pRGKL-1448). Cells were treated with their respective ligands. Normalized luciferase activities are shown as mean ± S.D. of three independent experiments in triplicate and are expressed as -fold increase relative to the basal activity. The expression of HA-SHP protein was validated by immunoblotting (A and B). Rat primary cultured hepatocytes were transduced by adenoviral expression of SHP. After 3 h, T1317 (1 μm)/9-CR (1 μm) (C) or rosiglitazone (1 μm)/9-CR (1 μm) (D) were treated for 24 h. The transcription levels of LGK and expression of SHP were quantitated by real-time PCR (C and D).
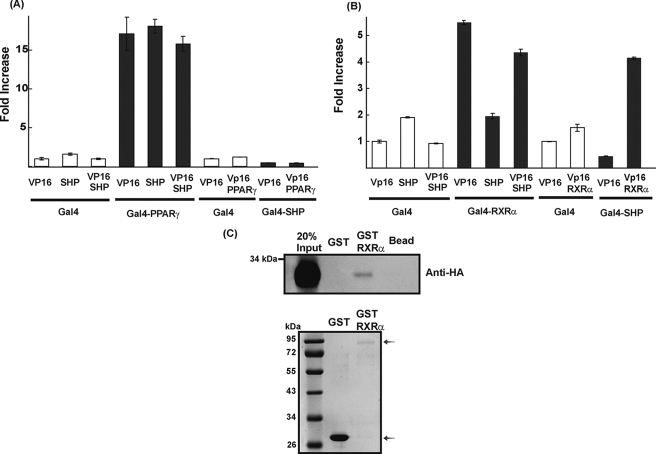
In order to know whether the inhibitory action of SHP on PPARγ is LGK-specific or not, we repeated the same experiment using reporter constructs that contain three copies of APRE or GPRE in front of the thymidine kinase minimal promoter (Fig. 7, B and C). The APRE reporter construct was activated by PPARγ/RXRα and suppressed by ectopic expression of SHP in both Alexander cells and CV-1 cells. However, activation of GPRE by PPARγ/RXRα was not affected by SHP in these cells. To explore a precise mechanism of how SHP inhibits the activation of the LGK promoter by LXRα and PPARγ, we performed a mammalian two-hybrid assay using a luciferase reporter that has upstream activating sequence in front of the thymidine kinase minimal promoter. We employed a system utilizing a GAL4-PPARγ fusion and SHP fused with or without the VP16 activation domain or the reciprocal combination of fusion proteins. As shown in Fig. 8A, expression of the Gal4-PPARγ fusion protein increased luciferase activity in the presence of rosiglitazone (1 μm). However, neither SHP nor VP16-SHP affected Gal4-PPARγ activity. We further confirmed the interaction of PPARγ and SHP in the reciprocal combination. Gal4-SHP expression decreased luciferase activity because of the intrinsic repression function of SHP.
FIGURE 8.
SHP interacts with RXRα but not with PPARγ. Alexander cells were cotransfected with expression vectors for Gal4DBD (100 ng) or fusion proteins Gal4DBD-PPARγ (100 ng), Gal4DBD-RXRα (100 ng), Gal4DBD-SHP (100 ng), VP16 (100 ng) or VP16-PPARγ (100 ng), VP16-RXRα (100 ng), VP16-SHP (100 ng), and the luciferase reporter construct (pUAS) containing three copies of the Gal4 binding site in front of the thymidine kinase minimal promoter. The cells were treated with rosiglitazone (1 μm) for PPARγ or 9-CR (1 μm) for RXRα. Normalized luciferase activities are shown as mean ± S.D. of three independent experiments in a triplicate and are expressed as -fold increase relative to the basal activity (A and B). In vitro translated SHP-HA protein was incubated with GST or GST-RXRα fusion protein bound to glutathione Q-Sepharose beads. Protein interaction was detected by immunoblotting using anti-HA antibody. The quantity ofGST and GST-RXRα fusion protein were confirmed by Coomassie Brilliant Blue staining (C).
