Abstract
In vertebrate visual pigments, a glutamic acid serves as a negative counterion to the positively charged chromophore, a protonated Schiff base of retinal. When photoisomerization leads to the Schiff base deprotonating, the anionic glutamic acid becomes protonated, forming a neutral species that activates the visual cascade. We show that in octopus rhodopsin, the glutamic acid has no anionic counterpart. Thus, the “counterion” is already neutral, so no protonated form of an initially anionic group needs to be created to activate. This helps to explain another observation—that the active photoproduct of octopus rhodopsin can be formed without its Schiff base deprotonating. In this sense, the mechanism of light activation of octopus rhodopsin is simpler than for vertebrates, because it eliminates one of the steps required for vertebrate rhodopsins to achieve their activating state.
Visual pigments contain 11-cis retinal, covalently linked to a lysine residue in the apoprotein via a protonated Schiff base, as their chromophore. The photoisomerization of the chromophore from 11-cis to all-trans form initiates a series of thermal transformations of the pigment, which leads to the activation of a G protein. The subsequent enzymatic cascade ultimately leads to the generation of an electrical signal in the photoreceptor cell (1, 2).
The role of negatively charged residues in controlling the absorption spectrum of bovine rhodopsin has been tested by mutating most aspartates and glutamates to asparagine and glutamine, respectively. These studies showed that Glu-113 serves as the counterion to the positively charged protonated Schiff base, and no other potentially charged residue in the transmembrane segments greatly influences the protonation state of the chromophore’s Schiff base (3–5). Because the glutamic acid residue at this position is conserved among all known vertebrate visual pigments, it has been accepted as the universal Schiff base counterion for these pigments (Fig. 1). However, Glu-113 is replaced by a tyrosine in all invertebrate rhodopsins with the exception of some UV-absorbing pigments that use a phenylalanine (Fig. 1) (6). Like bovine rhodopsin, the chromophore in octopus rhodopsin is a protonated Schiff base of retinal (7, 8). Oprian and coworkers (9) have hypothesized that in invertebrate rhodopsins, this tyrosine acts as the counterion to the protonated Schiff base, presumably by being in its anionic, deprotonated state. To elucidate whether the tyrosine residue is deprotonated in octopus rhodopsin, we performed a series of experiments. Our results show that there is no anionic group serving as a counterion in octopus rhodopsin. This finding greatly illuminates the observation that when light absorption leads to the active state of octopus rhodopsin, the pigment’s Schiff base does not deprotonate. Taken together, these findings allow us to posit that the steps required to achieve the activating state are different for vertebrate and invertebrate visual pigments. In vertebrates, the counterion must be neutralized after photoisomerization, whereas in invertebrates it already is neutralized.
Figure 1.
Alignment of the third transmembrane regions of vertebrate and invertebrate visual pigments. The position of the counterion to the protonated Schiff base of vertebrate visual pigments is denoted by ∗, where a glutamic acid residue is conserved in all known vertebrate visual pigments. This residue is replaced by a tyrosine or phenylalanine in invertebrate visual pigments. Rhod, rhodopsin; Bov, bovine; Hum, human; Lmp, lamprey; Chk, chicken; Gck, Gecko gecko; Oct, octopus; Lim, Limulus; Dro, Drosophila melanogaster.
MATERIALS AND METHODS
Preparation of Octopus Photoreceptor Membranes.
Microvillar membranes were prepared from octopus (Octopus dofleini) eyes as described (10). After isolation by sucrose flotation, the microvillar membranes were repeatedly washed with buffer A [400 mM KCl/10 mM MgCl2/10 mM Mes, pH 6.5/1 mM DTT/20 μM 4-(amidinophenyl)methanesulfonyl fluoride (APMSF)] and then with buffer B (10 mM MgCl2/10 mM Mes, pH 6.5/1 mM DTT/20 μM APMSF). Microvillar membranes were solubilized with a detergent [1% sucrose monolaurate (Dojin, Kumamoto, Japan) in 10 mM MOPS, pH 7.4] and centrifuged at 240,000 × g for 20 min to remove any insoluble material. The SDS/PAGE analysis of this sample showed essentially a single band, indicating that the membranes contain mostly octopus rhodopsin.
