Abstract
Brain-derived neurotrophic factor (BDNF) signaling through its receptor, TrkB, modulates survival, differentiation, and synaptic activity of neurons. Both full-length TrkB (TrkB-FL) and its isoform T1 (TrkB.T1) receptors are expressed in neurons; however, whether they follow the same endocytic pathway after BDNF treatment is not known. In this study we report that TrkB-FL and TrkB.T1 receptors traverse divergent endocytic pathways after binding to BDNF. We provide evidence that in neurons TrkB.T1 receptors predominantly recycle back to the cell surface by a “default” mechanism. However, endocytosed TrkB-FL receptors recycle to a lesser extent in a hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs)-dependent manner which relies on its tyrosine kinase activity. The distinct role of Hrs in promoting recycling of internalized TrkB-FL receptors is independent of its ubiquitin-interacting motif. Moreover, Hrs-sensitive TrkB-FL recycling plays a role in BDNF-induced prolonged mitogen-activated protein kinase (MAPK) activation. These observations provide evidence for differential postendocytic sorting of TrkB-FL and TrkB.T1 receptors to alternate intracellular pathways.
Brain-derived neurotrophic factor (BDNF)3 has been shown to play critical roles in vertebrate nervous system development and function (1–3). The actions of BDNF are dictated by two classes of cell surface receptors, the TrkB receptor and the p75 neurotrophin receptor. BDNF binding to TrkB receptors activates several signaling cascades, including phosphatidylinositol 3-kinase, phospholipase C, and Ras/mitogen-activated protein kinase (MAPK) pathways, that mediate growth and survival responses to BDNF (1, 4, 5). It has been established that upon binding neurotrophins, Trk receptors are rapidly endocytosed in a clathrin-dependent manner (6, 7). Postendocytic sorting of Trk receptors to diverse pathways after ligand binding has a significant impact on the physiological responses to neurotrophins because they also determine the strength and duration of intracellular signaling cascades initiated by activated Trk receptors (8). Three alternate endocytic pathways that Trk receptors can follow are trafficking to lysosomes for degradation, recycling back to the plasma membrane, or being retrogradely transported (9–13). The degradative pathway to lysosomes is characterized by down-regulation of the total number of receptors at the cell surface and a decreased response to ligand. Conversely, recycling of receptors back to the plasma membrane can lead to functional resensitization and prolongation of cell surface-specific signaling events. A recent study has shown that recycled and re-secreted BDNF plays an important role in mediating the maintenance of long term potentiation in hippocampal slices, which suggests a potential role of TrkB recycling in long term potentiation regulation (14).
Different TrkB isoforms, including the full-length TrkB (TrkB-FL) and three truncated isoforms named TrkB.T1, TrkB.T2, and TrkB.T-Shc, exist in the mammalian central nervous system because of alternative splicing (15–17). Truncated TrkB.T1 receptor lacks the kinase domain but contains short isoform-specific cytoplasmic domain in its place (15, 16). Many neuronal populations, including hippocampal and cortical neurons, express both full-length and truncated TrkB receptors (18, 19). TrkB.T1 is expressed at low levels in the prenatal rodent brain, but its expression increases postnatally, ultimately exceeding the level of full-length TrkB in adulthood (19–22). The physiological function of the TrkB.T1 receptor remains unclear, but it may serve as dominant-negative regulator of full-length TrkB receptors (23–25), may sequester ligand and limit diffusion (26, 27), may regulate cell morphology and dendritic growth (28, 29), and may even autonomously activate signaling cascades in a neurotrophin-dependent manner (30). TrkB-FL and TrkB.T1 are localized to both somatodendritic and axonal compartments in neurons (31); however, little is known about TrkB.T1 endocytic trafficking fate upon BDNF treatment.
In this study we conducted an analysis of the postendocytic fates (degradation and recycling) of TrkB-FL and TrkB.T1 receptors in PC12 cells and neurons. We have determined that, unlike TrkB-FL, TrkB.T1 receptors recycle more efficiently in a default pathway to plasma surface after internalization, which is independent of hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs). Conversely, Hrs could bind with TrkB-FL in a kinase activity-dependent manner and regulate TrkB-FL receptors postendocytic recycling. Hrs was identified as a tyrosine-phosphorylated protein in cells stimulated with growth factors and cytokines (32). Hrs is expressed in the cytoplasm of all cells and is predominantly localized to endosomes (33). Hrs has also been proposed to play a role in regulating cell surface receptor postendocytic trafficking (34). These observations provide evidence for differential postendocytic sorting to alternate intracellular pathways between TrkB-FL and TrkB.T1 receptors after internalization.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Human recombinant BDNF was obtained from PeproTech (Rocky Hill, NJ). Antibodies were purchased as follows: rabbit anti-TrkB antibody from Millipore; mouse anti-Hrs from Alexis Biochemicals, Switzerland; mouse anti-FLAG (M1, M2) and rabbit anti-HA and mouse anti-tubulin antibodies from Sigma; Alexa Fluor® 488- or 594-conjugated goat anti-mouse or rabbit IgG (H+L) and Alexa Fluor® 594-labeled-human transferrin from Invitrogen; horseradish peroxidase-conjugated goat anti-mouse or rabbit IgG from Calbiochem. The restriction enzymes were purchased from MBI Fermentas (Hanover, MD). Sulfo-NHS-biotin, Sulfo-NHS-S-S-biotin, and chemiluminescence were from Pierce. Vectashield mounting medium was obtained from Vector Laboratories(Burlingame, CA). The other reagents were from Sigma-Aldrich.
Plasmid Constructs and siRNA Oligos—Rat TrkB-FL and TrkB.T1 cDNAs were subcloned into pCDNA3.1Neo expression vector (Invitrogen) by using EcoRI and EcoRV sites. The amino-terminal FLAG epitope tag was added after the signal peptide of TrkB by PCR. TrkB kinase dead mutant was made by site-directed mutagenesis. All the TrkB deletion constructs and TrkB-FL and TrkB.T1 chimeric constructs were generated by means of two-step PCR. HA-tagged Hrs and its mutants (ΔUIM, ΔVHS, VHS-FYVE) were subcloned into pCDNA3.1 plasmid (Invitrogen). All of the constructs were confirmed by DNA sequence to exclude potential PCR-introduced mutations. The detailed information of all the constructs used in the study was described in supplemental Table 1. To knockdown the expression of Hrs, 19 nucleotides of Hrs sequence (GACGTGTGGGTTCTTGTCG for human (35) and CGACAAGAACCCACACGTC for rat) were targeted with small interfering RNA using pSuper mammalian RNA expression vector (OligoEngine, Seattle, WA) according to the manufacturer's instructions. The resulting Hrs siRNA and the control siRNA constructs were transfected into HEK293 or PC12 cells using Lipofectamine 2000 reagent (Invitrogen), and the levels of Hrs were determined by Western blotting with anti-Hrs antibodies.
Hippocampal Neuron Cultures and Transfection—Cultured hippocampal neurons from timed-pregnant Sprague-Dawley rats were prepared as previously described (36). In brief, hippocampi were dissected from embryonic day 18 rats, dissociated with 0.25% trypsin in Hanks' balanced salt solution without Ca2+ and Mg2+ at 37 °C for 15 min, triturated in Dulbecco's modified Eagle's medium, F12, 10% fetal bovine serum. For transfection, the neurons were seeded onto coverslips coated with 0.1 mg/ml poly-d-lysine in 6-well plates at a cell density of 5 × 105 cells/ml. Neurons were grown in Neurobasal media (Invitrogen) containing 2% B27 supplement (Invitrogen), 0.5 mm l-glutamine (Invitrogen), and 100 units/ml penicillin-streptomycin (Sigma) at 37 °C, 5% CO2, and 95% humidity. Cultures were grown for 8–12 days before being used for experiments, and the media were changed every 3 days. Neurons were transfected using Lipofectamine 2000 transfection reagent following the manufacturer's instructions (Invitrogen). Forty-eight hours after transfection, experiments were performed.