Coexpression of Gal4-SHP and VP16-PPARγ did not stimulate luciferase activity that was repressed in Gal4-SHP. This result suggested that PPARγ did not interact with SHP in hepatocytes. By contrast, SHP did repress the promoter activity that was activated by the Gal4-RXRα (Fig. 8B), and VP16-SHP restored the promoter activity that was repressed by SHP, suggesting that the VP16 activation domain compensated for the repressive effect of SHP on RXRα-dependent activation.
When we checked the interaction between RXRα and SHP with the reciprocal combination, Gal4-SHP decreased luciferase activity, and coexpression of VP16-RXRα with Gal4-SHP stimulated the luciferase activity from the repressed state. The interaction between RXRα and SHP was further confirmed by the GST pull-down assay using in vitro translated HA-SHP and GST-RXRα fusion protein (Fig. 8C). These data do not agree with earlier reports that SHP increases the transcriptional activity of PPARγ by directly interacting with PPARγ (26, 43). It is not clear whether repression of PPARγ by SHP is a gene-specific phenomenon or not. Based on our experiments using Alexander cells and CV-1 cells, it is likely that this phenomenon may not be cell-specific. We also tried coimmunoprecipitation to confirm the interaction of PPARγ and SHP in a different way, but interaction between PPARγ and SHP was not observed (data not shown). Therefore, our results suggested that SHP functioned as a negative modulator of the PPARγ and that LXRα mediated LGK gene transcription by interacting with RXRα.
DISCUSSION
Intracellular glucose 6-phosphate is the primary physiologic stimulus for the hepatic glucose metabolism, since it is a substrate for both glycolysis and glycogen synthesis (44), and small changes in the expression of the enzyme LGK, which produces glucose 6-phosphate from glucose, can have a profound impact on the blood glucose concentration (2, 3, 45). Here, we present data indicating that LXRα plays an orchestrated role in the regulation of LGK expression. We have identified an LXRE in the LGK promoter and shown that LXRα could activate LGK expression directly through binding to its promoter. We have also shown that LXRα activates the LGK promoter indirectly through a mechanism requiring PPARγ and SREBP-1c. In addition, we showed that SHP functions as a negative modulator of LGK transcription by inhibiting the transcriptional activity of LXRα and RXRα.
LXRα has an insulin-like effect on hepatic carbohydrate metabolism by stimulating genes involved in glucose storage and inhibiting gluconeogenesis, which would be consistent with a role in promoting energy storage in a model of diet-induced obesity and insulin resistance (37). Recently, Oosterveer et al. (46) reported defective induction of lipogenic gene expression after fasting/refeeding in LXRα KO mice. Thus, LXRα has an important role in the coordination of lipid and glucose metabolism, which suggests that LXRα ligands may have therapeutic potential through the modulation of glucose homeostasis. LXRα is known to activate SREBP-1c expression and thereby increases the activity of genes involved in glycolysis and lipogenesis (15, 47). However, there are still some questions of whether the relationship between SREBP-1c and LXRα exists, LXRα is involved directly in the physiologic effects of insulin, or the role of LXRα is solely to activate SREBP-1c, and these questions need to be explored.