Measurements.
All sample manipulations and measurements were carried out under dim red light. Absorption spectra of the solutions containing octopus rhodopsin were recorded with a Shimadzu Model MPS-2000 spectrophotometer equipped with a personal computer (PC9801, NEC, Tokyo) for processing the data. For all measurements, the temperature of the sample was maintained at 15°C with a circulating temperature bath (RM 6, Lauda, Königshofen, Germany). Denaturation of octopus rhodopsin was performed by addition of an aliquot of either 10 μl of 30% SDS (final concentration = 0.12%) or 4 μl of 0.5 M HCl (final pH 3.5) to 2.5 ml of rhodopsin solution (OD476 = 0.1, 0.5% sucrose monolaurate) in a quartz cuvette, where the solution was magnetically stirred. Immediately after mixing, the absorption spectrum was recorded. It took about 4 min to complete one scan, including data transfer time to the computer.
RESULTS
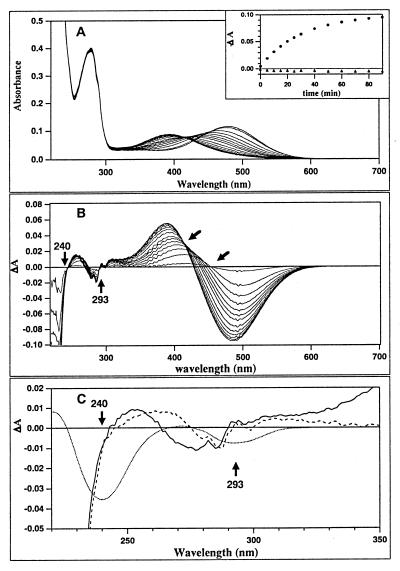
To determine whether invertebrate rhodopsin contains a tyrosinate as a counterion, we denatured octopus rhodopsin under conditions where a tyrosinate would be expected to be reprotonated to its usual neutral state (normal pKa = 10). The reprotonation of the tyrosinate can be detected by its characteristic difference spectra, shown in Fig. 2C. One way to denature octopus rhodopsin (solid curve in Fig. 2A) is to add SDS. There are two sets of spectral changes in the visible region: first, the absorption maximum shifts from 476 nm to 450 nm; this is complete within 5 min after addition of SDS (Fig. 2A). The absorption maximum then slowly shifts to 390 nm. The 450-nm absorption maximum suggests that the initial denaturation product is a protonated Schiff base, whereas the final (390 nm) product is not protonated. Fig. 2B shows the difference spectra at different times after the addition of SDS. Fig. 2C shows the difference spectra of the first and second transitions in the UV region. Also shown is the size and direction of the absorbance changes expected if one tyrosinate per rhodopsin was transformed to a tyrosine. Neither difference spectrum suggests that a tyrosinate has been reprotonated. The absorbance change at 275–285 nm is quite distinct from the tyrosine/tyrosinate difference spectrum, which has a strong band at 240 nm and a weaker one at 295 nm. Furthermore, if the assumption that a tyrosinate acts as the counterion to the protonated Schiff base is true, then its reprotonation should occur concurrently with the deprotonation of the Schiff base. However, no such correlation was seen (Fig. 2A Inset). The changes observed in the 230–280 nm region probably are caused by environmental changes near tryptophan (10 in octopus rhodopsin) and/or tyrosine (22 in octopus) residues.
Figure 2.