Surface Biotinylation and Receptor Degradation Assay—For biotinylation experiments, the cultured neurons were plated at a density of 1 × 106 cells/ml. Cell surface receptor biotinylation and degradation assays were performed as previously described (12). In brief, serum-starved neurons were washed twice with ice-cold phosphate-buffered saline (PBS) and incubated with 300 μg/ml sulfo-N-hydroxysuccinimide-biotin (Pierce) in PBS for 45 min at 4 °C to biotinylate surface proteins. Unreacted biotin was quenched and removed with Tris-buffered saline. Biotinylated cells were then transferred to prewarmed medium containing BDNF (50 ng/ml) for 1 h, and cells were immediately chilled on ice and lysed in extraction buffer (1.0% (v/v) Triton X-100, 10 mm Tris-HCl, pH 7.5, 120 mm NaCl, 25 mm KCl, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 2 μg/ml aprotinin, and 0.5 mm phenylmethylsulfonyl fluoride). Extracts were clarified by centrifugation (12,000 × g for 15 min), and then biotinylated proteins were isolated from cell extracts by precipitation with streptavidin-conjugated Sepharose beads (Pierce). Washed beads were eluted with SDS sample buffer, and eluted proteins were resolved by SDS-PAGE. For densitometric analyses, immunoreactive bands were scanned and quantitated using NIH Image J (Scion, Frederick, MD). All experiments were carried out at least in triplicate.
Cleavable Surface Biotinylation Assay—Cell surface receptor cleavable biotinylation, internalization, and recycling assays were performed as previously described (11). Neurons were preincubated with 100 μg/ml leupeptin for 90 min before biotinylation with sulfo-NHS-S-S-biotin. Leupeptin was included in all subsequent steps to inhibit lysosomal proteolysis. For the internalization assay neurons were then incubated at 37 °C for 15 min in media alone (lanes 1 and 3) or in media containing BDNF to allow for internalization of cell-surface receptors. The remaining cell surface biotin was cleaved by glutathione (for all samples except lane 1, which represents total biotinylated TrkB receptors). Lane 2 shows a control for the efficiency of the stripping procedure, in which cells were kept at 4 °C after treatment with biotin and then subjected to biotin cleavage. Neurons were subsequently lysed, biotinylated proteins were precipitated with streptavidin beads, and complexes were immunoblotted with anti-TrkB antibodies. For the recycling assay, after biotinylation with sulfo-NHS-S-S-biotin, neurons were incubated with 50 ng/ml BDNF for 30 min to drive internalization of biotinylated receptors. Afterward, the cells were cooled on ice, and the surface-exposed biotinylated proteins were stripped of their biotin tag by treatment with the reducing agent glutathione. Internalized biotinylated receptors were protected from biotin-stripping. Subsequently, prewarmed medium was added, and the cells were incubated at 37 °C for 45 min to allow recycling of internalized receptors. This incubation was followed by glutathione treatment on ice to remove biotin from the surface-exposed biotinylated receptors that had recycled back to the cell surface during the previous re-warming period. Un-recycled receptors were detected by streptavidin pulldown followed by anti-TrkB immunoblotting. For densitometric analysis, immunoreactive bands were scanned and quantitated using NIH Image J (Scion, Frederick, MD). All experiments were carried out at least in triplicate.
Surface TrkB Internalization Measurement—Neurons transfected with FLAG-TrkB-FL or FLAG-TrkB.T1 constructs were grown on glass coverslips. After serum starvation, cells were incubated with Alexa-488-conjugated M1 anti-FLAG antibody for 30 min at 4 °C to selectively label FLAG-tagged receptors on the plasma membrane at the beginning of the experiment. Cells were then incubated (at 37 °C for 15 min) in the presence of BDNF to drive receptor internalization. At the end of this incubation, cells were quickly washed 3 times with ice-cold PBS supplemented with 3 mm EDTA to dissociate fluorescent M1 antibody from residual cell surface receptors. Cells were then fixed with 4% paraformaldehyde, with the intracellular fluorescence representing the internalized receptors. In each experiment, two parallel control coverslips were included, one in which cells were fixed after a 300-min incubation in the absence of ligand and without EDTA stripping step (100% surface receptor control), and one in which cells were stripped immediately after the feeding step (0% Internalization control). Cells were examined by fluorescence microscopy by using appropriate filter sets to selectively detect Alexa-488, and staining intensities of each fluor in individual cells were integrated using MetaMorph software (Molecular Devices, Sunnyvale, CA). Each experimental condition was repeated at least three times. In each experiment more than 30 cells were examined at random.
Analysis of TrkB Receptor Recycling by Using Fluorescence Ratio Microscopy—To quantify the extent of recycling observed after ligand removal in individual cells, a receptor recycling method was used as described previously (12). In brief, cells transiently transfected with TrkB receptors were grown on glass coverslips and incubated with Alexa-488-conjugated M1 anti-FLAG antibody after serum starvation. Cells were then incubated (at 37 °C for 30 min) in the presence of BDNF (50 ng/ml) to drive internalization. At the end of this incubation, cells were quickly washed 3 times with ice-cold PBS/EDTA to dissociate FLAG antibody bound to residual surface receptors remaining in the plasma membrane, thereby leaving fluorescent antibody bound only to the internalized pool of receptors. EDTA-stripped cells were then incubated (at 37 °C for 45 min) in the presence of Alexa-594-anti-mouse IgG to label all receptors that returned back to the cell surface, and then cells were fixed with 4% paraformaldehyde. In each experiment and for each receptor construct examined, two parallel control coverslips were included, one in which cells were fixed after a 30-min incubation in the absence of ligand and without EDTA stripping step (100% surface receptor control) and one in which cells were fixed immediately after the EDTA-mediated stripping step (0% recycled control). Cells were examined by epifluorescence microscopy by using appropriate filter sets to selectively detect Alexa-488 or Alexa-594, and staining intensities of each fluor in individual cells were integrated using MetaMorph software (Molecular Devices). This analysis indicated that the efficiency of the EDTA strip (reduction of Alexa-594 staining intensity in the 0% recycled control relative to the 100% surface receptor control) was >95%. The percentage of receptors recycled in individual cells after ligand washout was then calculated from the red/green ratios determined from the control conditions according to the following formula (E - Z)/(C - Z) × 100, where E is the mean ratio for the experimental coverslip, Z is the mean ratio for the zero surface control, and C is the mean ratio for the 100% surface control. More than 30 cells/construct/condition were analyzed at random in this manner for each experiment.
Establishment of HEK293 Stable Cell Line—HEK293 cells were transfected with FLAG-TrkB-FL construct using Lipofectamine 2000. To obtain stable transfectants, cells were treated with 800 μg/ml G418. After 15–20 days of G418 selection, resistant clones were subjected to Western blot analysis using FLAG antibodies to detect TrkB protein expression. Positive clones were maintained in 200 μg/ml G418. HEK293 cells have been widely used in studying Hrs-regulated G protein-coupled receptors (GPCRs) endocytic trafficking (37, 38). Here we chose a HEK293 TrkB-FL stable cell line to evaluate the TrkB degradation rate under different conditions due to the high transfection efficiency with HEK293 cells.
Immunocytochemical Staining and Fluorescence Microscopy—Hippocampal neurons were grown on glass coverslips coated with poly-d-lysine (Sigma) and transfected with FLAG tagged TrkB-FL or TrkB.T1 constructs. Forty-eight hours after transfection, internalized FLAG receptors were visualized by Alexa-488-M1 antibody feeding (1:1000) followed by BDNF treatment. Cells were then fixed with a 4% paraformaldehyde solution in PBS and permeabilized by brief treatment with cold methanol (5 min) followed by Hrs immunostaining with primary and secondary antibodies. Transferrin receptors were labeled with Alexa-594-transferrin (Invitrogen). Confocal fluorescence microscopy was performed on hippocampal neuron specimens using a Zeiss LSM510 microscope. Quantitative analysis was carried out with MetaMorph software. All experiments were carried out at least in triplicate.
Statistical Analysis—Statistical significance was determined using Student's t test or analysis of variance followed by post hoc tests. Differences were considered significant at p < 0.05.