Insulin is known to increase LXRα-dependent SREBP-1c expression by producing endogenous LXRα ligands (47). Recently, Hegarty et al. (41) showed that conversion of the membrane-bound SREBP-1c precursor into the mature soluble transcription factor is enhanced after acute exposure to insulin, and LXRα increases LGK expression in primary cultured hepatocytes by increasing SREBP-1c gene expression in the presence of insulin. Administration of the LXRα ligand GW3965 for 3 days increased LGK expression in mouse liver. In addition, treatment of mice with another synthetic LXR ligand, T1317, for 12 h did not increase LGK gene expression in the SREBP-1c null mouse (38, 39). These data suggested that LGK induction by LXRα is mediated by SREBP-1c. But there is a caveat to be considered. When Liang et al. (39) tested GK gene expression in the liver of the SREBP-1c null mouse, they treated mice with GW3695 instead of T1317 only for 12 h in a chow diet. This treatment protocol seemed to be less effective than 3 days' gavage feeding of T1317, which potentially makes it more difficult to observe changes in gene expression that is not strongly activated by LXRα. In addition, we used rat instead of mouse LGK genes and rat primary cultured hepatocytes. We compared the 3-kb upstream region from exon 1 of the mouse LGK gene, but we could not find a similar sequence with LGK-LXRE of rats. Thus, there might be a species difference in the regulation mechanism of LGK expression by LXRα. We have also observed that LXRα activates the LGK promoter through a PPRE. This is likely to be indirect, because it has been reported that LXRα could increase the production of endogenous PPARγ agonists through the up-regulation of SREBP-1c gene as well as the expression of PPARγ (19, 48). In these results, a role for ligand activation of RXRα, a permissive partner of LXRα, cannot be ruled out. However, overexpression of RXRα alone did not activate LGK-PPRE in the artificial promoter context (data not shown). Based on all of our observations, we propose a novel mechanism of activation for LGK by LXRα, which involves both SREBP-1c and PPARγ.
Finally, we also showed that SHP inhibited the transcriptional activation of the LGK promoter by LXRα and PPARγ. We also showed that adenoviral expression of SHP significantly decreased LGK expression in the primary hepatocytes. Our results suggest that the inhibitory effects of SHP on LXRα-or PPARγ-mediated LGK gene expression are supposed to occur through mutual interaction with RXRα, resulting in the decrease in LGK promoter activity. Considering that SHP is also known to work as a negative regulator for the transcriptional activators involved in the gluconeogenic genes as well as the LGK gene (49), we speculate that SHP plays a fine tuning role in the hepatic glucose metabolism by modulating transcription factors.
We provide a model summarizing the orchestrated role of PPARγ, LXRα, SREBP-1c, and the negative modulator SHP in the regulation of LGK gene transcription (Fig. 9). In this model, LXRα up-regulates the gene expression of LGK and SREBP-1c by direct binding to a cis-element in their promoters. Insulin activates the SREBP-1c gene as well as its maturation, resulting in the synergistic increase of LXRα-mediated LGK gene expression. In addition, we propose that LXRα increases LGK gene expression by increasing the transcriptional activity of PPARγ, possibly either by increasing the production of endogenous PPARγ ligand or by increasing PPARγ expression in hepatocytes (19, 48). LXRα and PPARγ increase the transcription of the SHP gene (40, 50), which in turn represses the transcription of the LXRα- or PPARγ-mediated LGK gene by interacting with RXRα.
FIGURE 9.
A model showing the interrelationship between LXRα, SREBP-1c, PPARγ, and SHP in the transcriptional regulation of the LGK gene. LXRα/RXRα binds the LXRE in the LGK gene promoter. LXRα-mediated LGK gene transcription is also increased by SREBP-1c or PPARγ in the indirect mechanism (shown as a dotted arrow). Up-regulation of the SHP gene by LXRα or PPARγ, in turn, plays a role in the fine tuning of the transcription of the LGK gene.
Acknowledgments
We thank Dr. Ronald M. Evans, Dr. Bruce M. Spiegelman, Dr. David J. Mangelsdorf, Dr. Heung-Sik Choi, Dr. In-Kyu Lee, and Dr. Jae Bum Kim for providing valuable clones and recombinant adenovirus and thank Dr. Sahng Wook Park and Dr. Kyung-Ah Kim for comments and suggestions. We thank Dr. Timothy Osborne (University of California, Irvine) for comments on the manuscript.
This work was supported by Korea Science and Engineering Foundation Grant R13-2002-054-03001-0 funded by the Korean government, Yonsei University College of Medicine faculty research grant for 2005 6-2005-0047, and Yonsei University Research Fund of 2005 Grant 6-2005-0084 (to H. K.).