Denaturation of octopus rhodopsin with SDS. (A) Absorption changes of octopus rhodopsin from 0–90 minutes after addition of SDS to a final concentration of 0.12%. Each spectrum was recorded every 5 min for the first 30 minutes and then every 10 minutes for the 30–90 minute time range. (Inset) The time courses of the difference absorbance at 480 and 293 nm. ● and ▴ represents the absorption changes at 480 nm and 293 nm, respectively. (B) The overlay of the transient difference spectra, with the slanted arrows indicating the isosbestic points of the first and second spectral transitions. Vertical arrows indicate the wavelengths at which negative peaks are expected to appear during tyrosinate reprotonation as shown in panel C. (C) The difference spectra in the UV of the first (dashed line, 5 min–0 min) and the second (solid line, 90 min–5 min) spectral transitions. The expected difference spectrum, if a single tyrosine residue is reprotonated, is drawn as dotted line. Neither spectrum suggests that a tyrosinate is reprotonated when octopus rhodopsin is denatured.
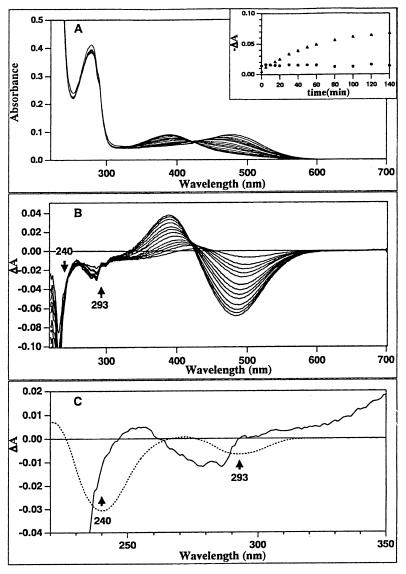
Acid denaturation of octopus rhodopsin also was studied (Fig. 3). If a tyrosinate were present, the denaturation of the pigment by acid should assure its reprotonation. Each panel is similar to the corresponding ones in Fig. 2. In this case, although the first spectral transition from 476 nm to 450 nm is not as clear as for the SDS-induced denaturation, the final product displayed a similar absorption maximum at 390 nm, suggesting an unprotonated Schiff base. No evidence for a spectral change indicating tyrosinate reprotonation was observed. Also, as in Fig. 2, the time courses of the difference absorbance at 480 nm and 293 nm were not synchronous (Fig. 3A Inset). These results also indicate that tyrosinate reprotonation does not occur during acid denaturation of octopus rhodopsin.
Figure 3.
Acid denaturation of octopus rhodopsin: 2.5 ml of rhodopsin solubilized in 0.5% solution of the detergent sucrose monolaurate (solid line) was denatured by the addition of 4 μl of 0.5 M HCl. Each panel is depicted in a similar way as Fig. 2. Each spectrum was recorded at interval of every 4 minutes for the first 20 minutes, then every 10 minutes for the 20–60 minute range, and finally every 20 minutes for the 80–140 minute time range. In this case, although the first phase is not as clear as with SDS denaturation, the final product similarly displayed an absorption maximum of 390 nm, suggesting an unprotonated Schiff base. The spectral changes in the UV expected for the reprotonation of tyrosinate are not observed.
DISCUSSION
We have found that Tyr-112, the putative counterion for the chromophore of octopus rhodopsin, is neutral; there is no anionic amino acid at the position of the Glu-113 in vertebrate pigments to act as the counterion to the positively charged protonated Schiff base of octopus rhodopsin. This result is consistent with our recent UV resonance Raman spectroscopy study (11) in which we showed that no Raman bands caused by a tyrosinate could be observed in octopus rhodopsin. The possibility that an inorganic ion such as chloride might act as the natural counterion also was excluded. Anion substitution from chloride to sulfate or nitrate did not lead to any detectable wavelength shift (data not shown), a shift that would be expected if a free anion were the counterion (3). Next, we asked whether any other group within the hydrophobic core of octopus rhodopsin could act as an anionic counterion. Asp-81 is the only carboxylic acid in the transmembrane region. However, our earlier experiments have indicated that Asp-81 is protonated (12). Moreover, this highly conserved position is also an aspartic acid in bovine rhodopsin but is protonated (13) and does not act as counterion there (3–5). Finally, we note that there are no other potentially anionic amino acid residues (Ser, Thr, and Cys) that are conserved in the invertebrate pigments (6). Thus, we conclude that there is no counterion for octopus rhodopsin’s protonated Schiff base.