RESULTS
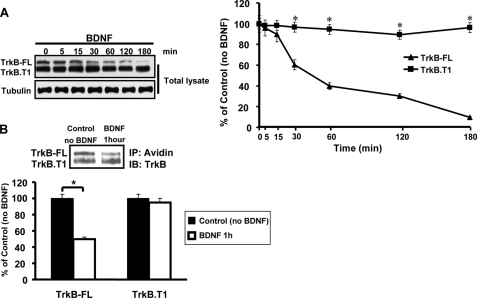
TrkB-FL and TrkB.T1 Receptors Have Different Degradation Kinetics upon BDNF Treatment—As it has been reported that both TrkB-FL and TrkB.T1 receptors could be endocytosed by BDNF (39), we sought to determine whether there were differences in the trafficking fate of the two receptors after endocytosis. To examine whether these two receptors were differentially degraded, we directly compared the rates of BDNF-dependent degradation of the TrkB-FL and TrkB.T1 receptors in primary cultured hippocampal neurons. Hippocampal neurons were serum-starved followed by cycloheximide (20 μg/ml) pretreatment to inhibit new protein synthesis, and BDNF (50 ng/ml) was added to the culture media for various times periods. Cell lysates were then collected at different time point to detect TrkB-FL and TrkB.T1 protein levels by Western blot. We found that TrkB-FL was degraded at a significantly quicker rate (<1 h for more than one-half of the receptors to be degraded) than TrkB.T1 receptors (Fig. 1A). To confirm this result, surface biotinylation assays were performed on hippocampal neurons to monitor surface receptor degradation. After 1 h of BDNF incubation, more than half of the surface TrkB-FL receptors were degraded; however, no obvious change in TrkB.T1 receptor levels was found (Fig. 1B).
FIGURE 1.
TrkB-FL and TrkB. T1 exhibit differential BDNF-induced trafficking to degradative pathway. A, cultured neurons were treated with BDNF for the indicated times in the presence of cycloheximide (20 μg/ml), and total TrkB was detected by anti-TrkB immunoblotting. The degradation was quantitated as described under “Experimental Procedures,” and the error bars represent the S.E. of three independent experiments. *, p < 0.05 by analysis of variance followed by post hoc tests. B, cultured neurons were surface-biotinylated and incubated at 37 °C for 1 h in the absence or presence of 50 ng/ml BDNF. Surface biotin-labeled receptors were detected by streptavidin pulldown followed by anti-TrkB immunoblotting (IB). IP, immunoprecipitation. The error bars represent the S.E. of three independent experiments. *, p < 0.05 by Student's t test.
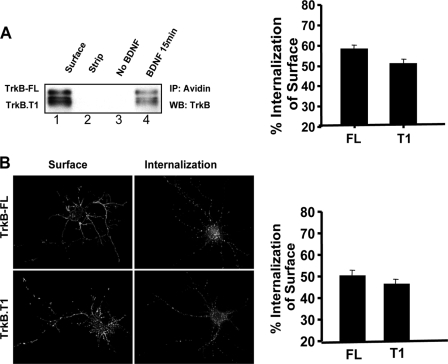
Both TrkB-FL and TrkB.T1 Receptors Are Efficiently Internalized after BDNF Treatment—We hypothesized that the differences in degradation rates may be due to alterations in TrkB-FL and TrkB.T1 receptor trafficking at certain steps after ligand addition, namely 1) at the initial internalization step and/or 2) at a postendocytic step in which Trk receptors can be sorted to either degradative or recycling pathways. To address these questions related to the TrkB endocytic pathways, we measured TrkB-FL and TrkB.T1 receptor internalization by a cleavable surface biotinylation assay. After 15 min of BDNF treatment, about 56 ± 2% TrkB-FL and 50 ± 2% TrkB.T1 receptor were internalized, respectively, in hippocampal neurons, with no significant difference between each other (Fig. 2A). To confirm the cleavable biotinylation result, quantitative fluorescence assay was employed to measure TrkB-FL and TrkB.T1 receptor internalization. Hippocampal neurons were transfected with FLAG epitope-tagged TrkB-FL and TrkB.T1 receptors. Respectively, and fed with Alexa488-conjugated FLAG antibodies. We took advantage of a novel feature of the amino-terminal FLAG epitope-tagged versions of Trk receptors, in which calcium-sensitive fluorescent anti-FLAG antibodies can be rapidly dissociated with PBS/EDTA (12). After ligand-induced surface receptor internalization, PBS/EDTA was used to strip the cell surface fluorescent antibodies. Any remaining intracellular fluorescence, therefore, represented the internalized receptors. We found both TrkB-FL and TrkB.T1 receptors were rapidly internalized in the presence of BDNF, and no significant differences were observed in their levels of internalization (Fig. 2B).
FIGURE 2.
TrkB-FL and TrkB. T1 exhibit similar ligand-induced internalization. A, internalization of cleavable biotinylated TrkB receptors in cultured neurons. As described under “Experimental Procedures,” lane 1 shows the total TrkB on cell surface, and lane 2 shows a control for the efficiency of the stripping procedure. Neurons were subsequently lysed, biotinylated proteins were precipitated (IP) with streptavidin beads, and complexes were immunoblotted (WB) with anti-TrkB antibodies. Data represent the mean ± S.E. from three independent experiments. B, internalization of receptors was measured by the fluorescence ratio of internalization/surface receptors. All of the data are presented as the mean ± S.E. determined from analysis of more than 3 independent experiments (n ≥ 30 cells for each condition per experiment).
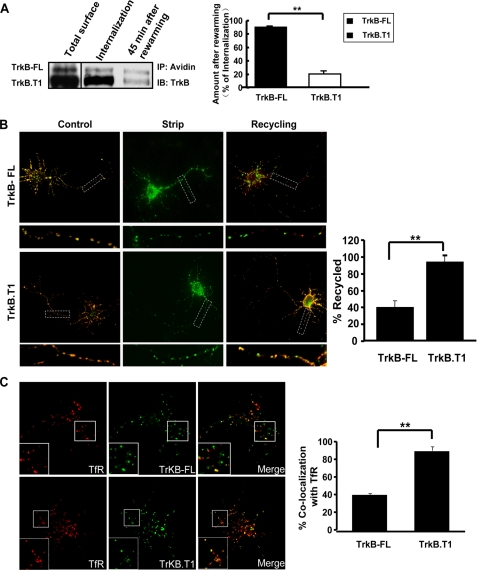
TrkB-FL and TrkB.T1 Receptors Behave Differently in Their Postendocytic Recycling—Because TrkB-FL and TrkB.T1 receptors have similar internalization levels but significantly different degradation kinetics, we sought to measure the degree of their postendocytic recycling. First, a cleavable biotinylation assay was used to detect receptor recycling after internalization in hippocampal neurons. We found a higher percentage of the remaining internalized TrkB-FL receptor after the re-warming process as compared with TrkB.T1 receptor (Fig. 3A), which suggested that more internalized TrkB.T1 receptor has been recycled back to the cell surface to be cleaved. A live cell ratiometric fluorescence-based recycling assay, which has been used in our previous study (12), was also employed to measure the TrkB-FL and TrkB.T1 receptor recycling (Fig. 3B). Quantification of these results by ratiometric analyses (see “Experimental Procedures”) confirmed that TrkB.T1 recycled in a BDNF-dependent manner significantly (90 ± 1%), whereas TrkB-FL recycled to a lesser degree (39 ± 1%) (Fig. 3B). Consistent with this finding is that compared with TrkB-FL, internalized TrkB.T1 receptor was colocalized significantly more with the transferrin receptor, a default recycling receptor, as determined by confocal microscopy (Fig. 3C). Together, these results suggest that TrkB.T1 receptor is predominantly sorted to the recycling pathway, which is consistent with its minimal BDNF-dependent degradation (Fig. 1A), whereas TrkB-FL receptor recycles to a lesser extent after BDNF treatment.
FIGURE 3.
TrkB-FL and TrkB.T1 recycles differently after BDNF-induced endocytosis. A, differential recycling of internalized TrkB-FL and TrkB.T1 measured by cleavable surface biotinylation. Total surface refers to the total biotinylated TrkB receptors on the cell surface; Internalization refers to the internalized biotinylated receptors before rewarming. Data represent the mean ± S.E. from three independent experiments (The double asterisk represents p < 0.01, Student's t test). B, TrkB recycling behavior was quantitated as described under “Experimental Procedures.” Control refers to the 100% surface TrkB receptor control, Strip refers to the 0% recycled control, and the error bars represent the S.E. of five independent experiments (n ≥ 30 cells for each condition per experiment; double asterisk represents p < 0.01, Student's t test). C, colocalization of internalized TrkB (green) with transferrin receptor (TfR, red) in hippocampal neurons visualized by confocal microscopy. The proportion of colocalization between internalized TrkB and transferrin receptors is presented as the mean ± S.E. determined from analysis of four independent experiments (n ≥ 30 cells for each condition per experiment; double asterisk represents p < 0.01, Student's t test).