Footnotes
The abbreviations used are: LGK, liver glucokinase; GK, glucokinase; SRE, sterol regulatory element; SREBP, SRE-binding protein; LXR, liver X receptor; LXRE, LXR response element; PPAR, peroxisome proliferator-activated receptor; PPRE, peroxisome proliferator-responsive element; SHP, small heterodimer partner; APRE, PPRE from the acyl-CoA oxidase promoter; GPRE, PPRE from the β cell-specific GK promoter; 9-CR, 9-cis-retinoic acid; His, polyhistidine; siRNA, small interfering RNA; HA, hemagglutinin; GST, glutathione S-transferase.
References
- 1.Nordlie, R. C., Foster, J. D., and Lange, A. J. (1999) Annu. Rev. Nutr. 19 379-406 [DOI] [PubMed] [Google Scholar]
- 2.Ferre, T., Riu, E., Bosch, F., and Valera, A. (1996) FASEB J. 10 1213-1218 [DOI] [PubMed] [Google Scholar]
- 3.Niswender, K. D., Shiota, M., Postic, C., Cherrington, A. D., and Magnuson, M. A. (1997) J. Biol. Chem. 272 22570-22575 [DOI] [PubMed] [Google Scholar]
- 4.Magnuson, M. A. (1990) Diabetes 39 523-527 [DOI] [PubMed] [Google Scholar]
- 5.Iynedjian, P. B., Pilot, P. R., Nouspikel, T., Milburn, J. L., Quaade, C., Hughes, S., Ucla, C., and Newgard, C. B. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 7838-7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iynedjian, P. B., Jotterand, D., Nouspikel, T., Asfari, M., and Pilot, P. R. (1989) J. Biol. Chem. 264 21824-21829 [PubMed] [Google Scholar]
- 7.Magnuson, M. A., Andreone, T. L., Printz, R. L., Koch, S., and Granner, D. K. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 4838-4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foretz, M., Guichard, C., Ferré, P., and Foufelle, F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12737-12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, S. Y., Kim, H. I., Kim, T. H., Im, S. S., Park, S. K., Lee, I. K., Kim, K. S., and Ahn, Y. H. (2004) J. Biol. Chem. 279 30823-30829 [DOI] [PubMed] [Google Scholar]
- 10.Janowski, B. A., Grogan, M. J., Jones, S. A., Wisely, G. B., Kliewer, S. A., Corey, E. J., and Mangelsdorf, D. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 266-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tontonoz, P., and Mangelsdorf, D. J. (2003) Mol. Endocrinol. 17 985-993 [DOI] [PubMed] [Google Scholar]
- 12.Shulman, A. I., and Mangelsdorf, D. J. (2005) N. Engl. J. Med. 353 604-615 [DOI] [PubMed] [Google Scholar]
- 13.Willy, P. J., Umesono, K., Ong, E. S., Evans, R. M., Heyman, R. A., and Mangelsdorf, D. J. (1995) Genes Dev. 9 1033-1045 [DOI] [PubMed] [Google Scholar]
- 14.Repa, J. J., and Mangelsdorf, D. J. (2002) Nat. Med. 8 1243-1248 [DOI] [PubMed] [Google Scholar]
- 15.Repa, J. J., Liang, G., Ou, J., Bashmakov, Y., Lobaccaro, J. M., Shimomura, I., Shan, B., Brown, M. S., Goldstein, J. L., and Mangelsdorf, D. J. (2000) Genes Dev. 14 2819-2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph, S. B., Laffitte, B. A., Patel, P. H., Watson, M. A., Matsukuma, K. E., Walczak, R., Collins, J. L., Osborne, T. F., and Tontonoz, P. (2002) J. Biol. Chem. 277 11019-11025 [DOI] [PubMed] [Google Scholar]