The lack of a counterion probably explains an unusual property of octopus rhodopsin, that the pKa of its Schiff base is much lower than that of bovine rhodopsin’s (14). It is well known that neutralization of the counterions of bovine rhodopsin (3–5) or bacteriorhodopsin (15) leads to very large decreases in the pKa of their Schiff bases. Thus, it would be expected that replacing the vertebrate’s Glu-113 with a neutral tyrosine would lead to a much lower pKa for the Schiff base, as is observed (14). A second consequence of the lack of a counterion is that it disproves the simple version of the charge-separation hypothesis for the storage of the photon’s energy in the primary photoproduct of rhodopsin (16). If there is no counterion, there can be no energy storage by charge separation.
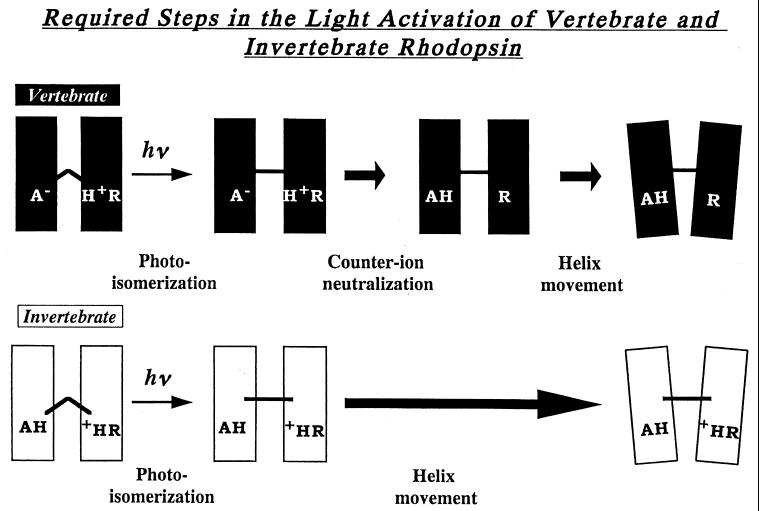
One of the most powerful concepts in understanding the molecular mechanisms underlying visual excitation is the finding of Oprian and coworkers (17) that most vertebrate visual pigments require the neutralization of the charge pair between the protonated Schiff base and the counterion to be able to activate their G proteins (Fig. 4). The deprotonated Schiff base and the protonated Glu-113 were considered a pair of “active” states that had to be generated from their “inactive” counterparts to allow rhodopsin to change its conformation so that it could activate a G protein (4, 5, 17). Later, it was found that light could activate several mutants of rhodopsin without their Schiff bases deprotonating, and so their counterions would not receive a proton from the protonated Schiff base (4, 5). However, in several of these cases, it was shown that in the initial state of the pigment, the amino acid at position 113 was already neutral and another residue was acting as the counterion. It was proposed that for activation, total ion pair neutralization was not required after photoisomerization, only the neutralization of Glu-113 (18). We propose that invertebrate rhodopsin acts just like these mutants; the only thing preventing the pigment from changing into its activating conformation is the inhibition by the 11-cis configuration of the chromophore. Whereas in native vertebrate rhodopsins light absorption leads to the deprotonation of Schiff base, with octopus, the Schiff base remains protonated while the pigment changes into its activating conformation, transient acid metarhodopsin (19). However, in both cases, the essential event seems to be that the position occupied by Glu-113 in vertebrate rhodopsins—which is negatively charged in that case and which is neutralized in the light-activated proton transfer that leads to the active conformation, Meta II—is already neutralized in octopus rhodopsin before light absorption. Hence, there is no need to transfer a proton to switch the pigment into its active state. By eliminating one of the steps that vertebrate pigments must undergo to attain the active state, invertebrate rhodopsins have a simpler mechanism of visual excitation.