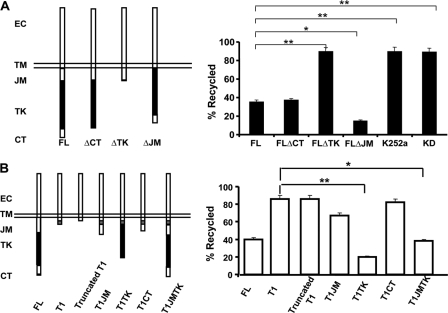
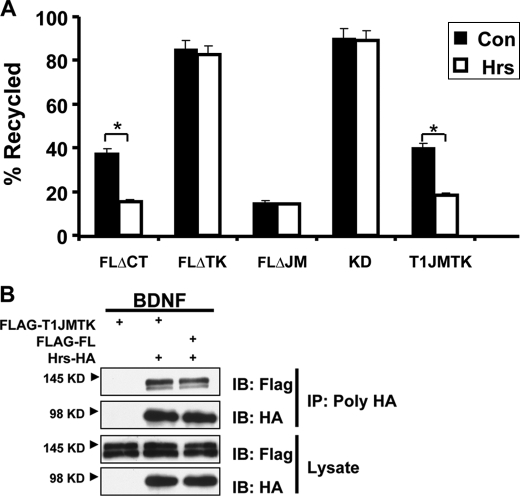
Kinase Domain Is the Key Motif Responsible for the Differential TrkB-FL and TrkB.T1 Endocytic Recycling—To define the potential region in TrkB-FL receptor that is responsible for the differences in TrkB-FL and TrkB.T1 recycling levels, various domains (juxtamembrane, kinase, and carboxyl terminus) were deleted from TrkB-FL, and ligand-dependent recycling in PC12 cells was assessed by the ratiometric recycling assay (Fig. 4A). In the ratiometric recycling assay, PC12 cells were used, as that is a well established neuron-like cell line.
FIGURE 4.
Identification of the domain in TrkB responsible for differential TrkB-FL and TrkB. T1 endocytic recycling. PC12 cells transiently transfected with the indicated FLAG-tagged mutants were subjected to the ratiometric recycling assay, which was described under “Experimental Procedures.” A, PC12 cells were transfected with TrkB-FL constructs lacking the following intracellular regions: carboxyl terminus, FLΔCT; tyrosine kinase, FLΔTK; juxtamembrane, FLΔJM and the TrkB kinase dead construct (TrkBKD). K252a was applied at 100 nm, a concentration that could inhibit Trk kinase activity. EC means extracellular. B, PC12 cells were transfected with truncated TrkB.T1 lack TrkB.T1 specific cytoplasmic tail or TrkB.T1 chimeras containing different regions of TrkB-FL (T1JM, T1TK, T1CT, T1JMTK). TrkB-FL and TrkB.T1 were also transfected as controls. In all experiments, error bars represent the S.E. of five independent experiments (n ≥ 30 cells for each condition per experiment; the asterisk represents p < 0.05; the double asterisk represents p < 0.01, Student's t test).
For all these TrkB mutants, there were no deficits in their plasma surface targeting or alterations in initial endocytosis rates.4 Only the TrkB-FL mutant lacking the kinase domain (TrkBΔTK) showed a significant increase in recycling as compared with wild type TrkB-FL receptor (Fig. 4A). This result is consistent with the subsequent finding that kinase activity, assessed by a point mutation in the kinase domain established to abolish kinase activity (K571A) or in the presence of specific Trk kinase inhibitor K252a, significantly reduced TrkB recycling levels (Fig. 4A). Thus, TrkB-FL kinase activity is necessary to maintain the TrkB-FL relatively low level of recycling. To be noted, the TrkB-FL mutant lacking juxtamembrane domain showed a significant reduction in recycling as compared with wild type receptor, suggesting this region serves as a “recycling signal,” which is consistent with our previous report that the juxtamembrane domain in TrkA receptor contains a postendocytic recycling signal (12). In contrast, the TrkB.T1 mutant lacking its specific cytoplasmic tail (truncated TrkB.T1) still efficiently recycled, suggesting that TrkB.T1 receptors recycle via the default pathway (Fig. 4B).
To address whether the kinase domain is sufficient to decrease TrkB.T1 recycling, we generated chimeras in which different domains from TrkB-FL were transplanted to the carboxyl terminus of TrkB.T1 receptor. From these chimeric receptors, we hoped to identify a domain that is sufficient to function in regulating differential TrkB-FL and TrkB.T1 recycling. We observed that chimeric receptors with TrkB-FL kinase domain transplanted into TrkB.T1 receptor led to significantly decreased recycling in PC12 cells (Fig. 4B). Immunoprecipitation experiments followed by p-Tyr (anti-phospho-Tyr) Western blot analyses confirmed that TrkB.T1TK receptor was phosphorylated upon BDNF treatment (data not shown). These observations, showing that the transplanted TrkB kinase domain can decrease TrkB.T1 receptor recycling, indicate that the kinase domain is sufficient to maintain ligand-dependent TrkB-FL recycling at a relatively low level.
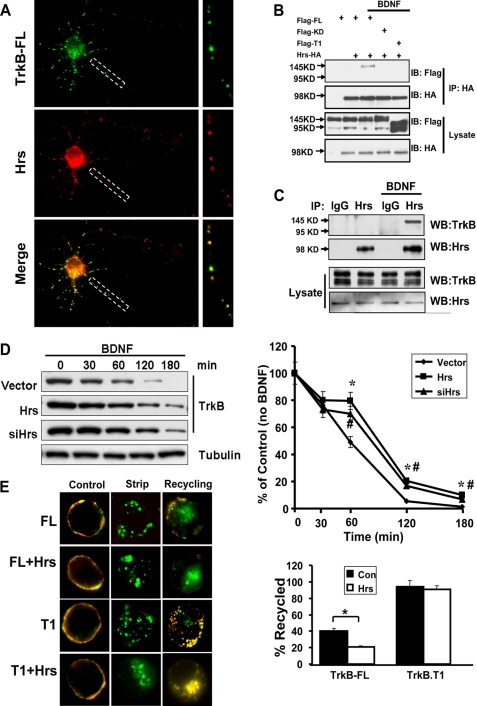
Hrs Regulates TrkB-FL but Not TrkB.T1 Endocytic Recycling—A recent study demonstrated that Hrs could be phosphorylated upon BDNF stimulation in primary cultured neurons (40). It has been reported that Hrs plays essential roles in both sorting activated receptors to the multivesicular body vesicles/lysosomal pathway and regulating some GPCR receptors recycling (37, 38, 41–43). We wanted to know whether Hrs was also involved in regulating TrkB degradation and recycling. As a first step toward examining the role of Hrs in post-endocytic trafficking of TrkB, we localized endogenous Hrs in neurons with internalized FLAG-TrkB-FL by immunofluorescence. We found that most endocytosed TrkB-FL was co-localized with Hrs in hippocampal neurons (Fig. 5A). We also determined whether these proteins had physiological interaction by co-immunoprecipitation studies. In HEK293 cells with Hrs and TrkB-FL, TrkB.T1, or TrkBKD overexpression, only TrkB-FL can be pulled down by Hrs in a BDNF-dependent manner, with no interaction between TrkB.T1/TrkBKD with Hrs (Fig. 5B). To exclude the possibility of ectopic overexpression artifact, the endogenous TrkB and Hrs interaction was examined in primary cultured hippocampal neurons. As shown in Fig. 5C, only the 145-kDa TrkB-FL proteins could be pulled down by Hrs, and this interaction was dependent on BDNF.
FIGURE 5.