- 17.Schultz, J. R., Tu, H., Luk, A., Repa, J. J., Medina, J. C., Li, L., Schwendner, S., Wang, S., Thoolen, M., Mangelsdorf, D. J., Lustig, K. D., and Shan, B. (2000) Genes Dev. 14 2831-2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao, G., Liang, Y., Broderick, C. L., Oldham, B. A., Beyer, T. P., Schmidt, R. J., Zhang, Y., Stayrook, K. R., Suen, C., Otto, K. A., Miller, A. R., Dai, J., Foxworthy, P., Gao, H., Ryan, T. P., Jiang, X. C., Burris, T. P., Eacho, P. I., and Etgen, G. J. (2003) J. Biol. Chem. 278 1131-1136 [DOI] [PubMed] [Google Scholar]
- 19.Seo, J. B., Moon, H. M., Kim, W. S., Lee, Y. S., Jeong, H. W., Yoo, E. J., Ham, J., Kang, H., Park, M. G., Steffensen, K. R., Stulnig, T. M., Gustafsson, J. A., Park, S. D., and Kim, J. B. (2004) Mol. Cell. Biol. 24 3430-3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seol, W., Choi, H. S., and Moore, D. D. (1996) Science 272 1336-1339 [DOI] [PubMed] [Google Scholar]
- 21.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schütz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., and Evans, R. M. (1995) Cell 83 835-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Y., and Dufau, M. L. (2004) Vitam. Horm. 68 1-48 [DOI] [PubMed] [Google Scholar]
- 23.Lee, H. K., Lee, Y. K., Park, S. H., Kim, Y. S., Park, S. H., Lee, J. W., Kwon, H. B., Soh, J., Moore, D. D., and Choi, H. S. (1998) J. Biol. Chem. 273 14398-14402 [DOI] [PubMed] [Google Scholar]
- 24.Sinal, C. J., Tohkin, M., Miyata, M., Ward, J. M., Lambert, G., and Gonzalez, F. J. (2000) Cell 102 731-744 [DOI] [PubMed] [Google Scholar]
- 25.Brendel, C., Schoonjans, K., Botrugno, O. A., Treuter, E., and Auwerx, J. (2002) Mol. Endocrinol. 16 2065-2076 [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa, H., Yamagata, K., Shimomura, I., Takahashi, M., Kuriyama, H., Kishida, K., Hotta, K., Nagaretani, H., Maeda, N., Matsuda, M., Kihara, S., Nakamura, T., Nishigori, H., Tomura, H., Moore, D. D., Takeda, J., Funahashi, T., and Matsuzawa, Y. (2002) J. Biol. Chem. 277 1586-1592 [DOI] [PubMed] [Google Scholar]
- 27.Kim, S. Y., Kim, H. I., Park, S. K., Im, S. S., Li, T., Cheon, H. G., and Ahn, Y. H. (2004) Diabetes 53 Suppl. 1, S66-70 [DOI] [PubMed] [Google Scholar]
- 28.Kim, H. I., Kim, J. W., Kim, S. H., Cha, J. Y., Kim, K. S., and Ahn, Y. H. (2000) Diabetes 49 1517-1524 [DOI] [PubMed] [Google Scholar]
- 29.Kim, J. Y., Chu, K., Kim, H. J., Seong, H. A., Park, K. C., Sanyal, S., Takeda, J., Ha, H., Shong, M., Tsai, M. J., and Choi, H. S. (2004) Mol. Endocrinol. 18 776-790 [DOI] [PubMed] [Google Scholar]
- 30.Peet, D. J., Turley, S. D., Ma, W., Janowski, B. A., Lobaccaro, J. M., Hammer, R. E., and Mangelsdorf, D. J. (1998) Cell 93 693-704 [DOI] [PubMed] [Google Scholar]
- 31.Kim, H. I., Cha, J. Y., Kim, S. Y., Kim, J. W., Roh, K. J., Seong, J. K., Lee, N. T., Choi, K. Y., Kim, K. S., and Ahn, Y. H. (2002) Diabetes 51 676-685 [DOI] [PubMed] [Google Scholar]
- 32.Dalen, K. T., Ulven, S. M., Bamberg, K., Gustafsson, J. A., and Nebb, H. I. (2003) J. Biol. Chem. 278 48283-48291 [DOI] [PubMed] [Google Scholar]
- 33.Latasa, M. J., Griffin, M. J., Moon, Y. S., Kang, C., and Sul, H. S. (2003) Mol. Cell. Biol. 23 5896-5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Im, S. S., Kim, J. W., Kim, T. H., Song, X. L., Kim, S. Y., Kim, H. I., and Ahn, Y. H. (2005) Exp. Mol. Med. 37 101-110 [DOI] [PubMed] [Google Scholar]