Figure 4.
Schematic diagram for the light-induced changes that vertebrate and invertebrate visual pigments must undergo to be able to activate a G protein. Besides photoisomerization (20) and “counterion” neutralization (3–5), there is good evidence that in bovine rhodopsin, the helices must move apart to achieve the activating state (21, 22).
Acknowledgments
We thank Jim Hurley, Akio Maeda, Kris Palczewski, and Bill Parson for many helpful discussions. We thank Yiannis Koutalos and Jie Liang for preliminary experiments. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, and Science (07408019, 09833005, 09680654, and 09780611), Hyogo Science and Technology Association, and the U.S. National Institutes of Health (National Institutes of Health EY01323).
References
- 1.Tsuda M. Photochem Photobiol. 1987;45:915–931. [Google Scholar]
- 2.Stryer L. Annu Rev Neurosci. 1986;9:87–119. doi: 10.1146/annurev.ne.09.030186.000511. [DOI] [PubMed] [Google Scholar]
- 3.Nathans J. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 4.Rao V R, Oprian D D. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 5.Sakmar T P. Prog Nucleic Acid Res Mol Biol. 1998;59:1–34. doi: 10.1016/s0079-6603(08)61027-2. [DOI] [PubMed] [Google Scholar]
- 6.Gartner W, Towner P. Photochem Photobiol. 1995;62:1–16. doi: 10.1111/j.1751-1097.1995.tb05231.x. [DOI] [PubMed] [Google Scholar]
- 7.Kitagawa T, Tsuda M. Biochim Biophys Acta. 1980;624:211–217. doi: 10.1016/0005-2795(80)90240-8. [DOI] [PubMed] [Google Scholar]
- 8.Pande C, Pande A, Yue K T, Callender R H, Ebrey T G, Tsuda M. Biochemistry. 1987;26:4941–4947. doi: 10.1021/bi00390a009. [DOI] [PubMed] [Google Scholar]
- 9.Zhukovsky E, Oprian D. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 10.Tsuda M. Biochim Biophys Acta. 1979;545:537–546. doi: 10.1016/0005-2728(79)90162-2. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto S, Takeuchi H, Nakagawa M, Tsuda M. FEBS Lett. 1996;398:239–242. doi: 10.1016/s0014-5793(96)01250-1. [DOI] [PubMed] [Google Scholar]
- 12.Masuda S, Morita E, Tasumi M, Iwasa T, Tsuda M. FEBS Lett. 1993;317:223–227. doi: 10.1016/0014-5793(93)81280-d. [DOI] [PubMed] [Google Scholar]
- 13.Siebert F. Isr J Chem. 1995;35:309–323. [Google Scholar]
- 14.Koutalos Y, Ebrey T G, Gilson H R, Honig B. Biophys J. 1990;58:493–501. doi: 10.1016/S0006-3495(90)82394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramaniam S, Greenhalgh D A, Khorana H G. J Biol Chem. 1992;267:25730–25733. [PubMed] [Google Scholar]
- 16.Honig B, Ebrey T, Callender R H, Dinur U, Ottolenghi M. Proc. Natl Acad Sci USA. 1979;76:2503–2507. doi: 10.1073/pnas.76.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen G B, Yang T, Robinson P R, Oprian D D. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 18.Fahmy K, Siebert F, Sakmar T P. Biochemistry. 1994;33:13700–13705. doi: 10.1021/bi00250a021. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa M, Kikkawa S, Tominaga K, Tsugi N, Tsuda M. FEBS Lett. 1998;436:259–262. doi: 10.1016/s0014-5793(98)01138-7. [DOI] [PubMed] [Google Scholar]
- 20.Mao B, Tsuda M, Ebrey T G, Akita H, Balogh-Nair V, Nakanishi K. Biophys J. 1981;35:543–546. doi: 10.1016/S0006-3495(81)84809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 22.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]