Hrs regulates TrkB-FL but not TrkB. T1 endocytic recycling. A, subcellular colocalization of internalized TrkB (green) with Hrs (red) in hippocampal neurons visualized by confocal microscopy. B, coimmunoprecipitation (IP) of Hrs with TrkB-FL, TrkB-KD, and TrkB.T1. For TrkB-FL, the coimmunoprecipitation was performed in HEK293 cell stably expressing TrkB-FL; others were by transient transfection. All the cells were transfected with HA-tagged Hrs, and 20 min of BDNF treatment was performed. Lysates were immunoprecipitated with HA antibodies and immunoblotted with FLAG antibodies (top). Immunoprecipitation of HA-Hrs was confirmed by immunoblotting with HA antibodies (middle). C, endogenous association of Hrs with TrkB was performed in cultured hippocampal neurons. Lysates were subjected to immunoprecipitation with anti-Hrs antibodies or normal goat IgG and immunoblotted (WB) with TrkB and Hrs antibodies. D, HEK293 cells stably expressing FLAG-tagged TrkB-FL were transfected with Hrs or Hrs siRNA and then treated with 50 ng/ml BDNF for the indicated times at 37 °C in the presence of cycloheximide (20 μg/ml). Total TrkB receptors were detected by anti-TrkB immunoblotting. Data shown are the mean ± S.E. of three separate experiments (the asterisk represents p < 0.05 between control and Hrs transfection group; the number symbol (#) represents p < 0.05 between control and siHrs transfection group by analysis of variance followed by post hoc tests). E, PC12 cells were transfected with TrkB-FL/T1 and Hrs, and BDNF-dependent recycling was measured in these cells as described in Fig. 3. Data shown are the mean ± S.E. of five separate experiments (n ≥ 30 cells for each condition per experiment; the asterisk represents p < 0.05, Student's t test).
Next we found Hrs overexpression or knockdown of Hrs by siRNA could significantly decrease TrkB-FL degradation in HEK293 TrkB-FL stable cell lines (supplemental Figs. 1 and 5D), which suggests that Hrs regulates TrkB-FL degradation. To our surprise, Hrs also plays a significant role in TrkB-FL recycling. Overexpression of Hrs did not detectably perturb BDNF-induced endocytosis of TrkB-FL, but this manipulation significantly inhibited recycling of TrkB-FL by “trapping” internalized receptors in enlarged early endosomes (Fig. 5E). In contrast, recycling of TrkB.T1 receptor was not detectably inhibited in cells overexpressing Hrs. This result is similar to previous studies indicating that Hrs overexpression produced no detectable effect on either endocytosis or recycling of transferrin receptors (44). The discrepancy in the dependence on Hrs of TrkB-FL and TrkB.T1 receptors recycling is consistent with our previous result that only TrkB-FL can bind with Hrs in a BDNF-dependent manner. We hypothesized that endocytosed TrkB-FL receptor is sorted into a “regulated” recycling pathway, which undergoes sequence-directed recycling and depends on Hrs. TrkB.T1, however, recycles in a default pathway that is independent of Hrs. To address this hypothesis, we investigated the effect of Hrs overexpression on recycling of different TrkB mutants after BDNF-induced endocytosis (Fig. 6A). The TrkB-FL mutants, which have a kinase domain deletion or are kinase dead, could recycle more efficiently in a default pathway, which is independent of Hrs. On the contrary, transplanting the juxtamembrane and kinase domain of TrkB-FL receptor to TrkB.T1 receptor (T1JMTK) not only gained the capacity to bind to Hrs (Fig. 6B) but also became sensitive to Hrs effects on recycling (Fig. 6A). This result suggests the TrkB-FL kinase domain could switch TrkB.T1 receptor recycling from default to regulated pathway.
FIGURE 6.
Identification of a sequence in TrkB-FL responsible for Hrs controlled recycling. A, PC12 cells were co-transfected with TrkB-FL mutants and T1-TrkB chimeras containing the juxtamembrane and kinase region of TrkB-FL (T1JMTK) and HA-Hrs, and BDNF-dependent recycling was measured in these cells as described in Fig. 3. Data shown are the mean ± S.E. of five separate experiments (n ≥ 30 cells for each condition per experiment; the asterisk represents p < 0.05, Student's t test). Con, control. B, coimmunoprecipitation (IP) of Hrs with TrkB-FL and T1JMTK. Cells were co-transfected with HA-tagged Hrs with TrkB-FL and T1JMTK, and a 20-min BDNF treatment was performed. Lysates were immunoprecipitated with HA antibodies and immunoblotted (IB) with FLAG antibodies (top).
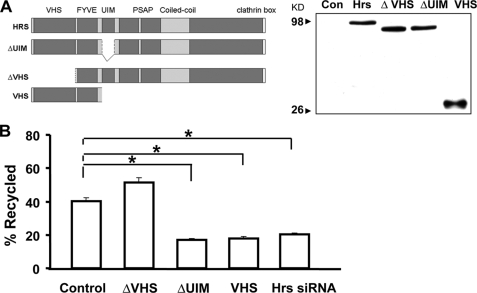
The VHS Domain of Hrs Is Required for TrkB-FL Recycling—Hrs contains several conserved domains that are present in proteins implicated in signal transduction and/or membrane trafficking. The VHS (Vps27p, Hrs, STAM) domain is present at the amino terminus of Hrs, which has been reported to be required for Hrs-dependent recycling of the β2-adrenergic receptor (37). The FYVE (Fab1p, YOYB, Vac1p, and EEA1) is a zinc finger domain present in more than 40 proteins and is required for membrane localization (45, 46). Ubiquitin-interacting motif (UIM) serves as the major ubiquitin-binding site in Hrs (47). The PSAP domain contains a tetrapeptide P(S/T)AP motif and has been known to be essential for the budding of viruses from host cells (48).
To determine which domain in Hrs is required for regulating TrkB-FL recycling, we constructed a series of deletion mutants of HA-tagged Hrs and tested their ability to inhibit recycling of the TrkB-FL when overexpressed. All of the mutant Hrs constructs were expressed at similarly high levels as Hrs wild type (Fig. 7A). Truncation of the amino-terminal VHS domain produced a mutant Hrs protein (ΔVHS) that was defective in its ability to inhibit TrkB-FL recycling when overexpressed. In contrast, deletion of the ubiquitin-interacting motif (ΔUIM) which is essential in Hrs-dependent ubiquitin-directed lysosomal sorting function, was still capable of strongly inhibiting TrkB-FL recycling (Fig. 7B). Moreover, overexpression of VHS and FYVE domain (named VHS for short) was sufficient to inhibit TrkB-FL recycling. These results suggest that the VHS domain of Hrs may be specifically required for the function of Hrs in regulating TrkB-FL recycling.
FIGURE 7.
The VHS domain of Hrs is required for its effect on TrkB recycling. A, schematic diagram of the domain organization of full-length Hrs and the truncated mutants used as indicated. HEK293 cells were transfected with HA-Hrs truncated mutants as indicated, and a Western blot was performed to verify similar expression levels of all proteins in cell lysates. Con, control. B, PC12 cells were co-transfected with Hrs constructs lacking different regions and TrkB-FL, and BDNF-dependent recycling was measured in these cells as described in Fig. 3. Data shown are the mean ± S.E. of five separate experiments (n ≥ 30 cells for each condition per experiment; the asterisk represents p < 0.05, Student's t test).
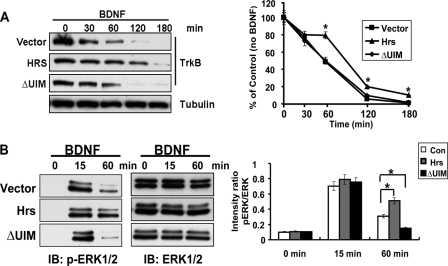
Functional Requirement of the Hrs-sensitive Recycling for BDNF-induced Signaling—To assess the functional consequences of Hrs-sensitive TrkB-FL recycling, we investigated the effect of disrupting Hrs function on BDNF-induced MAP kinase activation in a HEK293 TrkB-FL stable cell line because of its high transfection efficiency. First, by total TrkB degradation assay, we found, in contrast to Hrs full-length overexpression, which blocks TrkB-FL degradation, overexpression of Hrs lacking the UIM motif has no effect on TrkB-FL degradation (Fig. 8A). Because overexpression of HrsΔUIM could inhibit TrkB-FL recycling while having no effect on TrkB-FL degradation, it could be used to study the role of TrkB-FL recycling in BDNF-induced signaling. In HrsΔUIM-transfected 293-TrkB cells, MAP kinase activation was monitored by immunoblotting with phospho-MAP kinase (p-MAPK)-specific antibodies. As shown in Fig. 8B, 293-TrkB cells expressing HrsΔUIM were stimulated with BDNF for 15 or 60 min, two time points that can discriminate between transient and sustained MAP kinase activation (49, 50). HrsΔUIM overexpression did not perturb the transient MAPK activation by BDNF, but prolonged MAPK activation was significantly reduced (Fig. 8B). In contrast, overexpression of full-length Hrs, which both inhibited TrkB-FL degradation and recycling, enhanced BDNF-induced sustained MAPK activation. These results suggest that the Hrs-sensitive TrkB-FL recycling may play a role in BDNF-induced prolonged MAPK activation. However, in addition to inhibiting TrkB-FL recycling, overexpression of HrsΔUIM may also interfere with other intracellular signaling pathways, which contribute to the altered MAPK activation triggered by BDNF.