- 35.Wagner, B. L., Valledor, A. F., Shao, G., Daige, C. L., Bischoff, E. D., Petrowski, M., Jepsen, K., Baek, S. H., Heyman, R. A., Rosenfeld, M. G., Schulman, I. G., and Glass, C. K. (2003) Mol. Cell. Biol. 23 5780-5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, K. G., Lee, K. M., Seo, H. Y., Suh, J. H., Kim, H. S., Wang, L., Won, K. C., Lee, H. W., Park, J. Y., Lee, K. U., Kim, J. G., Kim, B. W., Choi, H. S., and Lee, I. K. (2007) Diabetes 56 431-437 [DOI] [PubMed] [Google Scholar]
- 37.Kalaany, N. Y., Gauthier, K. C., Zavacki, A. M., Mammen, P. P., Kitazume, T., Peterson, J. A., Horton, J. D., Garry, D. J., Bianco, A. C., and Mangelsdorf, D. J. (2005) Cell Metab. 1 231-244 [DOI] [PubMed] [Google Scholar]
- 38.Laffitte, B. A., Chao, L. C., Li, J., Walczak, R., Hummasti, S., Joseph, S. B., Castrillo, A., Wilpitz, D. C., Mangelsdorf, D. J., Collins, J. L., Saez, E., and Tontonoz, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5419-5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L., and Brown, M. S. (2002) J. Biol. Chem. 277 9520-9528 [DOI] [PubMed] [Google Scholar]
- 40.Goodwin, B., Watson, M. A., Kim, H., Miao, J., Kemper, J. K., and Kliewer, S. A. (2003) Mol. Endocrinol. 17 386-394 [DOI] [PubMed] [Google Scholar]
- 41.Hegarty, B. D., Bobard, A., Hainault, I., Ferré, P., Bossard, P., and Foufelle, F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 791-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, Y. K., Dell, H., Dowhan, D. H., Hadzopoulou-Cladaras, M., and Moore, D. D. (2000) Mol. Cell. Biol. 20 187-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, Y. S., Han, C. Y., Kim, S. W., Kim, J. H., Lee, S. K., Jung, D. J., Park, S. Y., Kang, H., Choi, H. S., Lee, J. W., and Pak, Y. K. (2001) J. Biol. Chem. 276 33736-33740 [DOI] [PubMed] [Google Scholar]
- 44.Aiston, S., Trinh, K. Y., Lange, A. J., Newgard, C. B., and Agius, L. (1999) J. Biol. Chem. 274 24559-24566 [DOI] [PubMed] [Google Scholar]
- 45.Hariharan, N., Farrelly, D., Hagan, D., Hillyer, D., Arbeeny, C., Sabrah, T., Treloar, A., Brown, K., Kalinowski, S., and Mookhtiar, K. (1997) Diabetes 46 11-16 [DOI] [PubMed] [Google Scholar]
- 46.Oosterveer, M. H., van Dijk, T. H., Grefhorst, A., Bloks, V. W., Havinga, R., Kuipers, F., and Reijngoud, D. J. (2008) J. Biol. Chem. 283 25437-25445 [DOI] [PubMed] [Google Scholar]
- 47.Chen, G., Liang, G., Ou, J., Goldstein, J. L., and Brown, M. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11245-11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, J. B., Wright, H. M., Wright, M., and Spiegelman, B. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 4333-4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagata, K., Daitoku, H., Shimamoto, Y., Matsuzaki, H., Hirota, K., Ishida, J., and Fukamizu, A. (2004) J. Biol. Chem. 279 23158-23165 [DOI] [PubMed] [Google Scholar]
- 50.Kim, H. I., Koh, Y. K., Kim, T. H., Kwon, S. K., Im, S. S., Choi, H. S., Kim, K. S., and Ahn, Y. H. (2007) Biochem. Biophys. Res. Commun. 360 301-306 [DOI] [PubMed] [Google Scholar]