FIGURE 8.
The role of Hrs-sensitive TrkB-FL recycling in BDNF-induced MAP kinase activation. A, total TrkB degradation assay was performed as described in Fig. 5D to detect the role of HrsΔUIM on TrkB degradation. The data are presented as the means ± S.E., determined from analysis of more than three independent experiments. The asterisk represents p < 0.05, Student's t test. B, lysates from HEK293 TrkB-FL stable cell line with Hrs or HrsΔUIM overexpression were detected by p-Erk/Erk antibodies upon BDNF treatment. p-Erk/Erk immunoreactivity intensity is quantitated by densitometric analysis, and optical density values are expressed as a ratio between the p-Erk and Erk. Data shown are the mean ± S.E. of five separate experiments. IB, immunoblot; Con, control. The asterisk represents p < 0.05, Student's t test.
DISCUSSION
Endocytic trafficking of receptors to alternate intracellular pathways has been shown to lead to diverse biological consequences. In this study we have demonstrated that TrkB-FL receptor and its isoform, TrkB.T1, traverse divergent endocytic pathways after binding to BDNF. We provide direct evidence that TrkB.T1 predominantly recycles back to the cell surface after ligand treatment, whereas TrkB-FL predominantly is sorted to the degradative pathway. Moreover, Hrs differentially regulates TrkB-FL and TrkB.T1 postendocytic trafficking fates.
Our results provide several new insights into the mechanism of endocytic trafficking of TrkB-FL and TrkB.T1 receptors. In astrocytes or Schwann cells, which only express TrkB.T1 but not TrkB-FL receptor (51–53), iodinated BDNF could be internalized and released into culture medium, suggesting that TrkB.T1 receptor could be endocytosed by BDNF treatment (39). A recent study in hippocampal neuronal cultures using recombinant BDNF and yellow fluorescent protein (BDNF-YFP) noted that it could be internalized and re-secreted, which suggested that BDNF might be trafficked to the recycling pathway (14). However, it is not clear whether the recycling of BDNF is mediated by TrkB-FL or TrkB.T1 receptor. In our study, we found that BDNF could efficiently drive TrkB-FL or TrkB.T1 receptor internalization. Interestingly, the degree of BDNF-dependent recycling of TrkB.T1 receptor is much higher than TrkB-FL receptor by the use of a live cell fluorescence-based recycling assay. The level of ligand-dependent recycling of TrkB.T1 receptor (∼90%) is equivalent to the previously established recycling receptors such as the transferrin receptor. Accordingly, TrkB-FL was degraded more quickly than TrkB.T1 receptor upon BDNF stimulation. Thus, TrkB-FL and TrkB.T1 receptors are sorted into different trafficking pathway after BDNF-triggered endocytosis.
Second, we show that Hrs plays an essential function in the recycling of TrkB-FL but not TrkB.T1 receptors. Hrs is well known to terminate cell signaling by sorting activated ubiquitinated receptors to the multivesicular body vesicles/lysosomal pathway (41, 42, 54). TrkA and TrkB receptors are ubiquitinated in response to neurotrophins and subsequently degraded by the lysosomal pathway (9, 55–57). In our study, we found overexpression or siRNA knock down of Hrs could attenuate TrkB degradation upon BDNF treatment, suggesting that Hrs plays a role in TrkB lysosomal sorting and down-regulation of signaling receptors via VPS/ESCRT pathway. Our results are consistent with a recent report showing that Hrs overexpression disrupts TrkB degradation in hippocampal neurons (40). Furthermore, we found Hrs could be associated with TrkB upon BDNF treatment, which is dependent on the TrkB tyrosine kinase activity.
To our surprise, we found Hrs could also mediate TrkB-FL recycling, which depends on its VHS domain. Recently a distinct role of Hrs in promoting rapid recycling of endocytosed signaling receptors to the plasma membrane has been found (37, 38, 43). There exist two pathways for membrane proteins recycling back to the cell surface; one is the default pathway, which occurs via a process of bulk membrane flow that requires no specific targeting signals (58, 59), the other is the sequence-directed recycling, which depends on a diverse set of cytoplasmic targeting sequences (60–64) that appear to bind to distinct cytoplasmic proteins (64, 65). An increasing number of signaling receptors is being found to undergo this “active” sorting to the recycling pathway. Our previous study showed that endocytosed TrkA receptor recycles to the plasma membrane by its juxtamembrane sequence (12). In the present study we found the juxtamembrane domain is also essential in mediating internalized TrkB-FL receptor recycling, which suggests that TrkB-FL is sorted into sequence-directed regulated recycling after endocytosis. In contrast, TrkB.T1 receptors recycle more efficiently after internalization via the default pathway compared with TrkB-FL. In particular, TrkB.T1 mutants, which lack its specific cytoplasmic tail, could still efficiently recycle. The requirement of Hrs in TrkB-FL recycling was first indicated by the ability of Hrs overexpression to specifically inhibit recycling of TrkB-FL but not TrkB.T1 receptors. Depletion of endogenous Hrs using siRNA produced a phenotype identical to overexpression, establishing an essential role of Hrs in sequence-directed recycling. The observation that Hrs overexpression and knockdown both have inhibitory effects on trafficking has been previously reported (35, 44, 66). As Hrs has multiple interaction domains and is thought to exist in a large protein complex (67), overexpression could titrate out the associated proteins present in limiting amounts and inhibit their function (68), whereas knockdown could prevent the formation of such protein complexes that may be required for cargo to be transported. Thus, not only for GPCRs (37), but also for TrkB receptor, Hrs regulates sequence-directed, but not default recycling. The present results implicate the amino-terminal VHS domain of Hrs in the sequence-directed recycling mechanism. Overexpression of an Hrs mutant lacking the VHS domain could not inhibit TrkB-FL recycling, whereas overexpression of only the VHS and FYVE domain was sufficient to inhibit TrkB-FL recycling. Moreover, the UIM domain of Hrs is not required for its dominant-negative effect on TrkB-FL recycling. Our data are consistent with previous studies showing that VHS domain of Hrs plays essential role in sequence directed GPCR recycling (37).
Third, we found the tyrosine kinase domain in the TrkB-FL was sufficient to confer Hrs-dependent recycling when fused to a TrkB.T1 receptor that recycles efficiently by default and independently of Hrs. We proposed that the recycling of internalized TrkB-FL receptor needs two steps which require two different sorting sequences; first, it is sorted to the sequence-directed (Hrs-dependent) regulated recycling pathway, which depends on the kinase domain, and then is transported back to the cell surface, which requires its juxtamembrane domain. Deletion of the TrkB-FL kinase domain produced a dramatically different phenotype (default recycling) than mutation of the juxtamembrane domain, which greatly inhibits recycling when deleted. This suggests that the kinase domain functions upstream of the recycling sequence, mediating the initial sorting of endocytosed TrkB-FL receptors to the specialized recycling pathway (Fig. 9). This hypothesis accounts for the ability of receptors with a deleted kinase domain to effectively recycle, bypassing the Hrs requirements and, in turn, also explaining why the TrkB-FL recycles to a lesser extent.
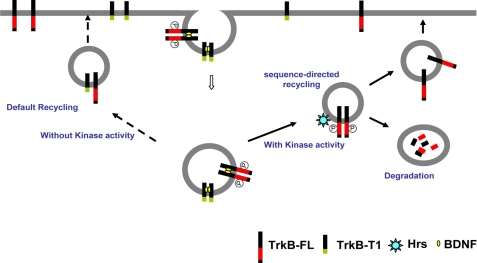
FIGURE 9.
Simplified model depicting differential TrkB-FL and TrkB.T1 postendocytic recycling pathway. Both TrkB-FL and TrkB.T1 receptors are efficiently internalized after binding to BDNF (yellow dots). Endocytic TrkB-FL receptors traverse sequence-directed recycling pathway, in which process the TrkB kinase activity plays an essential role (heavy lines). This sequence-directed recycling pathway requires both Hrs and the juxtamembrane domain of TrkB-FL. Therefore, depletion of cellular Hrs or deletion of the juxtamembrane domain prevents TrkB-FL recycling. Conversely, internalized TrkB.T1 receptors, which lack kinase activity, are not sorted to the Hrs-dependent pathway and, thus, recycle by the default pathway (dashed lines).
Finally, we also demonstrated that Hrs-dependent recycling may be functionally important for producing prolonged MAPK signaling after activation of the TrkB-FL, a typical signaling characteristic for Trk receptor activation. Overexpression of Hrs mutant lacking UIM domain, which inhibits TrkB-FL recycling without affecting its degradation, attenuates BDNF-induced prolonged MAPK signaling. In contrast, overexpression of full-length Hrs could prolong MAPK signaling triggered by BDNF. We inferred that the dominant effect of overexpression of Hrs full-length was to block TrkB-FL degradation because TrkB-FL is predominantly degraded and less efficiently recycled, which as a result enhances the prolonged MAPK signaling stimulated by BDNF. Thus, different domains in Hrs may differentially regulate receptor endocytic trafficking and, as a result, alter receptor-mediated signaling. A previous study showed for the efficiently regulated recycled β2-adrenergic receptor, knockdown of Hrs expression strongly inhibited β2-adrenergic receptor resensitization (37). Therefore, the physiological function of Hrs may depend on which endocytic trafficking pathway (degradation or recycling) represents a major regulatory pathway. Considering that efficient recycling can occur without a requirement for any known sorting signals (as for transferrin receptor and TrkB.T1 receptor), why does such complexity exist in the recycling of the TrkB-FL? One possibility is to provide flexibility of receptor regulation. Relatively low levels of TrkB-FL recycling, although disadvantageous for functional recovery of signaling upon ligand treatment, could be flexibly reprogrammed under different physiological conditions.
In conclusion, the present results demonstrate that TrkB-FL and its isoform TrkB.T1 receptor traverse divergent endocytic pathways after binding with BDNF. Recycling of endocytic TrkB-FL receptors occurs by a specifically sorted mechanism that requires the endosome-associated protein Hrs. In contrast, TrkB.T1 receptors recycle by a default mechanism that does not require Hrs. Our study provides novel insights into the diverse cellular functions of Hrs, and suggests a new mechanistic link between processes of receptor membrane trafficking and intracellular signaling.
Supplementary Material
Acknowledgments
We thank Francis S. Lee and Siobhan Pattwell for helpful discussions.
This work was supported by Natural Science Foundation of China Grants 30671050, 30725020, 30700258, and 90713016, National 973 Basic Research Program of China Grants 2009CB941403 and 2009CB526507, National High-Tech Research and Development Program of China Grant 2006AA02A406, Cultivation Fund of the Key Science and Technology Innovation Project from Chinese Ministry of Education Grant 707040, Fok Ying Tong Education Foundation Grant 111044, China Postdoctoral Science Foundation Grants 20070421083, 200702028, and 200801406, Research Fund for the Doctoral Program of Higher Education of China Grant 200804221070, and the National Alliance for Research on Schizophrenia and Depression.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: BDNF, brain-derived neurotrophic factor; MAPK, mitogen-activated protein (MAP) kinase; Hrs, hepatocyte growth factor-regulated tyrosine kinase substrate; UIM, ubiquitin-interacting motif; siRNA, small interfering RNA; HA, hemagglutinin; PBS, phosphate-buffered saline; FL, full-length; GPCR, G protein-coupled receptor.
S.-H. Huang, L. Zhao, Z.-P. Sun, X.-Z. Li, Z. Geng, K.-D. Zhang, M. V. Chao, and Z.-Y. Chen, unpublished data.
References
- 1.Chao, M. V. (2003) Nat. Rev. Neurosci. 4 299-309 [DOI] [PubMed] [Google Scholar]
- 2.Huang, E. J., and Reichardt, L. F. (2001) Annu. Rev. Neurosci. 24 677-736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tessarollo, L. (1998) Cytokine Growth Factor Rev. 9 125-137 [DOI] [PubMed] [Google Scholar]
- 4.Kaplan, D. R., and Miller, F. D. (2000) Curr. Opin. Neurobiol. 10 381-391 [DOI] [PubMed] [Google Scholar]
- 5.Huang, E. J., and Reichardt, L. F. (2003) Annu. Rev. Biochem. 72 609-642 [DOI] [PubMed] [Google Scholar]
- 6.Grimes, M. L., Beattie, E., and Mobley, W. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 9909-9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng, J., Shen, W. H., Lu, T. J., Zhou, Y., Chen, Q., Wang, Z., Xiang, T., Zhu, Y. C., Zhang, C., Duan, S., and Xiong, Z. Q. (2008) J. Biol. Chem. 283 13280-13288 [DOI] [PubMed] [Google Scholar]
- 8.Heerssen, H. M., and Segal, R. A. (2002) Trends Neurosci. 25 160-165 [DOI] [PubMed] [Google Scholar]
- 9.Sommerfeld, M. T., Schweigreiter, R., Barde, Y. A., and Hoppe, E. (2000) J. Biol. Chem. 275 8982-8990 [DOI] [PubMed] [Google Scholar]
- 10.Jullien, J., Guili, V., Reichardt, L. F., and Rudkin, B. B. (2002) J. Biol. Chem. 277 38700-38708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena, S., Howe, C. L., Cosgaya, J. M., Steiner, P., Hirling, H., Chan, J. R., Weis, J., and Krüttgen, A. (2005) Mol. Cell. Neurosci. 28 571-587 [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z. Y., Ieraci, A., Tanowitz, M., and Lee, F. S. (2005) Mol. Biol. Cell 16 5761-5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginty, D. D., and Segal, R. A. (2002) Curr. Opin. Neurobiol. 12 268-274 [DOI] [PubMed] [Google Scholar]
- 14.Santi, S., Cappello, S., Riccio, M., Bergami, M., Aicardi, G., Schenk, U., Matteoli, M., and Canossa, M. (2006) EMBO J. 25 4372-4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middlemas, D. S., Lindberg, R. A., and Hunter, T. (1991) Mol. Cell. Biol. 11 143-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, R., Conway, D., Parada, L. F., and Barbacid, M. (1990) Cell 61 647-656 [DOI] [PubMed] [Google Scholar]
- 17.Stoilov, P., Castren, E., and Stamm, S. (2002) Biochem. Biophys. Res. Commun. 290 1054-1065 [DOI] [PubMed] [Google Scholar]
- 18.Armanini, M. P., McMahon, S. B., Sutherland, J., Shelton, D. L., and Phillips, H. S. (1995) Eur. J. Neurosci. 7 1403-1409 [DOI] [PubMed] [Google Scholar]
- 19.Escandón, E., Soppet, D., Rosenthal, A., Mendoza-Ramírez, J. L., Szönyi, E., Burton, L. E., Henderson, C. E., Parada, L. F., and Nikolics, K. (1994) J. Neurosci. 14 2054-2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allendoerfer, K. L., Cabelli, R. J., Escandón, E., Kaplan, D. R., Nikolics, K., and Shatz, C. J. (1994) J. Neurosci. 14 1795-1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fryer, R. H., Kaplan, D. R., Feinstein, S. C., Radeke, M. J., Grayson, D. R., and Kromer, L. F. (1996) J. Comp. Neurol. 374 21-40 [DOI] [PubMed] [Google Scholar]
- 22.Knüsel, B., Rabin, S. J., Hefti, F., and Kaplan, D. R. (1994) J. Neurosci. 14 1542-1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eide, F. F., Vining, E. R., Eide, B. L., Zang, K., Wang, X. Y., and Reichardt, L. F. (1996) J. Neurosci. 16 3123-3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ninkina, N., Adu, J., Fischer, A., Piñón, L. G., Buchman, V. L., and Davies, A. M. (1996) EMBO J. 15 6385-6393 [PMC free article] [PubMed] [Google Scholar]
- 25.Dorsey, S. G., Renn, C. L., Carim-Todd, L., Barrick, C. A., Bambrick, L., Krueger, B. K., Ward, C. W., and Tessarollo, L. (2006) Neuron 51 21-28 [DOI] [PubMed] [Google Scholar]
- 26.Biffo, S., Offenhäuser, N., Carter, B. D., and Barde, Y. A. (1995) Development 121 2461-2470 [DOI] [PubMed] [Google Scholar]
- 27.Fryer, R. H., Kaplan, D. R., and Kromer, L. F. (1997) Exp. Neurol. 148 616-627 [DOI] [PubMed] [Google Scholar]
- 28.Haapasalo, A., Saarelainen, T., Moshnyakov, M., Arumäe, U., Kiema, T. R., Saarma, M., Wong, G., and Castrén, E. (1999) Oncogene 18 1285-1296 [DOI] [PubMed] [Google Scholar]
- 29.Yacoubian, T. A., and Lo, D. C. (2000) Nat. Neurosci. 3 342-349 [DOI] [PubMed] [Google Scholar]
- 30.Baxter, G. T., Radeke, M. J., Kuo, R. C., Makrides, V., Hinkle, B., Hoang, R., Medina-Selby, A., Coit, D., Valenzuela, P., and Feinstein, S. C. (1997) J. Neurosci. 17 2683-2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kryl, D., Yacoubian, T., Haapasalo, A., Castren, E., Lo, D., and Barker, P. A. (1999) J. Neurosci. 19 5823-5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komada, M., and Kitamura, N. (1995) Mol. Cell. Biol. 15 6213-6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komada, M., Masaki, R., Yamamoto, A., and Kitamura, N. (1997) J. Biol. Chem. 272 20538-20544 [DOI] [PubMed] [Google Scholar]
- 34.Raiborg, C., Rusten, T. E., and Stenmark, H. (2003) Curr. Opin. Cell Biol. 15 446-455 [DOI] [PubMed] [Google Scholar]
- 35.Bache, K. G., Raiborg, C., Mehlum, A., and Stenmark, H. (2003) J. Biol. Chem. 278 12513-12521 [DOI] [PubMed] [Google Scholar]
- 36.Chen, Z. Y., Patel, P. D., Sant, G., Meng, C. X., Teng, K. K., Hempstead, B. L., and Lee, F. S. (2004) J. Neurosci. 24 4401-4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanyaloglu, A. C., McCullagh, E., and von Zastrow, M. (2005) EMBO J. 24 2265-2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanyaloglu, A. C., and von Zastrow, M. (2007) J. Biol. Chem. 282 3095-3104 [DOI] [PubMed] [Google Scholar]
- 39.Alderson, R. F., Curtis, R., Alterman, A. L., Lindsay, R. M., and DiStefano, P. S. (2000) Brain Res. 871 210-222 [DOI] [PubMed] [Google Scholar]
- 40.Spellman, D. S., Deinhardt, K., Darie, C. C., Chao, M. V., and Neubert, T. A. (2008) Mol. Cell. Proteomics 7 1067-1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babst, M., Odorizzi, G., Estepa, E. J., and Emr, S. D. (2000) Traffic 1 248-258 [DOI] [PubMed] [Google Scholar]
- 42.Bilodeau, P. S., Urbanowski, J. L., Winistorfer, S. C., and Piper, R. C. (2002) Nat. Cell Biol. 4 534-539 [DOI] [PubMed] [Google Scholar]
- 43.Hasdemir, B., Bunnett, N. W., and Cottrell, G. S. (2007) J. Biol. Chem. 282 29646-29657 [DOI] [PubMed] [Google Scholar]
- 44.Raiborg, C., Bremnes, B., Mehlum, A., Gillooly, D. J., D'Arrigo, A., Stang, E., and Stenmark, H. (2001) J. Cell Sci. 114 2255-2263 [DOI] [PubMed] [Google Scholar]
- 45.Stenmark, H., Aasland, R., Toh, B. H., and D'Arrigo, A. (1996) J. Biol. Chem. 271 24048-24054 [DOI] [PubMed] [Google Scholar]
- 46.Christoforidis, S., McBride, H. M., Burgoyne, R. D., and Zerial, M. (1999) Nature 397 621-625 [DOI] [PubMed] [Google Scholar]
- 47.Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D., and Hicke, L. (2002) Nat. Cell Biol. 4 389-393 [DOI] [PubMed] [Google Scholar]
- 48.Pornillos, O., Garrus, J. E., and Sundquist, W. I. (2002) Trends Cell Biol. 12 569-579 [DOI] [PubMed] [Google Scholar]
- 49.Qui, M. S., and Green, S. H. (1992) Neuron 9 705-717 [DOI] [PubMed] [Google Scholar]
- 50.Chao, M., Casaccia-Bonnefil, P., Carter, B., Chittka, A., Kong, H., and Yoon, S. O. (1998) Brain Res. Brain Res. Rev. 26 295-301 [DOI] [PubMed] [Google Scholar]
- 51.Frisén, J., Verge, V. M., Fried, K., Risling, M., Persson, H., Trotter, J., Hökfelt, T., and Lindholm, D. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 4971-4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudge, J. S., Li, Y., Pasnikowski, E. M., Mattsson, K., Pan, L., Yancopoulos, G. D., Wiegand, S. J., Lindsay, R. M., and Ip, N. Y. (1994) Eur. J. Neurosci. 6 693-705 [DOI] [PubMed] [Google Scholar]
- 53.Wetmore, C., and Olson, L. (1995) J. Comp. Neurol. 353 143-159 [DOI] [PubMed] [Google Scholar]
- 54.Marchese, A., Raiborg, C., Santini, F., Keen, J. H., Stenmark, H., and Benovic, J. L. (2003) Dev. Cell 5 709-722 [DOI] [PubMed] [Google Scholar]
- 55.Arévalo, J. C., Waite, J., Rajagopal, R., Beyna, M., Chen, Z. Y., Lee, F. S., and Chao, M. V. (2006) Neuron 50 549-559 [DOI] [PubMed] [Google Scholar]
- 56.Geetha, T., Jiang, J., and Wooten, M. W. (2005) Mol. Cell 20 301-312 [DOI] [PubMed] [Google Scholar]
- 57.Jadhav, T., Geetha, T., Jiang, J., and Wooten, M. W. (2008) Biochem. Biophys. Res. Commun. 371 521-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruenberg, J., and Stenmark, H. (2004) Nat. Rev. Mol. Cell Biol. 5 317-323 [DOI] [PubMed] [Google Scholar]
- 59.Maxfield, F. R., and McGraw, T. E. (2004) Nat. Rev. Mol. Cell Biol. 5 121-132 [DOI] [PubMed] [Google Scholar]
- 60.Cong, M., Perry, S. J., Hu, L. A., Hanson, P. I., Claing, A., and Lefkowitz, R. J. (2001) J. Biol. Chem. 276 45145-45152 [DOI] [PubMed] [Google Scholar]
- 61.Gage, R. M., Kim, K. A., Cao, T. T., and von Zastrow, M. (2001) J. Biol. Chem. 276 44712-44720 [DOI] [PubMed] [Google Scholar]
- 62.Li, J. G., Chen, C., and Liu-Chen, L. Y. (2002) J. Biol. Chem. 277 27545-27552 [DOI] [PubMed] [Google Scholar]
- 63.Galet, C., Min, L., Narayanan, R., Kishi, M., Weigel, N. L., and Ascoli, M. (2003) Mol. Endocrinol. 17 411-422 [DOI] [PubMed] [Google Scholar]
- 64.Vargas, G. A., and Von Zastrow, M. (2004) J. Biol. Chem. 279 37461-37469 [DOI] [PubMed] [Google Scholar]
- 65.Tanowitz, M., and von Zastrow, M. (2003) J. Biol. Chem. 278 45978-45986 [DOI] [PubMed] [Google Scholar]
- 66.Hislop, J. N., Marley, A., and Von Zastrow, M. (2004) J. Biol. Chem. 279 22522-22531 [DOI] [PubMed] [Google Scholar]
- 67.Chin, L. S., Raynor, M. C., Wei, X., Chen, H. Q., and Li, L. (2001) J. Biol. Chem. 276 7069-7078 [DOI] [PubMed] [Google Scholar]
- 68.Morino, C., Kato, M., Yamamoto, A., Mizuno, E., Hayakawa, A., Komada, M., and Kitamura, N. (2004) Exp. Cell Res. 297 380-391 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.